An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer
Abstract
1. Introduction
- i.
- A fully automated framework for the five-class diagnosis of most occurring lung and colon cancer subtypes is proposed using EffcientNetV2-large (L), medium (M), and small (S) models based on histopathology images.
- ii.
- These existing pretrained models are finetuned and tested using a large, openly available lung and colon cancer histopathology image dataset called LC25000.
- iii.
- Visual saliency maps are provided using the gradCAM method to understand the model decisions during testing better.
Related Work
| Study | Year | Method | Dataset | Interpretability | Performance (%) |
|---|---|---|---|---|---|
| Chehade A. H. et al. [25] | 2022 | ML classifiers | LC25000 | No | Accuracy: 99.0 F1-score: 98.80 |
| Masud M. et al. [24] | 2021 | ML classifiers | LC25000 | No | Accuracy: 96.33 |
| Ali M. et al. [26] | 2021 | Multi-input capsule neural network | LC25000 | No | Accuracy: 99.58 |
| Togacar M. [28] | 2021 | DarkNet-19 and SVM | LC25000 | No | Accuracy: 99.69 |
| Mehmood S. et al. [27] | 2022 | Image enhancement and AlexNet | LC25000 | No | Accuracy: 98.40 |
| Teramoto A. et al. [21] | 2017 | Custom CNN model | Private dataset (298 microscopic images) | No | Accuracy: 71.10 (Only lung cancer) |
| Attallah O. et al. [31] | 2022 | Custom CNN + PCA, FWHT, DWT | LC25000 | No | Accuracy: 99.60 |
| Hatuwal B. K. et al. [22] | 2020 | Custom CNN | LC25000 | No | Accuracy: 97.20 (Only lung cancer) |
| Mangal S. et al. [32] | 2020 | Custom CNN | LC25000 | No | Accuracy: 96.50 |
| Talukder Md. A. et al. [30] | 2022 | Hybrid ensemble learning | LC25000 | No | Accuracy: 99.30 |
| Kumar N. et al. [29] | 2022 | DenseNet121 and RF | LC25000 | No | Accuracy: 98.60 F1-score: 98.50 |
| Hasan Md. I. et al. [23] | 2022 | Custom CNN and PCA | LC25000 | No | Accuracy: 99.80 (Only colon cancer) |
| Present study | 2023 | EffcientNetV2 | LC25000 | Yes | Accuracy: 99.97 F1-score: 99.97 BA: 99.97 AUC: 99.99 MCC: 99.96 |
2. Methods
2.1. Dataset
2.2. Physiological Mechanims of Lung and Colon Cancers
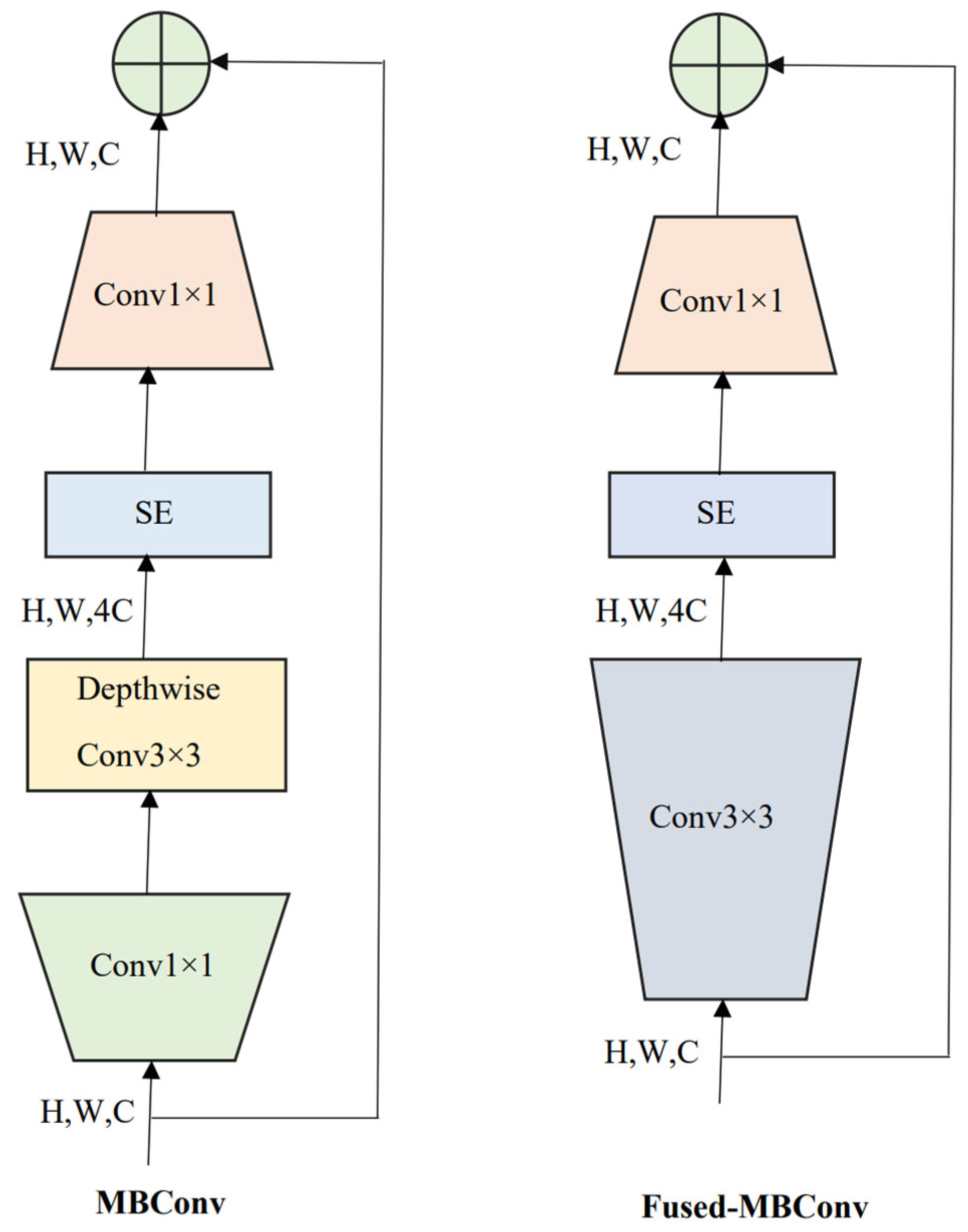
2.3. EffcientNetV2 and Compound Scaling
2.4. Model Training and Validation
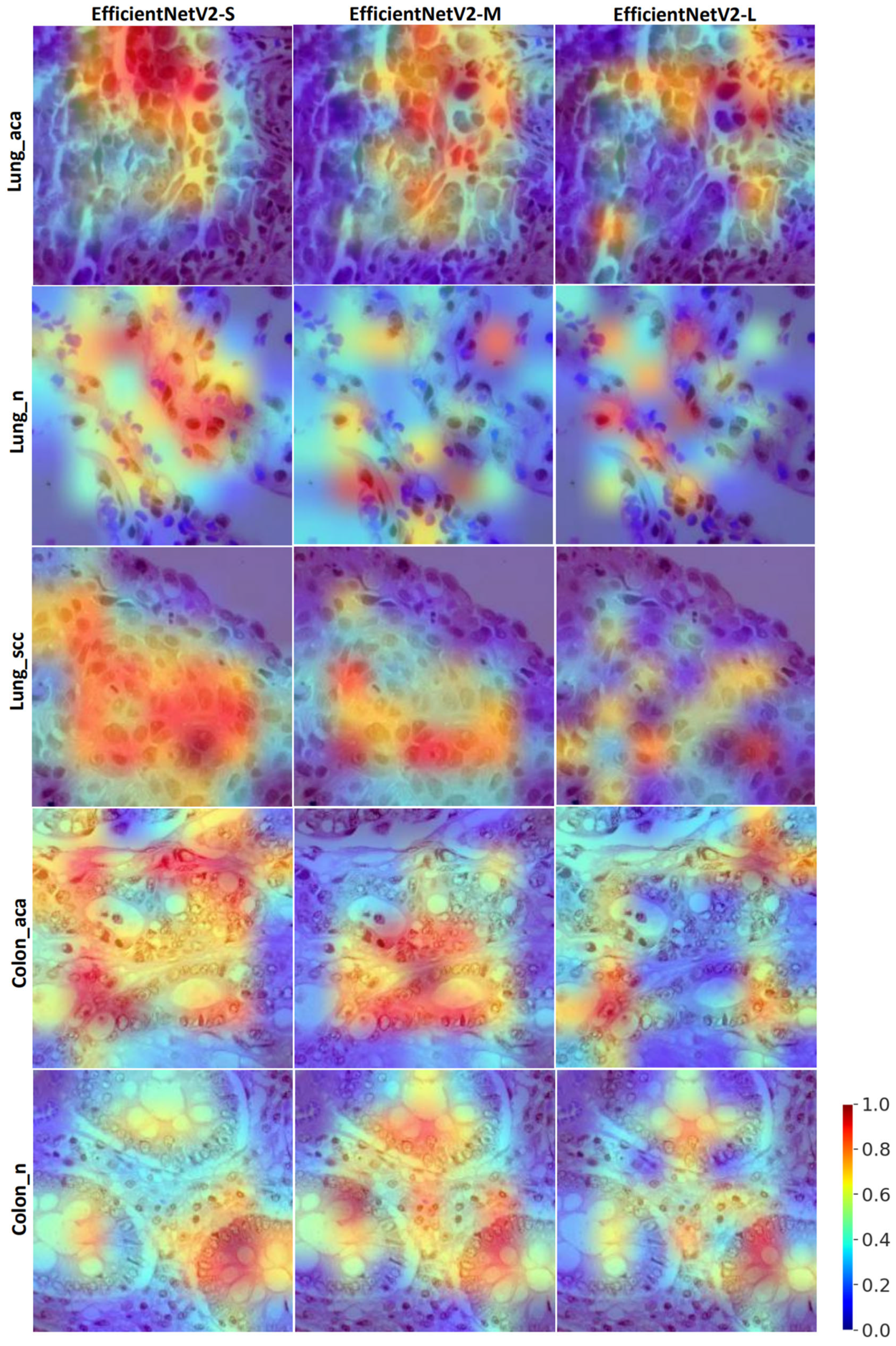
2.5. Visual Saliency Maps
2.6. Evaluation Metrics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global Burden of Cancer Attributable to Infections in 2018: A Worldwide Incidence Analysis. Lancet. Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kurishima, K.; Miyazaki, K.; Watanabe, H.; Shiozawa, T.; Ishikawa, H.; Satoh, H.; Hizawa, N. Lung Cancer Patients with Synchronous Colon Cancer. Mol. Clin. Oncol. 2018, 8, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning ICML, Long Beach, CA, USA, 9–15 June 2019; pp. 10691–10700. [Google Scholar]
- Tan, M.; Le, Q.V. EffcientNetV2: Smaller Models and Faster Training. arXiv 2021, arXiv:2104.00298. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, New Orleans, LA, USA, 18–24 June 2022; pp. 9992–10002. [Google Scholar] [CrossRef]
- Tummala, S. Deep Learning Framework Using Siamese Neural Network for Diagnosis of Autism from Brain Magnetic Resonance Imaging. In Proceedings of the 2021 6th International Conference for Convergence in Technology (I2CT), Maharashtra, India, 2 April 2021; pp. 1–5. [Google Scholar]
- Yousef, R.; Gupta, G.; Yousef, N.; Khari, M. A Holistic Overview of Deep Learning Approach in Medical Imaging. Multimed. Syst. 2022, 28, 881. [Google Scholar] [CrossRef]
- Tummala, S.; Kim, J.; Kadry, S. BreaST-Net: Multi-Class Classification of Breast Cancer from Histopathological Images Using Ensemble of Swin Transformers. Mathematics 2022, 10, 4109. [Google Scholar] [CrossRef]
- Galib, S.M.; Lee, H.K.; Guy, C.L.; Riblett, M.J.; Hugo, G.D. A Fast and Scalable Method for Quality Assurance of Deformable Image Registration on Lung CT Scans Using Convolutional Neural Networks. Med. Phys. 2020, 47, 99–109. [Google Scholar] [CrossRef]
- Tummala, S.; Thadikemalla, V.S.G.; Kadry, S.; Sharaf, M.; Rauf, H.T. EffcientNetV2 Based Ensemble Model for Quality Estimation of Diabetic Retinopathy Images from DeepDRiD. Diagnostics 2023, 13, 622. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 December 2016; pp. 2921–2929. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2016, 128, 336–359. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Improved Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks. Biomed Res. Int. 2017, 2017, 4067832. [Google Scholar] [CrossRef] [PubMed]
- Hatuwal, B.K.; Thapa, H.C. Lung Cancer Detection Using Convolutional Neural Network on Histopathological Images. Int. J. Comput. Trends Technol. 2020, 68, 21–24. [Google Scholar] [CrossRef]
- Hasan, I.; Ali, S.; Rahman, H.; Islam, K. Automated Detection and Characterization of Colon Cancer with Deep Convolutional Neural Networks. J. Healthc. Eng. 2022, 2022, 5269913. [Google Scholar] [CrossRef]
- Masud, M.; Sikder, N.; Nahid, A.A.; Bairagi, A.K.; Alzain, M.A. A Machine Learning Approach to Diagnosing Lung and Colon Cancer Using a Deep Learning-Based Classification Framework. Sensors 2021, 21, 748. [Google Scholar] [CrossRef]
- Hage Chehade, A.; Abdallah, N.; Marion, J.M.; Oueidat, M.; Chauvet, P. Lung and Colon Cancer Classification Using Medical Imaging: A Feature Engineering Approach. Phys. Eng. Sci. Med. 2022, 45, 729–746. [Google Scholar] [CrossRef]
- Ali, M.; Ali, R. Multi-Input Dual-Stream Capsule Network for Improved Lung and Colon Cancer Classification. Diagnostics 2021, 11, 1485. [Google Scholar] [CrossRef]
- Mehmood, S.; Ghazal, T.M.; Khan, M.A.; Zubair, M.; Naseem, M.T.; Faiz, T.; Ahmad, M. Malignancy Detection in Lung and Colon Histopathology Images Using Transfer Learning with Class Selective Image Processing. IEEE Access 2022, 10, 25657–25668. [Google Scholar] [CrossRef]
- Toğaçar, M. Disease Type Detection in Lung and Colon Cancer Images Using the Complement Approach of Inefficient Sets. Comput. Biol. Med. 2021, 137, 104827. [Google Scholar] [CrossRef]
- Kumar, N.; Sharma, M.; Singh, V.P.; Madan, C.; Mehandia, S. An Empirical Study of Handcrafted and Dense Feature Extraction Techniques for Lung and Colon Cancer Classification from Histopathological Images. Biomed. Signal Process. Control 2022, 75, 103596. [Google Scholar] [CrossRef]
- Talukder, M.A.; Islam, M.M.; Uddin, M.A.; Akhter, A.; Hasan, K.F.; Moni, M.A. Machine Learning-Based Lung and Colon Cancer Detection Using Deep Feature Extraction and Ensemble Learning. Expert Syst. Appl. 2022, 205, 117695. [Google Scholar] [CrossRef]
- Attallah, O.; Aslan, M.F.; Sabanci, K. A Framework for Lung and Colon Cancer Diagnosis via Lightweight Deep Learning Models and Transformation Methods. Diagnostics 2022, 12, 2926. [Google Scholar] [CrossRef]
- Mangal Engineerbabu, S.; Chaurasia Engineerbabu, A.; Khajanchi, A. Convolution Neural Networks for Diagnosing Colon and Lung Cancer Histopathological Images. arXiv 2020, arXiv:2009.03878. [Google Scholar]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and Colon Cancer Histopathological Image Dataset (LC25000). arXiv 2019, arXiv:1912.12142. [Google Scholar]
- Ihde, D.C.; Minna, J.D. Non-Small Cell Lung Cancer. Part I: Biology, Diagnosis, and Staging. Curr. Probl. Cancer 1991, 15, 65–104. [Google Scholar] [CrossRef]
- Cappell, M.S. Pathophysiology, Clinical Presentation, and Management of Colon Cancer. Gastroenterol. Clin. N. Am. 2008, 37, 1–24. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Tummala, S.; Suresh, A.K. Few-Shot Learning Using Explainable Siamese Twin Network for the Automated Classification of Blood Cells. Med. Biol. Eng. Comput. 2023, 1, 1–15. [Google Scholar] [CrossRef] [PubMed]
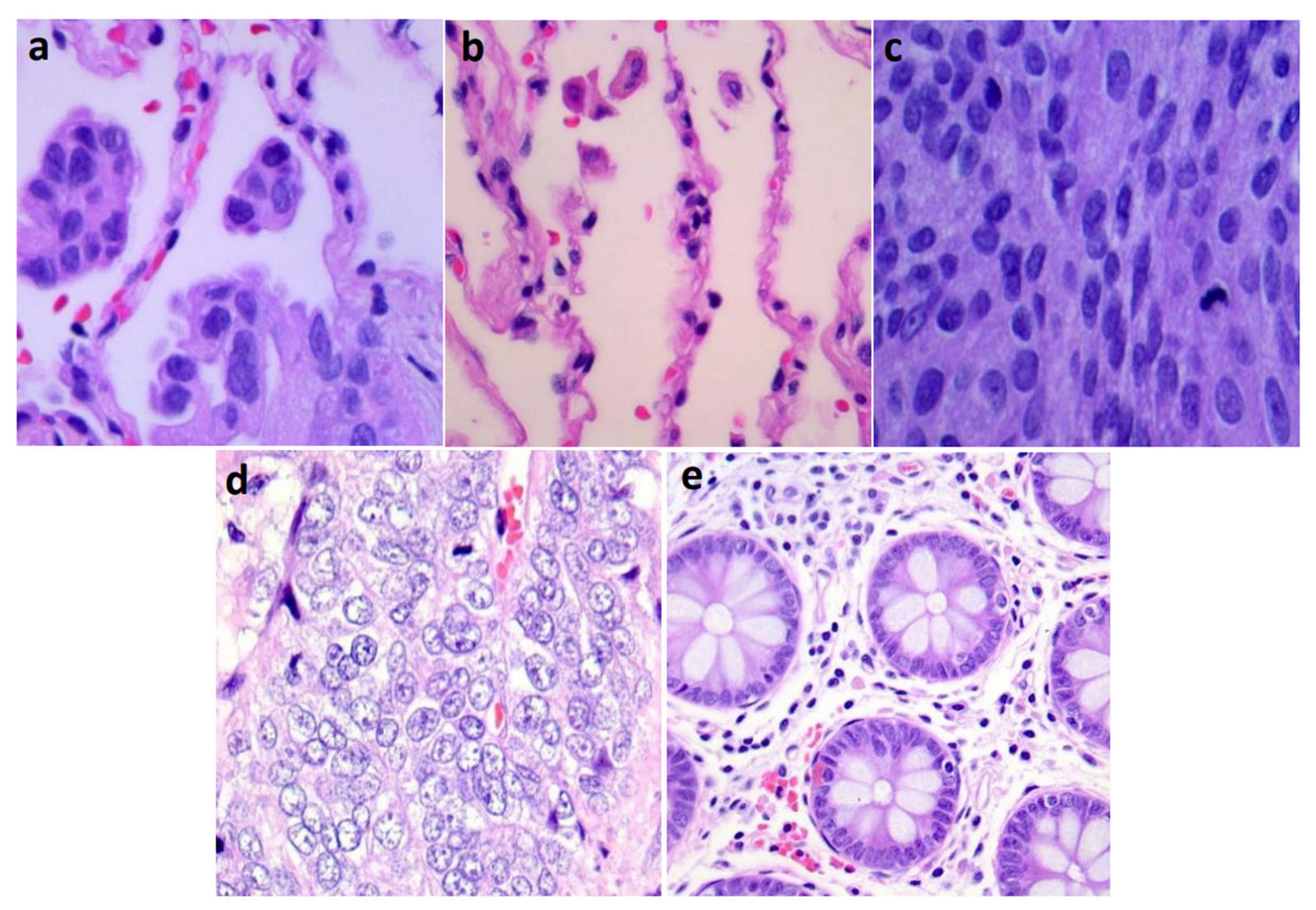

| Lung-aca | Lung-n | Lung-scc | Colon-aca | Colon-n | |
|---|---|---|---|---|---|
| Training | 3600 | 3600 | 3600 | 3600 | 3600 |
| Validation | 400 | 400 | 400 | 400 | 400 |
| Testing | 1000 | 1000 | 1000 | 1000 | 1000 |
| EffcientNetV2-S | EffcientNetV2-M | EffcientNetV2-L | |
|---|---|---|---|
| Accuracy | 99.90 | 99.96 | 99.97 |
| AUC | 99.99 | 99.99 | 99.99 |
| F1-Score | 99.90 | 99.96 | 99.97 |
| BA | 99.90 | 99.97 | 99.97 |
| MCC | 99.87 | 99.94 | 99.96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tummala, S.; Kadry, S.; Nadeem, A.; Rauf, H.T.; Gul, N. An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer. Diagnostics 2023, 13, 1594. https://doi.org/10.3390/diagnostics13091594
Tummala S, Kadry S, Nadeem A, Rauf HT, Gul N. An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer. Diagnostics. 2023; 13(9):1594. https://doi.org/10.3390/diagnostics13091594
Chicago/Turabian StyleTummala, Sudhakar, Seifedine Kadry, Ahmed Nadeem, Hafiz Tayyab Rauf, and Nadia Gul. 2023. "An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer" Diagnostics 13, no. 9: 1594. https://doi.org/10.3390/diagnostics13091594
APA StyleTummala, S., Kadry, S., Nadeem, A., Rauf, H. T., & Gul, N. (2023). An Explainable Classification Method Based on Complex Scaling in Histopathology Images for Lung and Colon Cancer. Diagnostics, 13(9), 1594. https://doi.org/10.3390/diagnostics13091594









