Skeletal Fracture Detection with Deep Learning: A Comprehensive Review
Abstract
:1. Introduction
2. Background
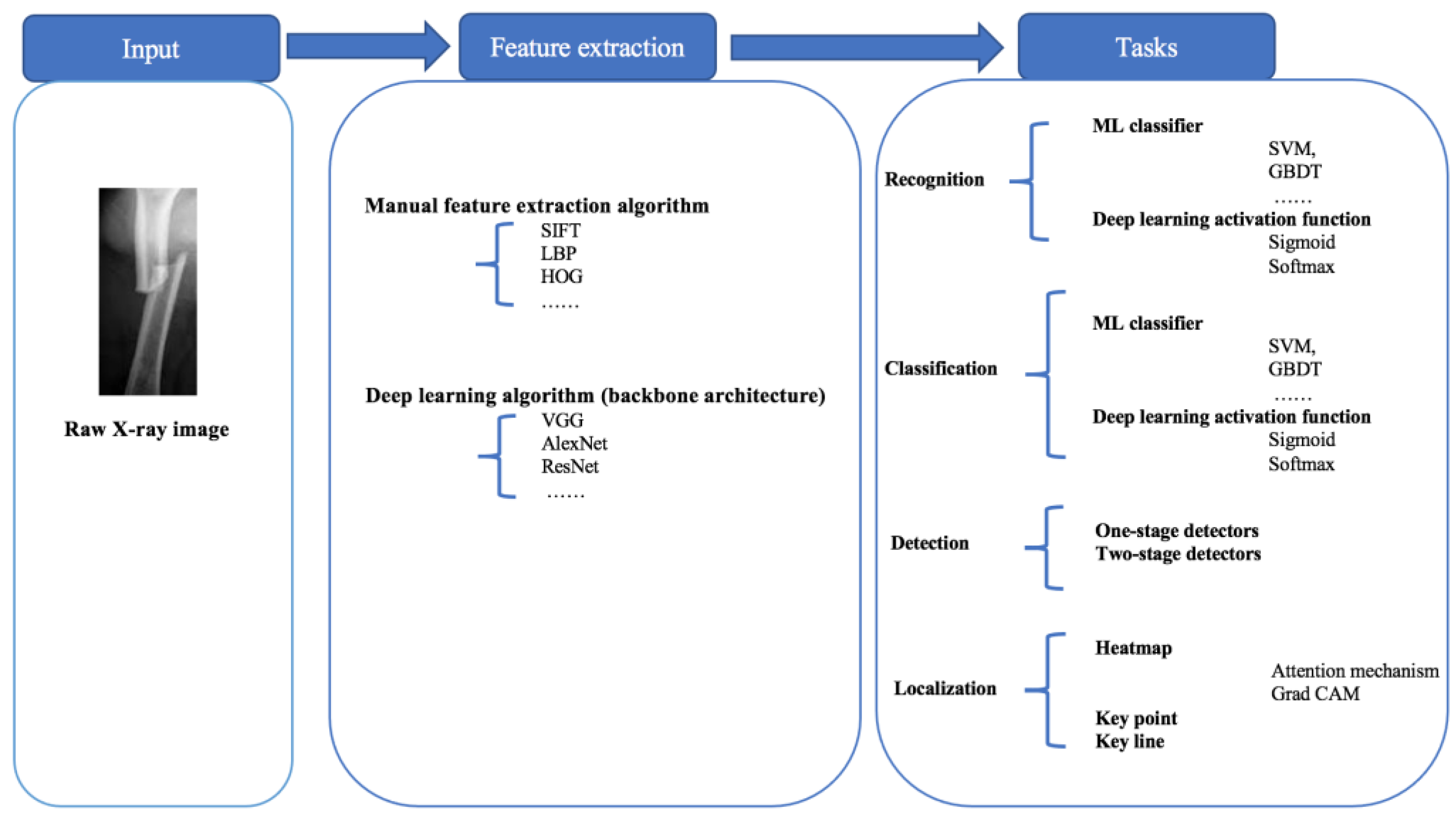
2.1. Task Definition
2.2. Basic Knowledge
3. Methods
4. Modeling Tasks
- Recognition: the recognition task is to identify whether the X-ray image is fractured or non-fractured.
- Classification: the classification task is to not only recognize fractures and non-fractures but also classify the fracture types.
- Detection: the detection task is to find the fracture position and engage the suitable bounding boxes to surround bone fracture parts completely and identify them with the help of bounding boxes.
- Localization: the localization task is to localize the fracture position directly with key points, key lines, or heat maps instead of bounding boxes, according to the entire semantic information of the X-ray images.
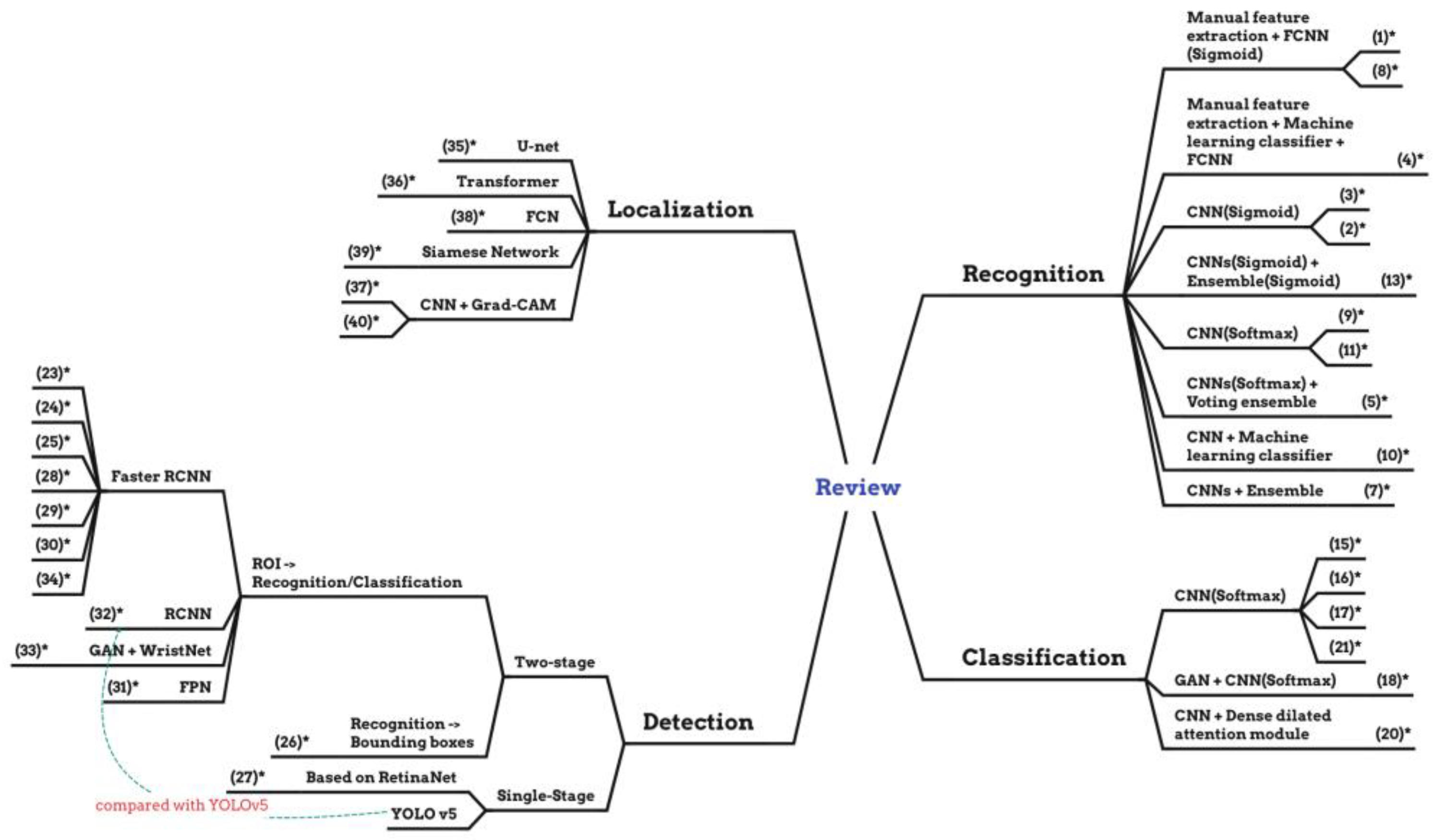
4.1. Recognition
4.1.1. Full-Connected Neural Network (FCNN)
4.1.2. Convolutional Neural Network (CNN)
4.2. Classification
4.2.1. Convolutional Neural Network (CNN)
4.2.2. Generative Adversarial Network (GAN)
4.3. Detection
4.3.1. Region Convolutional Neural Network (R-CNN)
4.3.2. Faster-Region Convolutional Neural Network (Faster R-CNN)
4.3.3. Feature Pyramid Network (FPN)
4.3.4. You Only Look Once (YOLO)
4.3.5. RetinaNet
4.4. Localization
4.4.1. U-Net
4.4.2. Fully Convolutional Network (FCN)
4.4.3. Spatial Transformer
4.4.4. Gradient-Weighted Class Activation Mapping (Grad-CAM)
5. Discussion
6. Solutions following the Three Issues
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med. 2020, 8, 713. [Google Scholar] [CrossRef]
- Latif, J.; Xiao, C.; Imran, A.; Tu, S. Medical imaging using machine learning and deep learning algorithms: A review. In Proceedings of the 2019 2nd International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, 30–31 January 2019; pp. 1–5. [Google Scholar]
- Fourcade, A.; Khonsari, R.H. Deep learning in medical image analysis: A third eye for doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sengupta, S. Lakshminarayanan, Explainable deep learning models in medical image analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Khalid, H.; Hussain, M.; Al Ghamdi, M.A.; Khalid, T.; Khalid, K.; Khan, M.A.; Fatima, K.; Masood, K.; Almotiri, S.H.; Farooq, M.S.; et al. A comparative systematic literature review on knee bone reports from mri, x-rays and ct scans using deep learning and machine learning methodologies. Diagnostics 2020, 10, 518. [Google Scholar] [CrossRef]
- Minnema, J.; Ernst, A.; van Eijnatten, M.; Pauwels, R.; Forouzanfar, T.; Batenburg, K.J.; Wolff, J. A review on the application of deep learning for ct reconstruction, bone segmentation and surgical planning in oral and maxillofacial surgery. Dentomaxillofacial Radiol. 2022, 51, 20210437. [Google Scholar] [CrossRef]
- Wong, S.; Al-Hasani, H.; Alam, Z.; Alam, A. Artificial intelligence in radiology: How will we be affected? Eur. Radiol. 2019, 29, 141–143. [Google Scholar] [CrossRef]
- Bujila, R.; Omar, A.; Poludniowski, G. A validation of spekpy: A software toolkit for modeling X-ray tube spectra. Phys. Medica 2020, 75, 44–54. [Google Scholar] [CrossRef]
- Lim, S.E.; Xing, Y.; Chen, Y.; Leow, W.K.; Howe, T.S.; Png, M.A. Detection of femur and radius fractures in X-ray images. In Proceedings of the 2nd International Conference on Advances in Medical Signal and Information Processing, Valletta, Malta, 5–8 September 2004; Volume 65. [Google Scholar]
- Linda, C.H.; Jiji, G.W. Crack detection in X-ray images using fuzzy index measure. Appl. Soft Comput. 2011, 11, 3571–3579. [Google Scholar] [CrossRef]
- Umadevi, N.; Geethalakshmi, S. Multiple classification system for fracture detection in human bone X-ray images. In Proceedings of the 2012 Third International Conference on Computing, Communication and Networking Technologies (ICCCNT’12), Coimbatore, India, 26–28 July 2012; pp. 1–8. [Google Scholar]
- Al-Ayyoub, M.; Hmeidi, I.; Rababah, H. Detecting hand bone fractures in X-ray images. J. Multimed. Process. Technol. 2013, 4, 155–168. [Google Scholar]
- He, J.C.; Leow, W.K.; Howe, T.S. Hierarchical classifiers for detection of fractures in X-ray images. In Computer Analysis of Images and Patterns, Proceedings of the 12th International Conference, CAIP 2007, Vienna, Austria, 27–29 August 2007; Proceedings 12; Springer: Cham, Switzerland, 2007; pp. 962–969. [Google Scholar]
- Lum, V.L.F.; Leow, W.K.; Chen, Y.; Howe, T.S.; Png, M.A. Combining classifiers for bone fracture detection in X-ray images. In Proceedings of the IEEE International Conference on Image Processing 2005, Genova, Italy, 14 September 2005; Volume 1, p. I-1149. [Google Scholar]
- Ghaderzadeh, M.; Aria, M.; Hosseini, A.; Asadi, F.; Bashash, D.; Abolghasemi, H. A fast and efficient CNN model for B-ALL diagnosis and its subtypes classification using peripheral blood smear images. Int. J. Intell. Syst. 2022, 37, 5113–5133. [Google Scholar] [CrossRef]
- Hosseini, A.; Eshraghi, M.A.; Taami, T.; Sadeghsalehi, H.; Hoseinzadeh, Z.; Ghaderzadeh, M.; Rafiee, M. A mobile application based on efficient lightweight CNN model for classification of B-ALL cancer from non-cancerous cells: A design and implementation study. Inform. Med. Unlocked 2023, 39, 101244. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, H.; Moradi, M.; Prasanna, P.; Syeda-Mahmood, T.F. Fracture detection in X-ray images through stacked random forests feature fusion. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 801–805. [Google Scholar]
- Myint, W.W.; Tun, K.S.; Tun, H.M. Analysis on leg bone fracture detection and classification using X-ray images. Mach. Learn. Res. 2018, 3, 49–59. [Google Scholar] [CrossRef]
- Langerhuizen, D.W.; Janssen, S.J.; Mallee, W.H.; Van Den Bekerom, M.P.; Ring, D.; Kerkhoffs, G.M.; Jaarsma, R.L.; Doornberg, J.N. What are the applications and limitations of artificial intelligence for fracture detection and classification in orthopaedic trauma imaging? a systematic review. Clin. Orthop. Relat. Res. 2019, 477, 2482. [Google Scholar] [CrossRef] [PubMed]
- Adam, A.; Rahman, A.H.A.; Sani, N.S.; Alyessari, Z.A.A.; Mamat, N.J.Z.; Hasan, B. Epithelial layer estimation using curvatures and textural features for dysplastic tissue detection. CMC-Comput. Mater. Contin. 2021, 67, 761–777. [Google Scholar] [CrossRef]
- LTanzi; Vezzetti, E.; Moreno, R.; Moos, S. X-ray bone fracture classification using deep learning: A baseline for designing a reliable approach. Appl. Sci. 2020, 10, 1507. [Google Scholar]
- Joshi, D.; Singh, T.P. A survey of fracture detection techniques in bone X-ray images. Artif. Intell. Rev. 2020, 53, 4475–4517. [Google Scholar] [CrossRef]
- Rainey, C.; McConnell, J.; Hughes, C.; Bond, R.; McFadden, S. Artificial intelligence for diagnosis of fractures on plain radiographs: A scoping review of current literature. Intell.-Based Med. 2021, 5, 100033. [Google Scholar] [CrossRef]
- Hassaballah, M.; Awad, A.I. Deep Learning in Computer Vision: Principles and Applications; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Dimililer, K. Ibfds: Intelligent bone fracture detection system. Procedia Comput. Sci. 2017, 120, 260–267. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Ansari, M.S.; Aslam, A.; Kanwal, N.; Asghar, M.; Lee, B. A survey of modern deep learning based object detection models. Digit. Signal Process. 2022, 126, 103514. [Google Scholar] [CrossRef]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef]
- Ebsim, R.; Naqvi, J.; Cootes, T.F. Automatic detection of wrist fractures from posteroanterior and lateral radiographs: A deep learning-based approach. In Computational Methods and Clinical Applications in Musculoskeletal Imaging, Proceedings of the 6th International Workshop, MSKI 2018, Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers 6; Springer: Cham, Switzerland, 2019; pp. 114–125. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Bagul, A.; Ding, D.; Duan, T.; Mehta, H.; Yang, B.; Zhu, K.; Laird, D.; Ball, R.L.; et al. Mura: Large dataset for abnormality detection in musculoskeletal radiographs. arXiv 2017, arXiv:1712.06957. [Google Scholar]
- Basha, C.Z.; Padmaja, T.M.; Balaji, G. An effective and reliable computer automated technique for bone fracture detection. EAI Endorsed Trans. Pervasive Health Technol. 2019, 5, e2. [Google Scholar] [CrossRef]
- Kitamura, G.; Chung, C.Y.; Moore, B.E. Ankle fracture detection utilizing a convolutional neural network ensemble implemented with a small sample, de novo training, and multiview incorporation. J. Digit. Imaging 2019, 32, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Badgeley, M.A.; Zech, J.R.; Oakden-Rayner, L.; Glicksberg, B.S.; Liu, M.; Gale, W.; McConnell, M.V.; Percha, B.; Snyder, T.M.; Dudley, J.T. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit. Med. 2019, 2, 31. [Google Scholar] [CrossRef]
- Derkatch, S.; Kirby, C.; Kimelman, D.; Jozani, M.J.; Davidson, J.M.; Leslie, W.D. Identification of vertebral fractures by convolutional neural networks to predict nonvertebral and hip fractures: A registry-based cohort study of dual X-ray absorptiometry. Radiology 2019, 293, 405–411. [Google Scholar] [CrossRef]
- Yang, A.Y.; Cheng, L.; Shimaponda-Nawa, M.; Zhu, H.-Y. Long-bone fracture detection using artificial neural networks based on line features of X-ray images. In Proceedings of the 2019 IEEE Symposium Series on Computational Intelligence (SSCI), Xiamen, China, 6–9 December 2019; pp. 2595–2602. [Google Scholar]
- Yadav, D.; Rathor, S. Bone fracture detection and classification using deep learning approach. In Proceedings of the 2020 International Conference on Power Electronics & IoT Applications in Renewable Energy and Its Control (PARC), Mathura, India, 28–29 February 2020; pp. 282–285. [Google Scholar]
- Yang, F.; Wei, G.; Cao, H.; Xing, M.; Liu, S.; Liu, J. Computer-assisted bone fractures detection based on depth feature. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2020; Volume 782, p. 022114. [Google Scholar]
- Beyaz, S.; Açıcı, K.; Sümer, E. Femoral neck fracture detection in X-ray images using deep learning and genetic algorithm approaches. Jt. Dis. Relat. Surg. 2020, 31, 175. [Google Scholar] [CrossRef]
- Tobler, P.; Cyriac, J.; Kovacs, B.K.; Hofmann, V.; Sexauer, R.; Paciolla, F.; Stieltjes, B.; Amsler, F.; Hirschmann, A. Ai-based detection and classification of distal radius fractures using low-effort data labeling: Evaluation of applicability and effect of training set size. Eur. Radiol. 2021, 31, 6816–6824. [Google Scholar] [CrossRef]
- Uysal, F.; Hardala, F.; Peker, O.; Tolunay, T.; Tokgöz, N. Classification of shoulder X-ray images with deep learning ensemble models. Appl. Sci. 2021, 11, 2723. [Google Scholar] [CrossRef]
- Kong, S.H.; Lee, J.-W.; Bae, B.U.; Sung, J.K.; Jung, K.H.; Kim, J.H.; Shin, C.S. Development of a spine X-ray-based fracture prediction model using a deep learning algorithm. Endocrinol. Metab. 2022, 37, 674–683. [Google Scholar] [CrossRef]
- Chung, S.W.; Han, S.S.; Lee, J.W.; Oh, K.-S.; Kim, N.R.; Yoon, J.P.; Kim, J.Y.; Moon, S.H.; Kwon, J.; Lee, H.-J.; et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. 2018, 89, 468–473. [Google Scholar] [CrossRef]
- Chen, Y. Classification of Atypical Femur Fracture with Deep Neural Networks. Master’s Thesis, KTH Royal Institute of Technology, Stockholm, Sweden, 2019. [Google Scholar]
- Lotfy, M.; Shubair, R.M.; Navab, N.; Albarqouni, S. Investigation of focal loss in deep learning models for femur fractures classification. In Proceedings of the 2019 International Conference on Electrical and Computing Technologies and Applications (ICECTA), Ras Al Khaimah, United Arab Emirates, 19–21 November 2019; pp. 1–4. [Google Scholar]
- Mutasa, S.; Varada, S.; Goel, A.; Wong, T.T.; Rasiej, M.J. Advanced deep learning techniques applied to automated femoral neck fracture detection and classification. J. Digit. Imaging 2020, 33, 1209–1217. [Google Scholar] [CrossRef]
- Lee, C.; Jang, J.; Lee, S.; Kim, Y.S.; Jo, H.J.; Kim, Y. Classification of femur fracture in pelvic X-ray images using meta-learned deep neural network. Sci. Rep. 2020, 10, 13694. [Google Scholar] [CrossRef] [PubMed]
- Yuxiang, K.; Jie, Y.; Zhipeng, R.; Guokai, Z.; Wen, C.; Yinguang, Z.; Qiang, D. Dense dilated attentive network for automatic classification of femur trochanteric fracture. Sci. Program. 2021, 2021, 1929800. [Google Scholar]
- Kang, Y.; Ren, Z.; Zhang, Y.; Zhang, A.; Xu, W.; Zhang, G.; Dong, Q. Deep scale-variant network for femur trochanteric fracture classification with hp loss. J. Healthc. Eng. 2022, 2022, 1560438. [Google Scholar] [CrossRef]
- Alzaid, A.; Wignall, A.; Dogramadzi, S.; Pandit, H.; Xie, S.Q. Automatic detection and classification of peri-prosthetic femur fracture. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 649–660. [Google Scholar] [CrossRef]
- Gan, K.; Xu, D.; Lin, Y.; Shen, Y.; Zhang, T.; Hu, K.; Zhou, K.; Bi, M.; Pan, L.; Wu, W.A. Artificial intelligence detection of distal radius fractures: A comparison between the convolutional neural network and professional assessments. Acta Orthop. 2019, 90, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Thian, Y.L.; Li, Y.; Jagmohan, P.; Sia, D.; Chan, V.E.Y.; Tan, R.T. Convolutional neural networks for automated fracture detection and localization on wrist radiographs. Radiol. Artif. Intell. 2019, 1, e180001. [Google Scholar] [CrossRef]
- Yahalomi, E.; Chernofsky, M.; Werman, M. Detection of distal radius fractures trained by a small set of X-ray images and faster r-cnn. In Intelligent Computing, Proceedings of the 2019 Computing Conference, London, UK, 16–17 July 2019; Springer: Cham, Switzerland, 2019; Volume 1, pp. 971–981. [Google Scholar]
- Jones, R.M.; Sharma, A.; Hotchkiss, R.; Sperling, J.W.; Hamburger, J.; Ledig, C.; O’Toole, R.; Gardner, M.; Venkatesh, S.; Roberts, M.M.; et al. Assessment of a deep-learning system for fracture detection in musculoskeletal radiographs. NPJ Digit. Med. 2020, 3, 144. [Google Scholar] [CrossRef]
- Krogue, J.D.; Cheng, K.V.; Hwang, K.M.; Toogood, P.; Meinberg, E.G.; Geiger, E.J.; Zaid, M.; McGill, K.C.; Patel, R.; Sohn, J.H.; et al. Automatic hip fracture identification and functional subclassification with deep learning. Radiol. Artif. Intell. 2020, 2, e190023. [Google Scholar] [CrossRef]
- Qi, Y.; Zhao, J.; Shi, Y.; Zuo, G.; Zhang, H.; Long, Y.; Wang, F.; Wang, W. Ground truth annotated femoral X-ray image dataset and object detection based method for fracture types classification. IEEE Access 2020, 8, 189436–189444. [Google Scholar] [CrossRef]
- Ma, Y.; Luo, Y. Bone fracture detection through the two-stage system of crack-sensitive convolutional neural network. Inform. Med. Unlocked 2021, 22, 100452. [Google Scholar] [CrossRef]
- Xue, L.; Yan, W.; Luo, P.; Zhang, X.; Chaikovska, T.; Liu, K.; Gao, W.; Yang, K. Detection and localization of hand fractures based on ga faster r-cnn. Alex. Eng. J. 2021, 60, 4555–4562. [Google Scholar] [CrossRef]
- Wu, H.-Z.; Yan, L.-F.; Liu, X.-Q.; Yu, Y.-Z.; Geng, Z.-J.; Wu, W.-J.; Han, C.Q.; Guo, Y.-Q.; Gao, B.-L. The feature ambiguity mitigate operator model helps improve bone fracture detection on X-ray radiograph. Sci. Rep. 2021, 11, 1589. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wang, H.; Chen, W.; Wang, Y.; Yang, B. An attention-based cascade r-cnn model for sternum fracture detection in X-ray images. CAAI Trans. Intell. Technol. 2022, 7, 658–670. [Google Scholar] [CrossRef]
- Wang, W.; Huang, W.; Lu, Q.; Chen, J.; Zhang, M.; Qiao, J.; Zhang, Y. Attention mechanism-based deep learning method for hairline fracture detection in hand X-rays. Neural Comput. Appl. 2022, 34, 18773–18785. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-H.; Horng, M.-H.; Li, R.-S.; Sun, Y.-N. Scaphoid fracture detection by using convolutional neural network. Diagnostics 2022, 12, 895. [Google Scholar] [CrossRef]
- Lindsey, R.; Daluiski, A.; Chopra, S.; Lachapelle, A.; Mozer, M.; Sicular, S.; Hanel, D.; Gardner, M.; Gupta, A.; Hotchkiss, R.; et al. Deep neural network improves fracture detection by clinicians. Proc. Natl. Acad. Sci. USA 2018, 115, 11591–11596. [Google Scholar] [CrossRef]
- Jim, A.; Kazi, A.; Albarqouni, S.; Kirchhoff, S.; Sträter, A.; Biberthaler, P.; Mateus, D.; Navab, N. Weakly-supervised localization and classification of proximal femur fractures. arXiv 2018, arXiv:1809.10692. [Google Scholar]
- Cheng, C.T.; Ho, T.Y.; Lee, T.Y.; Chang, C.C.; Chou, C.C.; Chen, C.C.; Chung, I.F.; Liao, C.H. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur. Radiol. 2019, 29, 5469–5477. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, L.; Cheng, C.-T.; Jin, D.; Harrison, A.P.; Xiao, J.; Liao, C.H.; Miao, S. Weakly supervised universal fracture detection in pelvic X-rays. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22; Springer: Cham, Switzerland, 2019; pp. 459–467. [Google Scholar]
- Chen, H.; Wang, Y.; Zheng, K.; Li, W.; Chang, C.-T.; Harrison, A.P.; Xiao, J.; Hager, G.D.; Lu, L.; Liao, C.-H.; et al. Anatomy-aware siamese network: Exploiting semantic asymmetry for accurate pelvic fracture detection in X-ray images. In Computer Vision–ECCV 2020, Proceedings of the 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part XXIII 16; Springer: Cham, Switzerland, 2020; pp. 239–255. [Google Scholar]
- Yoon, A.P.; Lee, Y.-L.; Kane, R.L.; Kuo, C.-F.; Lin, C.; Chung, K.C. Development and validation of a deep learning model using convolutional neural networks to identify scaphoid fractures in radiographs. JAMA Netw. Open 2021, 4, e216096. [Google Scholar] [CrossRef]
- AVoulodimos; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support vector machine versus random forest for remote sensing image classification: A meta-analysis and systematic review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Guo, R.; Fu, D.; Sollazzo, G. An ensemble learning model for asphalt pavement performance prediction based on gradient boosting decision tree. Int. J. Pavement Eng. 2022, 23, 3633–3646. [Google Scholar] [CrossRef]
- Guo, M.-H.; Xu, T.-X.; Liu, J.-J.; Liu, Z.-N.; Jiang, P.-T.; Mu, T.-J.; Zhang, S.-H.; Martin, R.R.; Cheng, M.-M.; Hu, S.-M. Attention mechanisms in computer vision: A survey. Comput. Vis. Media 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.-M. Bearing fault diagnosis using grad-cam and acoustic emission signals. Appl. Sci. 2020, 10, 2050. [Google Scholar] [CrossRef]
- Tanzi, L.; Audisio, A.; Cirrincione, G.; Aprato, A.; Vezzetti, E. Vision transformer for femur fracture classification. Injury 2022, 53, 2625–2634. [Google Scholar] [CrossRef]
- Huang, P.; He, P.; Tian, S.; Ma, M.; Feng, P.; Xiao, H.; Mercaldo, F.; Santone, A.; Qin, J. A ViT-AMC network with adaptive model fusion and multiobjective optimization for interpretable laryngeal tumor grading from histopathological images. IEEE Trans. Med. Imaging 2022, 42, 15–28. [Google Scholar] [CrossRef]
- Pan, H.; Peng, H.; Xing, Y.; Jiayang, L.; Hualiang, X.; Sukun, T.; Peng, F. Breast tumor grading network based on adaptive fusion and microscopic imaging. Opto-Electron. Eng. 2023, 50, 220158-1–220158-13. [Google Scholar]
- Zhou, X.; Tang, C.; Huang, P.; Tian, S.; Mercaldo, F.; Santone, A. ASI-DBNet: An adaptive sparse interactive resnet-vision transformer dual-branch network for the grading of brain cancer histopathological images. Interdiscip. Sci. Comput. Life Sci. 2023, 15, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, F.; Yang, X.; Wang, Q.; Sun, Y.; Tian, S.; Feng, P.; Huang, P.; Xiao, H. The Swin-Transformer network based on focal loss is used to identify images of pathological subtypes of lung adenocarcinoma with high similarity and class imbalance. J. Cancer Res. Clin. Oncol. 2023, 149, 8581–8592. [Google Scholar] [CrossRef]
- Huang, P.; Tan, X.; Zhou, X.; Liu, S.; Mercaldo, F.; Santone, A. FABNet: Fusion attention block and transfer learning for laryngeal cancer tumor grading in P63 IHC histopathology images. IEEE J. Biomed. Health Inform. 2021, 26, 1696–1707. [Google Scholar] [CrossRef]
- Huang, P.; Zhou, X.; He, P.; Feng, P.; Tian, S.; Sun, Y.; Mercaldo, F.; Santone, A.; Qin, J.; Xiao, H. Interpretable laryngeal tumor grading of histopathological images via depth domain adaptive network with integration gradient CAM and priori experience-guided attention. Comput. Biol. Med. 2023, 154, 106447. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, C.; Huang, P.; Mercaldo, F.; Santone, A.; Shao, Y. LPCANet: Classification of laryngeal cancer histopathological images using a CNN with position attention and channel attention mechanisms. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 666–682. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Tan, X.; Chen, C.; Lv, X.; Li, Y. AF-SENet: Classification of cancer in cervical tissue pathological images based on fusing deep convolution features. Sensors 2020, 21, 122. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28, 1–8. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Doll’ar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolo9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Doll´ar, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 May 2015; pp. 3431–3440. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial transformer networks. Adv. Neural Inf. Process. Syst. 2015, 28, 7–12. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-cam: Why did you say that? arXiv 2016, arXiv:1611.07450. [Google Scholar]
- Li, X.; Xiong, H.; Li, X.; Wu, X.; Zhang, X.; Liu, J.; Bian, J.; Dou, D. Interpretable deep learning: Interpretation, interpretability, trustworthiness, and beyond. Knowl. Inf. Syst. 2022, 64, 3197–3234. [Google Scholar] [CrossRef]
- Chen, K.; Stotter, C.; Klestil, T.; Nehrer, S. Artificial intelligence in orthopedic radiography analysis: A narrative review. Diagnostics 2022, 12, 2235. [Google Scholar] [CrossRef]
- Mazli, N.; Bajuri, M.Y.; Shapii, A.; Hassan, M.R. The semiautomated software (MyAnkle™) for preoperative templating in total ankle replacement surgery. J. Clin. Diagn. Res. 2019, 13, 7. [Google Scholar]
- Chua, S.K.; Singh, D.K.A.; Zubir, K.; Chua, Y.Y.; Rajaratnam, B.S.; Mokhtar, S.A. Relationship between muscle strength, physical performance, quality of life and bone mineral density among postmenopausal women at risk of osteoporotic fractures. Sci. Eng. Health Stud. 2020, 14, 8–21. [Google Scholar]
- Awang, N.; Ahmad, F.; Rahaman, R.A.; Sulaiman, R.; Shapi’i, A.; Rashid, A.H.A. Digital preoperative planning for high tibial osteotomy using 2d medical imaging. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 778–783. [Google Scholar] [CrossRef]
- Sharin, U.; Abdullah, S.; Omar, K.; Adam, A.; Sharis, S. Prostate cancer classification technique on pelvis ct images. Int. J. Eng. Technol. 2019, 8, 206–213. [Google Scholar]
- Nabil, S.; Nordin, R.; Rashdi, M.F. Are facial soft tissue injury patterns associated with facial bone fractures following motorcycle-related accident? J. Oral Maxillofac. Surg. 2022, 80, 1784–1794. [Google Scholar] [CrossRef]
- Gheisari, M.; Ebrahimzadeh, F.; Rahimi, M.; Moazzamigodarzi, M.; Liu, Y.; Dutta Pramanik, P.K.; Heravi, M.A.; Mehbodniya, A.; Ghaderzadeh, M.; Feylizadeh, M.R.; et al. Deep learning: Applications, architectures, models, tools, and frameworks: A comprehensive survey. CAAI Trans. Intell. Technol. 2023, 8, 581–606. [Google Scholar] [CrossRef]
- Li, Q.; Wang, F.; Chen, Y.; Chen, H.; Wu, S.; Farris, A.B.; Jiang, Y.; Kong, J. Virtual liver needle biopsy from reconstructed threedimensional histopathological images: Quantification of sampling error. Comput. Biol. Med. 2022, 147, 105764. [Google Scholar] [CrossRef]
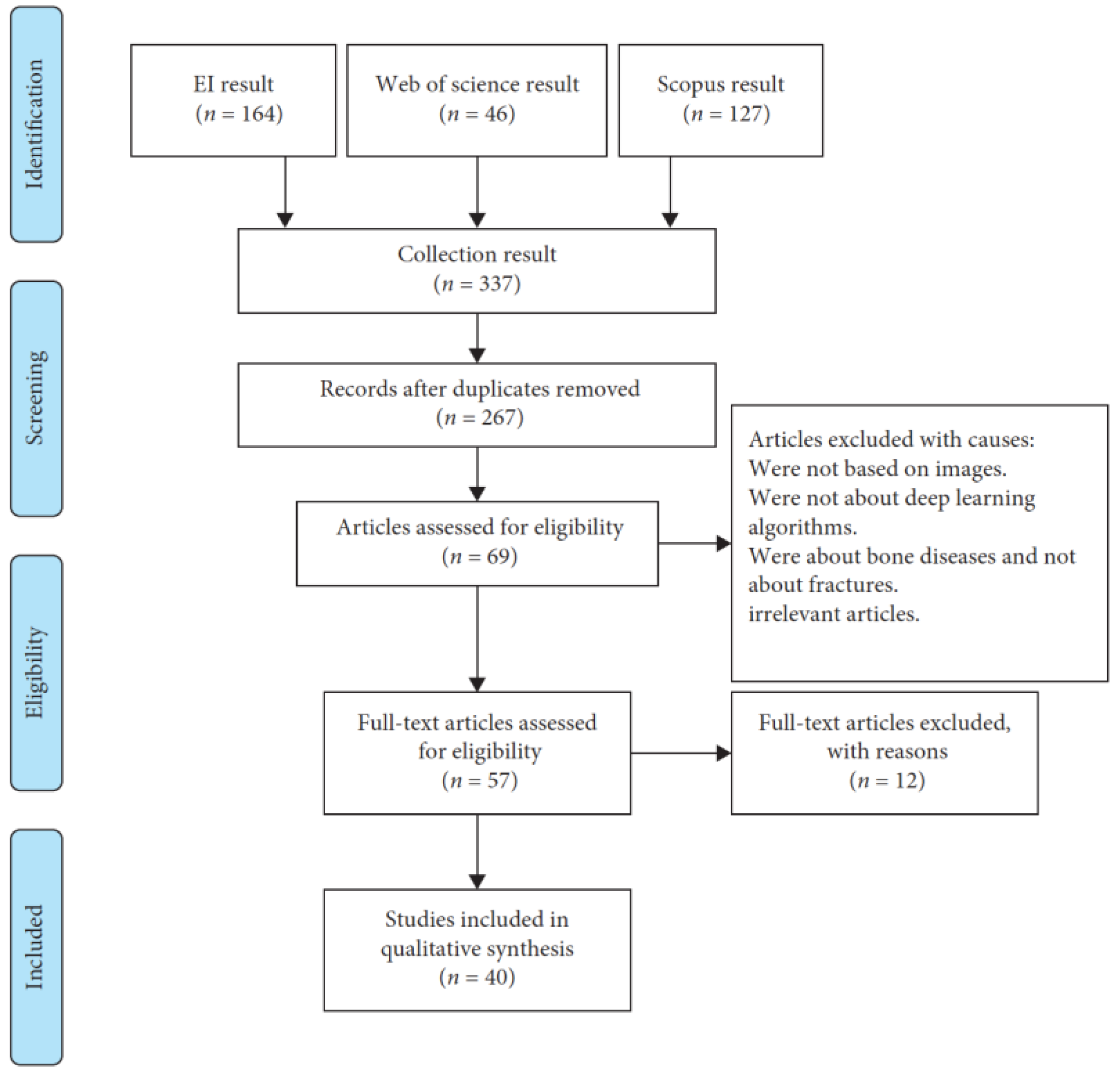

| No | Year | Paper | Task | Method | Dataset Size | Part of Bone |
|---|---|---|---|---|---|---|
| (1) | 2017 | [25] | Recognition | Original images → Haar wavelet applied and reduced size image → BPNN (A three-layer neural network with 1024 input neurons, Sigmoid) | Training set: 30 images; testing set: 70 images | Null |
| (2) | 2018 | [28] | Recognition | Random forest regression voting constrained local model (RF-CLM) → registered patches → CNN (Sigmoid) | 1010 pairs of wrist radiographs (PA, and LAT) | Wrist |
| (3) | 2018 | [29] | Recognition | View(s) → 169-layer convolutional neural network → arithmetic mean of output probabilities → probability of abnormality (Sigmoid) | MURA dataset | Shoulder, humerus, elbow, forearm, wrist, hand, and finger |
| (4) | 2019 | [30] | Recognition | Three steps: Haar wavelet transform and scale-invariant feature transform (SIFT) → k-means clustering based ‘bag of words’ methods → a classical back propagation neural network that contains 1024 neurons in 3 layers | Training set: 70 images; test set: 230 images | Null |
| (5) | 2019 | [31] | Recognition | A voting ensemble method (Inception V3, Resnet, and Xception convolutional neural networks → Softmax, two classes, normal, abnormal cases) | Validation and test sets: 240 views; training set: 1441 views | Ankle |
| (6) | 2019 | [32] | Recognition | Inception-v3 CNN architecture47 | 23,602 hip radiographs | Hip |
| (7) | 2019 | [33] | Recognition | Ensemble algorithm (InceptionResNetV2 CNN and a DenseNet CNN) | 12,742 routine clinical VFA images | Vertebral |
| (8) | 2019 | [34] | Recognition | Input → (1) Standard line-based fracture detection; (2) adaptive differential parameter optimized (ADPO) line-based fracture detection → PCA → 13 line features → ANN (FCNN, BPNN) → fractures and non-fractures | Training set: 20 X-ray images; test set: 23 X-ray images | Leg |
| (9) | 2020 | [35] | Recognition | Deep CNN → Softmax (healthy, fracture) | 4000 images | Null |
| (10) | 2020 | [36] | Recognition | AlexNet (extract features) → PCA → support vector machine (SVM), extreme learning machine (ELM) and random forest (RF) | MURA dataset | Shoulder, forearm, finger, humerus, elbow, hand and wrist |
| (11) | 2020 | [37] | Recognition | Convolutional neural network (CNN) architecture (five blocks, each containing a convolutional layer, batch normalization layer, rectified linear unit, and maximum pooling layer, Softmax, 2 classes, fractures, non-fractures) | 234 frontal pelvic X-ray images | Femoral neck |
| (12) | 2021 | [38] | Recognition | ResNet18 DCNN | 15,775 frontal and lateral radiographs | Distal radius |
| (13) | 2021 | [39] | Recognition | a new ensemble learning model (input → ResNet, ResNeXt, DenseNet, VGG, InceptionV3, and MobileNetV2 + Spinal FC layer → EL1, EL2, EL1, EL2 (spinal fully connected versions) models (Sigmoid) | The MURA dataset | Shoulder |
| (14) | 2022 | [40] | Recognition | A convolutional neural network (CNN)-based prediction algorithm called DeepSurv | Images and medical records from 7301 patients | Spine |
| (15) | 2018 | [41] | Classification | ResNet-152 | 1891 plain shoulder AP radiographs | Proximal humerus |
| (16) | 2019 | [42] | Classification | Convolutional neural network with two diagnostic pipelines | 796 images: 97 AFF images and 399 NFF images | Femur |
| (17) | 2019 | [43] | Classification | DenseNet | 750 images | Femur |
| (18) | 2020 | [44] | Classification | A customized residual network with Softmax classifier | 1444 hip radiographs from 1195 patients | Femoral neck |
| (19) | 2020 | [45] | Classification | An encoder-decoder structured neural network | 786 anterior–posterior pelvic X-ray images and 459 radiology reports acquired from 400 patients | Pelvic |
| (20) | 2021 | [46] | Classification | A novel dense dilated attentive (DDA) network | 390 X-ray images | Femur trochanteric |
| (21) | 2022 | [47] | Classification | A scale-variant network (ResNet + a scaled variant (SV) layer + a hybrid and progressive (HP) loss function) | 31/A1: 117 images; 31/A2: 125 images; 31/A3: 128 images | Femur trochanteric |
| (22) | 2022 | [48] | Classification + Detection + Localization | Vancouver Classification System (type A, B, C) (Densenet161, Resnet50, Inception, VGG and Faster R-CNN, RetinaNet) | 1272 X-ray images | Periprosthetic femur |
| (23) | 2019 | [49] | Detection | Faster-R-CNN → Inception-v4 | 2340 AP wrist radiographs from 2340 patients | Distal radius |
| (24) | 2019 | [50] | Detection | Faster R-CNN (Inception-ResNet) | 7356 wrist radiographic images | Wrist |
| (25) | 2019 | [51] | Detection | Faster R-CNN | 4476 images with labels and bounding boxes for each augmented image | Distal radius |
| (26) | 2020 | [52] | Detection | Dilated Residual Network | 16,019 unique radiographs | All |
| (27) | 2020 | [53] | Detection | RetinaNet object detection algorithm → ROIs (bounding boxes) → A densely connected convolutional neural network (DenseNet) | 3034 hip images | Hip |
| (28) | 2020 | [54] | Detection | An anchor-based Faster R-CNN (ResNet-50 + pyramid networks (FPN)) | 2333 X-ray images | Femoral |
| (29) | 2021 | [55] | Detection | Faster region with convolutional neural network (Faster R-CNN) → CrackNet | 3053 X-ray images | Null |
| (30) | 2021 | [56] | Detection | Faster R-CNN (GA module) | 3067 X-ray radiographs | Hand |
| (31) | 2021 | [57] | Detection | FAMO model (ResNext- 101 + FPN) | 1651 hand, 1302 wrist, 406 elbow, 696 shoulder, 1580 pelvic, 948 knee, 1180 ankle, and 1277 foot images | Hand, wrist, elbow, shoulder, pelvic, knee, ankle, and foot |
| (32) | 2022 | [58] | Detection | A deep convolutional neural network (cascade R-CNN + attention mechanism + atrous convolution) | 1227 labeled X-ray images | Sternum |
| (33) | 2022 | [59] | Detection | Generative adversative network (GAN) → WrisNet is composed of two components (a feature extraction module + a detection module) | 4346 X-rays | Hand |
| (34) | 2022 | [60] | Detection | Faster R-CNN network → ResNet | 167 fractured samples and 194 normal samples for detection task, and 166 fractured samples and 194 normal samples for classification task | Scaphoid |
| (35) | 2018 | [61] | Localization | An extension of the U-Net architecture (two outputs: (1) fracture probability; (2) conditional heat map) | 135,845 radiographs | Foot, elbow, shoulder, knee, spine, femur, ankle, humerus, pelvis, hip, and tibia |
| (36) | 2018 | [62] | Localization | Weakly supervised deep learning approach (spatial transformers (ST) + self-transfer learning (STL)) | 750 images from 672 patients taken | Proximal femur fractures |
| (37) | 2019 | [63] | Localization | DenseNet-121 → Grad-CAM | 25,505 limb radiographs + 3605 PXRs | Hip |
| (38) | 2019 | [64] | Localization | MIL-FCN (DenseNet-121) → mined localized ROIs | 4410 PXRs | Hip and pelvic |
| (39) | 2020 | [65] | Localization | A Siamese network (a spatial transformer layer) | 2359 PXRs | Pelvic |
| (40) | 2021 | [66] | Localization | DCNN (EfficientNetB3) → Grad-CAM | 8329 images | Scaphoid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Z.; Adam, A.; Nasrudin, M.F.; Ayob, M.; Punganan, G. Skeletal Fracture Detection with Deep Learning: A Comprehensive Review. Diagnostics 2023, 13, 3245. https://doi.org/10.3390/diagnostics13203245
Su Z, Adam A, Nasrudin MF, Ayob M, Punganan G. Skeletal Fracture Detection with Deep Learning: A Comprehensive Review. Diagnostics. 2023; 13(20):3245. https://doi.org/10.3390/diagnostics13203245
Chicago/Turabian StyleSu, Zhihao, Afzan Adam, Mohammad Faidzul Nasrudin, Masri Ayob, and Gauthamen Punganan. 2023. "Skeletal Fracture Detection with Deep Learning: A Comprehensive Review" Diagnostics 13, no. 20: 3245. https://doi.org/10.3390/diagnostics13203245
APA StyleSu, Z., Adam, A., Nasrudin, M. F., Ayob, M., & Punganan, G. (2023). Skeletal Fracture Detection with Deep Learning: A Comprehensive Review. Diagnostics, 13(20), 3245. https://doi.org/10.3390/diagnostics13203245







