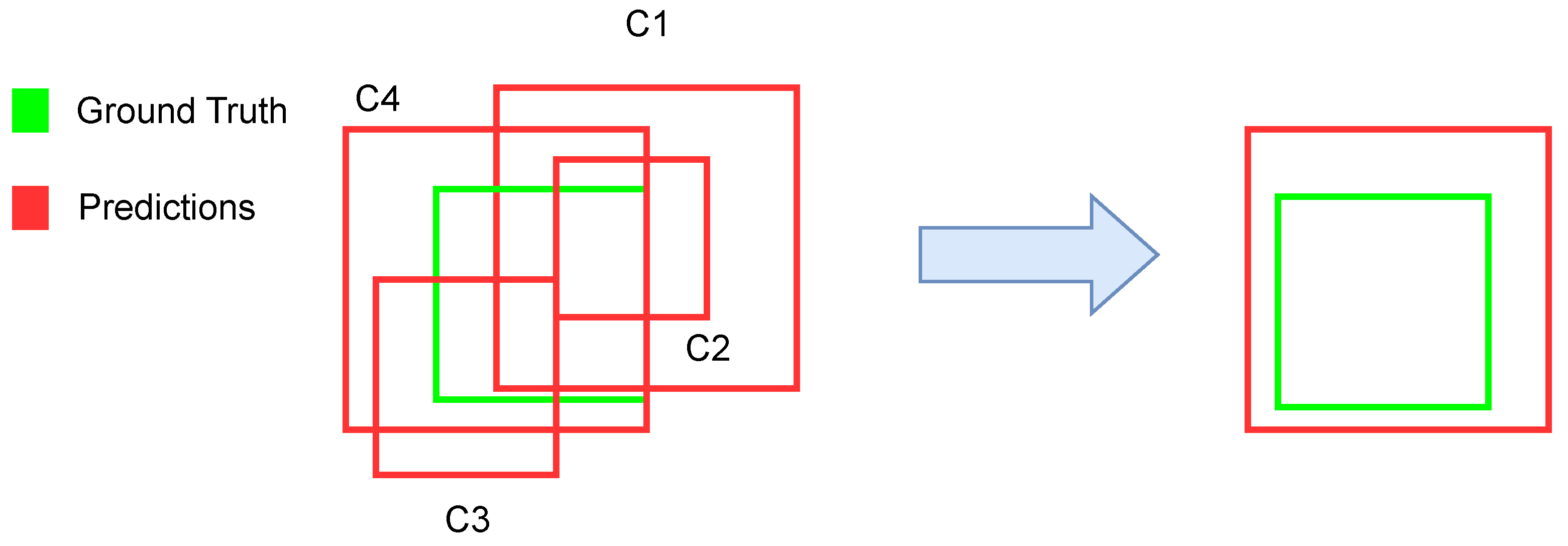
SARS-CoV-2 belongs to the same family of viruses as SARS-CoV, which causes Severe Acute Respiratory Syndrome (SARS), and MERS-CoV, responsible for the Middle East Respiratory Syndrome (MERS). These types of viruses are known as coronaviruses, and their main characteristic is that they contain a single-stranded RNA enclosed in a capsid (the protein shell of a virus) with spikes, therefore, resembling the corona-like appearance of the virus. These coronaviruses share many similarities, such as their manifestations of increased viral pneumonia or their survival decay.
1.1. Motivation
Plain chest radiography (CXR) and computer tomography (CT) are two radiological techniques that have been shown to have great clinical value in the diagnosis and evaluation of patients with COVID-19. These techniques are valuable in diagnosis, medical triage, and/or therapy. In [
8], it was shown that these imaging techniques were shown to be useful in clinical scenarios during the pandemic. For patients with mild symptoms of COVID-19, radiological images also provide a baseline for evaluating the patient’s condition. In the case of patients with moderate to severe symptoms, it also helps to identify lung and heart abnormalities and allows a more informed medical decision. CXR is also a great aid for fast triage. Finally, chest radiological tests are also recommended if the patient worsens so that he can be treated accordingly. In any case, radiological findings are not used as the primary screening tool to detect COVID-19, but rather to determine its severity and potential complications [
9].
Many investigations report a variety of appearances in CXR of COVID-19 patients such as: [
8,
10,
11,
12,
13]. The most typical reported appearances are multifocal bilateral and peripheral lung opacities, usually round-shaped and predominant in the lower part of the lungs [
14], although there might be other appearances, such as central or upper lung opacities, and also pneumothoraxes, pleural effusions, pulmonary oedemas, lobar consolidations, lung nodules, and cavities [
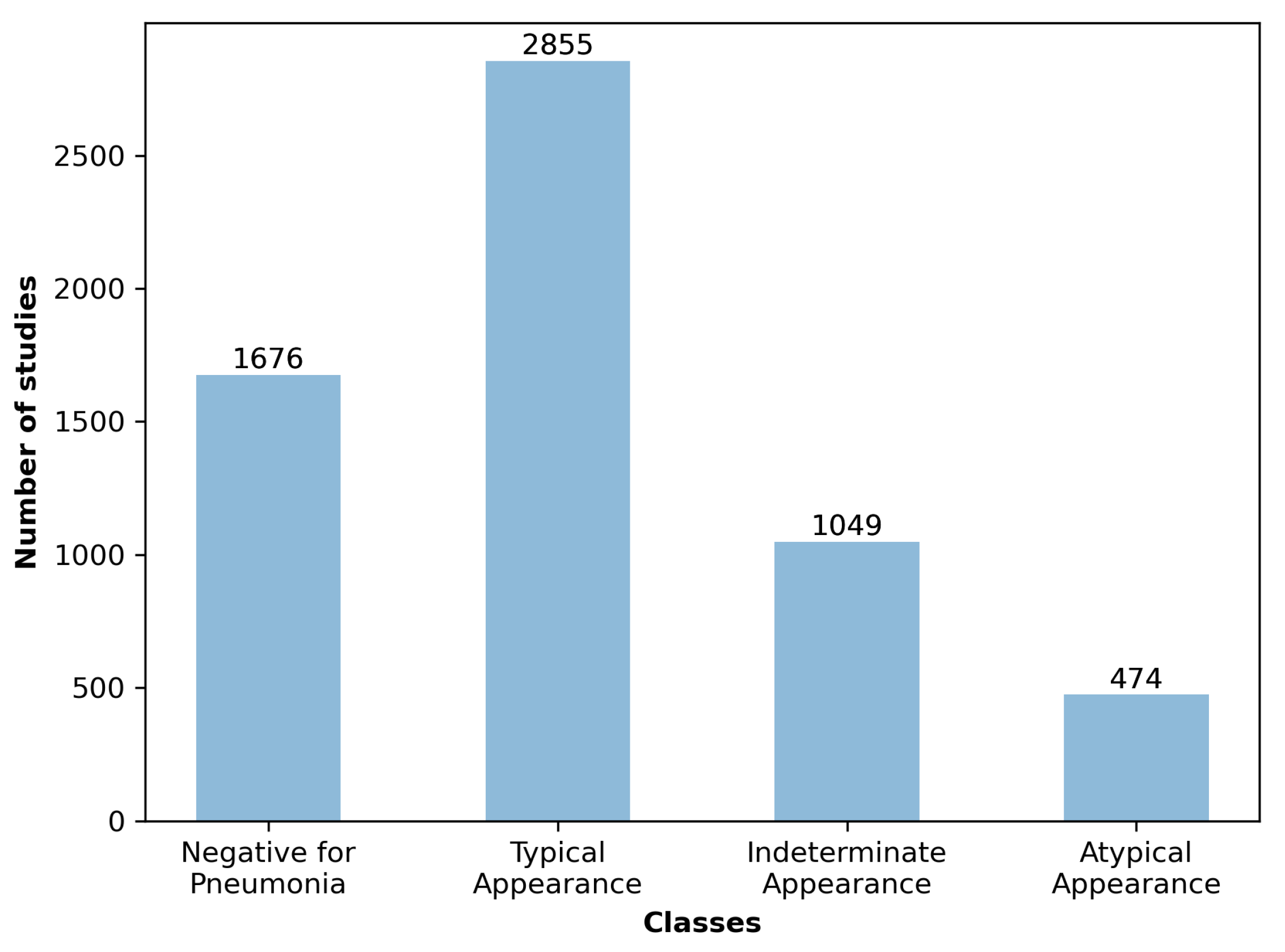
14]. In the same study, a classification of the type of appearance is made into four categories: typical appearance, atypical appearance, indeterminate appearance, and negative for pneumonia, depending on the type and location of the appearance.
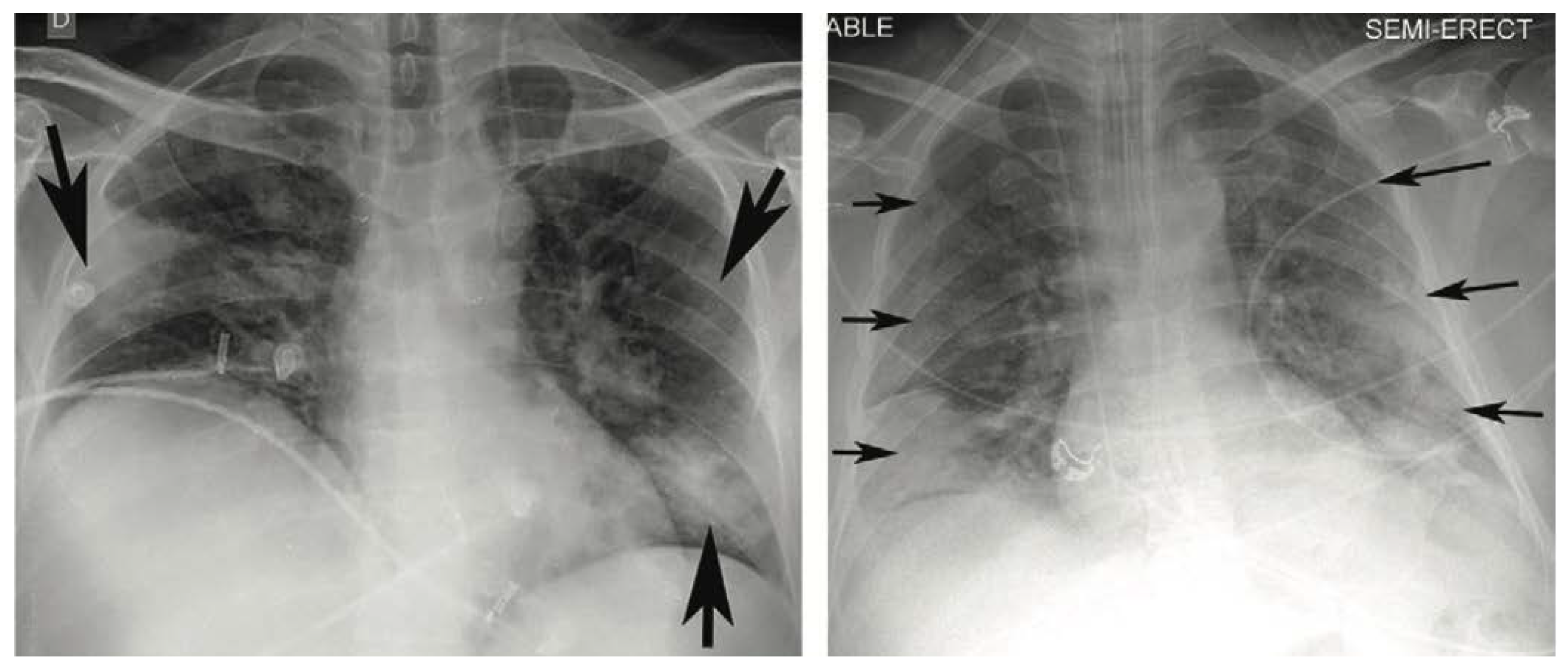
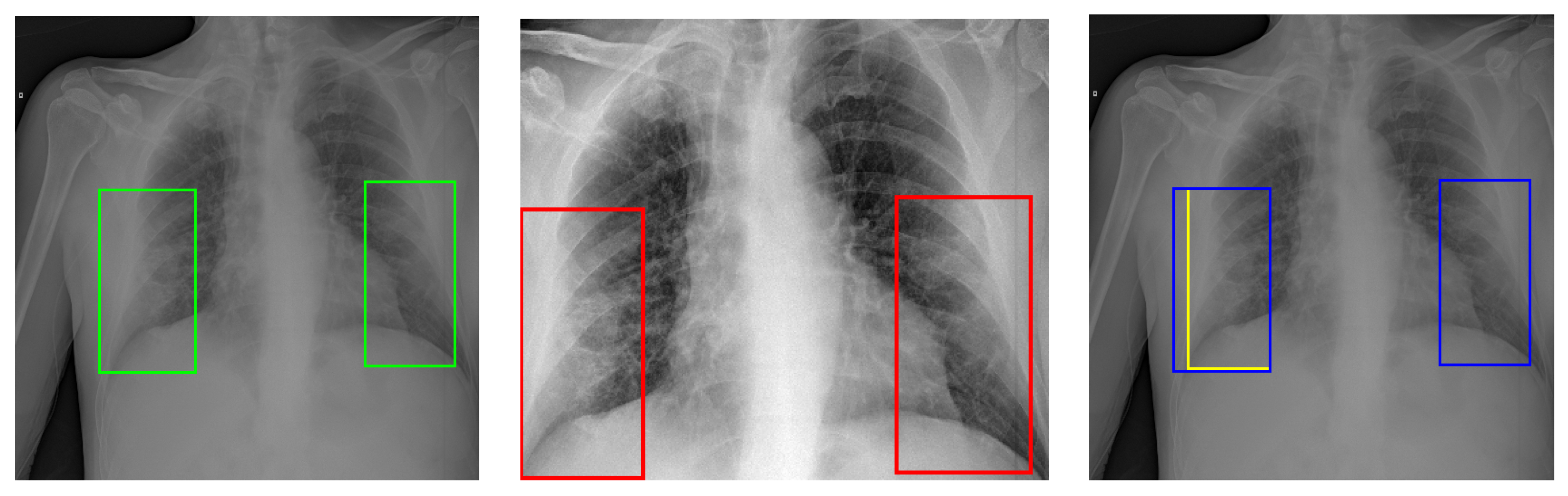
Figure 1 presents two examples of CXR images showing typical appearances in the form of lung opacities.
CXR images have been shown to be less sensitive to COVID-19 manifestations than those obtained with CT. In fact, one of the main limitations of using CXR as a diagnostic tool is the absence of appearances in the lungs in the early stages of the disease and, in certain cases, throughout the course of the disease. In addition to this, breast size, poor positioning, and lack of patient inspiration can introduce artefacts in CXR, often leading to false positives. In general, the sensitivity of CXR in combination with a human expert has been estimated to be around 69%, while for CT is around 98% [
15].
The individual study of the radiography of every patient is a difficult and time-consuming task, as it requires specialised personnel. Therefore, the design and implementation of automatic decision support systems (DSS) that can identify lesions due to COVID-19 are of interest not only to alleviate the workload of sanitation personnel, but also to possibly detect hard-to-find lung lesions that could be missed in a first rushed look at a CXR.
1.2. State of the Art
Since the pandemic began, much research has been carried out on using methods based on artificial intelligence (AI) to develop DSS for screening and evaluating COVID-19 using CXR images. In this context, the area of research that has received more attention in the literature is the design of automatic screening tools to identify the patient’s condition. Work has also been done to identify and segment associated lung lesions, but in a smaller number and mostly using CT as the main data source, leaving CXR in the second plane. The following review of the state-of-the-art focuses on work using artificial intelligence techniques to identify COVID-19 lesions from CXR images.
Most approaches developed previously to identify COVID-19-related lung lesions from CXR images were proposed in the context of the SIIM-FISABIO-RSNA COVID-19 Detection Challenge [
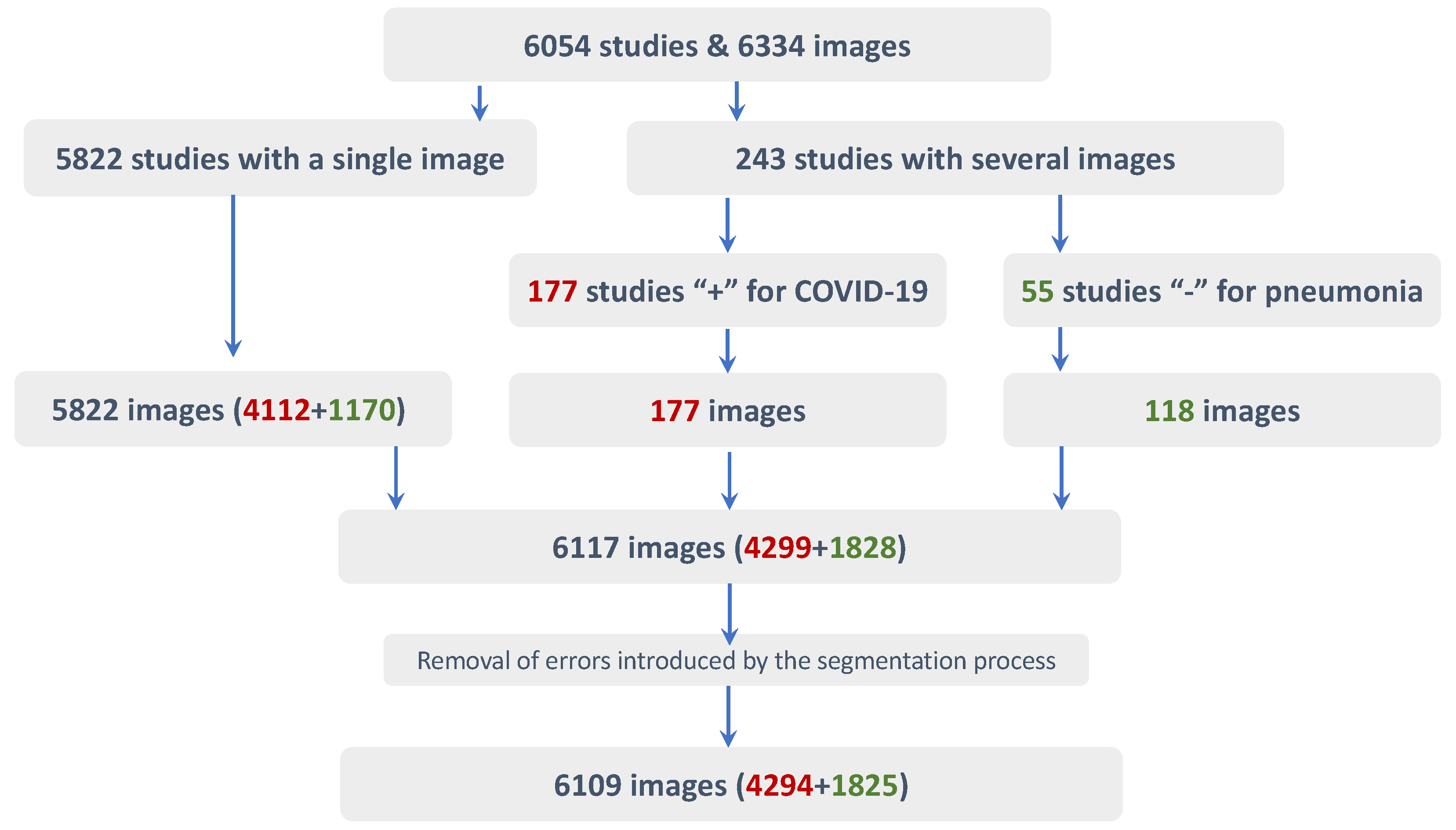
16]. The challenge’s goal was twofold, including lesion detection and classification into four categories. Participants trained their models using the SIIM-FISABIO-RSNA COVID-19 Detection data set [
17], which contains 6334 CXR images. The data set was manually annotated by experts with the corresponding bounding boxes for the lesions observed in the images.
The winner of the challenge [
18] used an ensemble of four models (YoloV5-x6, EfficientDet D7, Faster Region-Based Convolutional Neural Network Feature Pyramid Network [Faster-R-CNN-FPN] with a ResNet-101 backbone, and a Faster-R-CNN-FPN with a ResNet-200 backbone) fed with images of 768 × 768, 768 × 768, 1024 × 1024, and 768 × 768 pixels, respectively. Models were initially trained to predict pneumonia bounding boxes using an external data set. Then the pre-trained models were further trained with the data set provided by the challenge. Strong data augmentation along with Test Time Augmentation (TTA) were used. In this solution, the models were used to extract pseudo-labels that predict the bounding boxes in the test set and in three additional COVID-19 data sets (PadChest, Pneumothorax, and VinDr-CXR). Those boxes with a confidence greater than 0.7 were accepted as pseudo-labels. Finally, all labels were used for the final training of the models. The obtained Mean Average Precision (mAP) with an Intersection over Union (IoU) > 0.5 (mAP@50) for the four mentioned models was 0.580, 0.590, 0.592, and 0.596, respectively. The mAP@50 of the ensemble was not reported. The solution that scored second in the challenge [
18] used a set of YoloV5, YoloX, and EfficientDet. The images were resized to 384 × 384 and 512 × 512, respectively, and the mAP@50 of the ensemble reached 0.59. The solution in the third place [
18] used EfficientDet D7X, EfficientDet D6 and SWIN RepPoints. The models were pre-trained on an external pneumonia data set using five folds. To improve their results, the authors used the predictions of their classification model to complement the training/validation data set. No results in terms of mAP were provided for this solution. In fourth place, [
18] used a variation of YoloV5 grounded on a transformer backbone, although the authors did not give the implementation details. This model was ensembled with YoloV5-x6 and YoloV3 models and trained using images of size 512 × 512. The models were initially pre-trained on an external pneumonia data set, and to improve their results, the authors applied a post-processing technique based on the geometric mean to re-rank their bounding boxes scores. The reported mAP@50 was 0.562. In fifth place, ref. [
18] proposes an ensemble of five models (EfficientDet D3, EfficientDet D5, YoloV5-l, YoloV5-x, and RetinaNet-101), which was built and combined with the results of an additional classification model trained for binary predictions between the classes ‘none’ and ‘COVID-19’. No mAP@50 was reported for this solution. In sixth place, ref. [
18] used a Faster-R-CNN-FPN with an EfficientNet B7 backbone, which was trained using images of 800 × 800 pixels. The model was initially pre-trained with an external pneumonia data set. The authors used several training techniques: stochastic weight averaging, sharpness-aware minimisation, attentional-guided context FPN, and fixed feature attention using the feature pyramid from their classification model into the detection model for attention. Their solution achieved a validation mAP@50 of 0.585. The solution in the seventh place [
18] used an ensemble of 3 models (DetectoRS50, UniverseNet50, UniverseNet101) trained using pseudo-labelled images with 800 × 800 pixels. The models were ensembled using the non-Maximun Weighted (NMW) technique instead of the Weighted Box Fusion (WBF) [
19] used by all other solutions. The obtained mAP@50 was 0.553. In eighth place [
18], the solution used an ensemble of 3 YoloV5 models (YoloV5-x6 with input size of 640 × 640, YoloV5 with input size 1280 × 1280, and YoloV5x with input size 512 × 512), along with a Cascade R-CNN model using inputs of 640 × 640. In this case, background images were used to reduce the false positive rate. No performance metrics were reported. In the ninth place solution [
18], eight different models were grouped: four variations of EfficientDet (EfficientDet D0, EfficientDet D0, EfficientDet D3, EfficientDet Q2) and four variations of YoloV5 (YoloV5-s, YoloV5-m, YoloV5-x, 2-class YoloV5-s). The authors did not report metrics. In the tenth place, ref. [
18] used an ensemble of YoloV5 and EfficientDet. To improve training, the authors used an alternative data set (the RICORD COVID-19 data set [
10]) to create pseudo-labels. No validation metrics were reported for this solution.
In addition to the approaches of the SIIM-FISABIO-RSNA COVID-19 Detection Challenge, in [
20] authors developed an ensemble of YoloV5 and EfficientNet to locate ground glass opacities from radiological images also obtained from the SIIM-FISABIO-RSNA COVID-19 Detection data set. No image pre-processing was used, and mosaic data enhancement was used to train the YoloV5 model. The images were resized to 512 × 512, and the results reported 0.6 as mAP@50, without cross-validation. In addition, in [
21], a YoloV5 and a RetinaNet-101 model were trained with the same data set used in previous works, and ensembled using a WBF algorithm, resizing all images to 1333 × 800. The obtained mAP@50 was 0.552 after stratified cross-validation of K. The authors in [
22] ensembled a YoloV5 model with YoloV5m and YoloV5x backbones, a YoloX model with YoloX-m and YoloX-d backbones, and an EfficientDet B5 net to detect lung opacities in CXR images of 512 × 512 pixels. Histogram equalisation was used as a preprocessing technique, and strong data augmentation was performed for the YoloV5 model using mosaic augmentation, HSV (for hue, saturation, lightness) scaling, random shearing, and rotation; image mixup, random scaling, shearing, and rotation for the YoloX; and brightness, contrast, shearing, scaling and cropping for the Efficientdet B5. The ensemble of these three models reported mAP@50 of 0.62 after a stratified cross-validation of K times. In [
23] YoloV5 was used with the YoloV5s, YoloV5m, YoloV5l and YoloV5x backbones. The images were converted to.jpeg format, and no cross-validation was used. The best results reported 57% true positives and 48% false negative rate.
Table 1 presents a summary of the main characteristics of previous works reported in the state-of-the-art for the automatic identification of COVID-19 lesions using deep learning techniques along with their main characteristics. A common characteristic of all existing works is the use of the SIIM-FISABIO-RSNA COVID-19 Detection data set. This is the largest open data set currently available, and its use makes comparing results easier by establishing a common framework. However, comparing the results using the architectures that participated in the challenge with later proposals is not straightforward since, for the challenge, performance results were calculated using a test data set with labels that were not made open by the organisers. This means that the available data do not allow for a direct comparison of the results provided with the new architectural proposals. Furthermore, most of the proposed solutions to the challenge do not provide detailed information about their implementation, resulting in lack of reproducibility.
However, the remaining works reported in the state-of-the-art used all available 6334 images from the SIIM-FISABIO-RSNA COVID-19 detection data set. However, the data set contains a large number of studies with several images per patient, and even some of them are identical. It means that a fair comparison would require a common inclusion criterion before properly comparing the results. Furthermore, two of these schemes were tested without cross-validation, which could bias the results towards optimistic values.
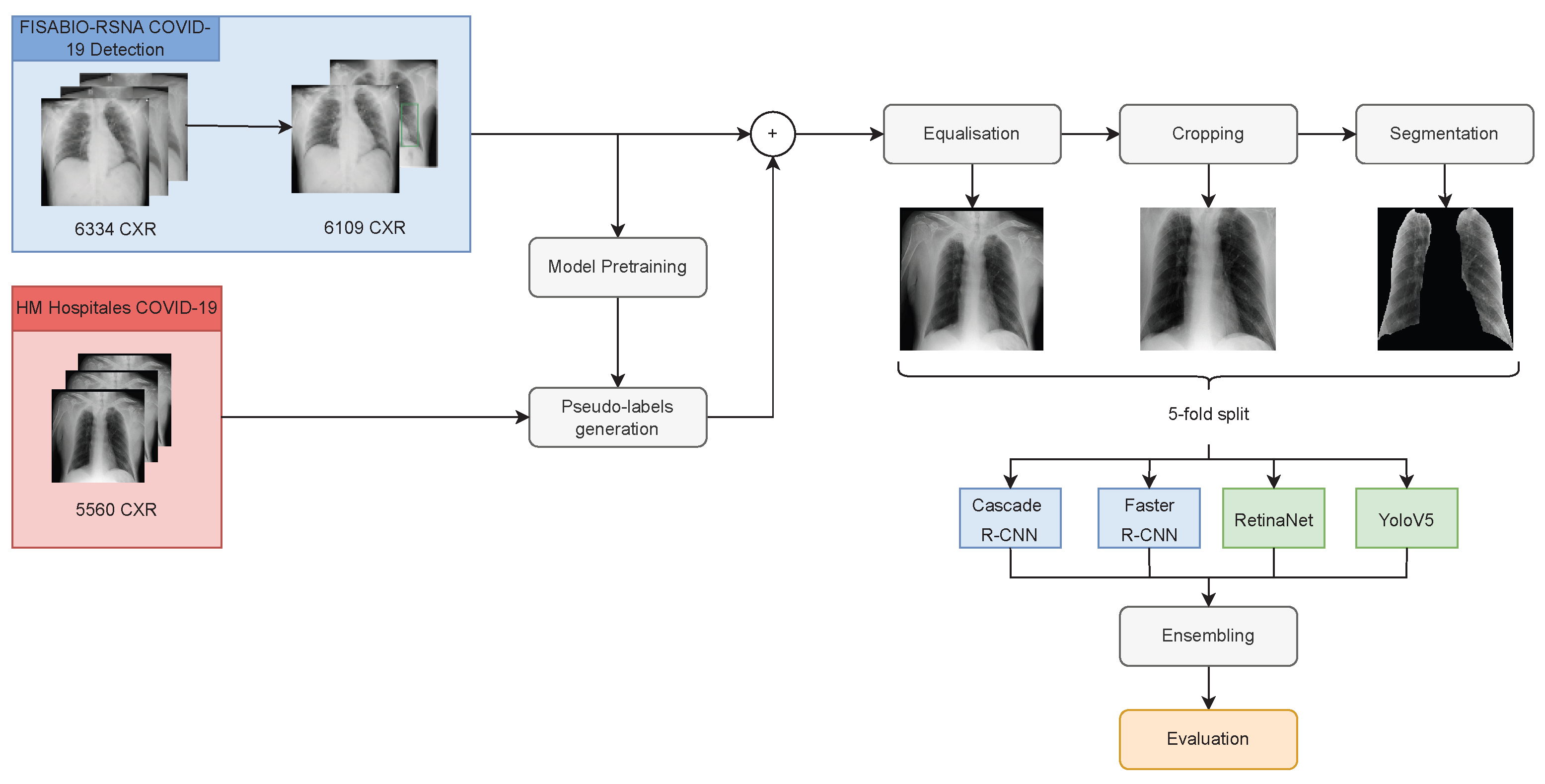
Following the pre-processing methodology proposed in [
24] for COID-19 detection, this article explores the effect of cropping radiological images in the lungs area and the semantic segmentation of the regions of interest (that is, the lungs area) on the identification of the bounding boxes containing the associated lesions. The goal is to guide the attention of the model by reducing the search space and also reducing the distracting artefacts that could confuse the networks. For this purpose, the paper evaluates four different off-the-shelf artificial neural networks (ANN) commonly used for object detection purposes. Two two-stage detectors: Faster R-CNN, Cascade R-CNN; and two one-stage detectors: RetinaNet, and YoloV5. In addition, different combinations of ensembles of these models were also tested according to common strategies in the state-of-the-art. The selection of these simple architectures aims to promote manageable designs that could be trained and deployed easier in medical practice and whose generalisation capabilities are also easier to evaluate, although similar pre-processing strategies can also be incorporated into more complex architectures.
This article also analyses the effect of image cropping and semantic segmentation of the lungs on estimating pseudo-labels to carry out a co-training-based [
25,
26] semi-supervised strategy, also following similar methods to those used in the best works presented at the FISABIO-RSNA COVID-19 Detection Challenge [
16]. As noted above, the state-of-the-art typically uses an arbitrary confidence score of 0.7 as a threshold to decide whether to accept the estimated boxes as pseudo-labels or not [
18]. However, the use of pseudo-labels have several drawbacks. First, other works have pointed out that the scores provided by modern ANNs are poorly calibrated [
27]; therefore, the scores obtained are difficult to compare, so applying the same threshold to two models with similar accuracies can perform quite differently in detecting the bounding boxes. Second, the scores provided by object detectors are related to the estimation of the category associated with the detected object, but many previous works have shown that the classification scores are not strongly correlated with the precision of box localisation (see [
28] and references therein). To evaluate this potential issue, this article presents two different experiments designed to evaluate the training using pseudo-labels for lung lesion detection, considering possible underconfident and overconfident models.
Table 1.
Summary of the literature available in the field.
Table 1.
Summary of the literature available in the field.
| Reference | Model Architecture | Image Size | Pretrained | mAP@50 | Corpus | Remarks |
|---|
| [29] | YoloV5, EfficientNet | 512 × 512 | No | 0.600 | SIIM-FISABIO-RSNA (only the training subset) | No crossvalidation |
| [21] | YoloV5, RetinaNet-101 | 1333 × 800 | No | 0.552 | SIIM-FISABIO-RSNA (only the training subset) | – |
| [22] | YoloV5, YoloX, EfficietNet-B5 | 384 × 384, 512 × 512 | No | 0.620 | SIIM-FISABIO-RSNA (only the training subset) | – |
| [23] | CNN-N | 256 × 256, 512 × 512 | No | – | SIIM-FISABIO-RSNA (only the training subset) | False Positive = 0.49, False Negative = 0.51, No crossvalidation |
| [18] 1st | YoloV5-x6, EfficientDet D7, Faster R-CNN FPN | 768 × 768, 1024 × 1024 | Yes | 0.596 | SIIM-FISABIO-RSNA | Pseudo labels |
| [18] 2nd | YoloV5, YoloX, EfficientDet D5 | 384 × 384, 512 × 512 | Yes | 0.590 | SIIM-FISABIO-RSNA | YoloV5 with 5 different backbones |
| [18] 3th | EfficientDet D7X, EfficientDet D6, SWN-RepPoints/EfficientNet | 640 × 640 | Yes | Not provided | SIIM-FISABIO-RSNA | – |
| [18] 4th | YoloV5-transformer, YoloV5-x6, YoloV3 | 512 × 512 | Yes | 0.562 | SIIM-FISABIO-RSNA | Geometric mean confidence postprocessing |
| [18] 5th | EfficientDet D3, EfficientDet D5, YoloV5-l, YoloV5-x, RetinaNet-101 | 512 × 512, 1333 × 800 | Yes | Not provided | SIIM-FISABIO-RSNA | – |
| [18] 6th | Faster R-CNN, EfficientNet B7 | 800 × 900 | Yes | 0.585 | SIIM-FISABIO-RSNA | – |
| [18] 7th | DetectoRS50, UniverseNet50, UniverseNet101 | 800 × 800 | No | 0.553 | SIIM-FISABIO-RSNA | Pseudo-labels, NMW |
| [18] 8th | YoloV5-x6, YoloV5-x, Cascade R-CNN | 640 × 640, 1280 × 1280 | No | 0.540 | SIIM-FISABIO-RSNA | – |
| [18] 9th | EfficientDet D0,D3,Q2, YoloV5-s,m,x,2-classs | 512 × 512, 640 × 640, 768 × 768 | No | Not provided | SIIM-FISABIO-RSNA | – |
| [18] 10th | YoloV5, EfficientDet | Not provided | Not provided | 0.620 | SIIM-FISABIO-RSNA | Pseudo labels |
Given the aforementioned, it is worth noting that this work is neither focussed on the development of new architectures nor on improving the accuracy of the state-of-the-art methods, but on discussing more interpretable approaches, and on identifying best methodological approaches and good practices.
The rest of the paper is organised as follows.
Section 2 introduces the material and methods used in this paper.
Section 3 mainly describes the results and an analysis of the different experiments.
Section 4 ends with a discussion and conclusions.