Evaluating Deep Q-Learning Algorithms for Controlling Blood Glucose in In Silico Type 1 Diabetes
Abstract
:1. Introduction
Structure of Paper
2. Methods
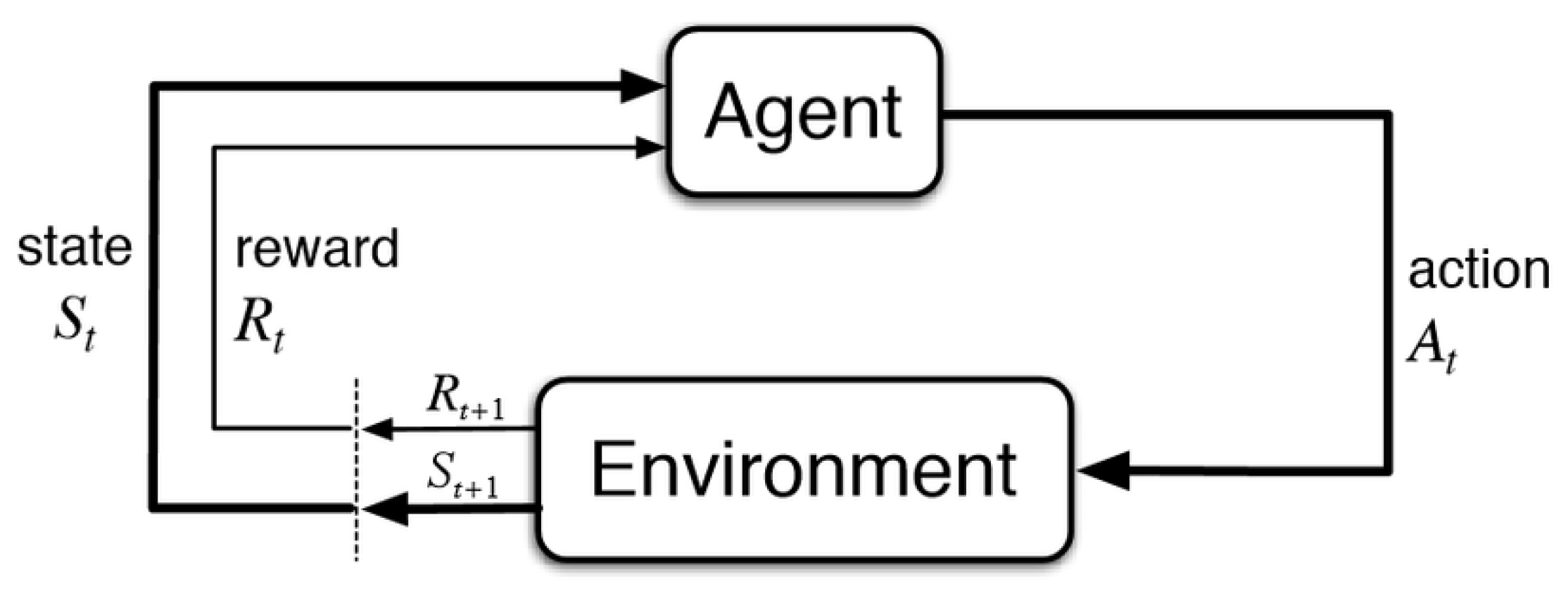
2.1. Reinforcement Learning
2.2. Q-Learning
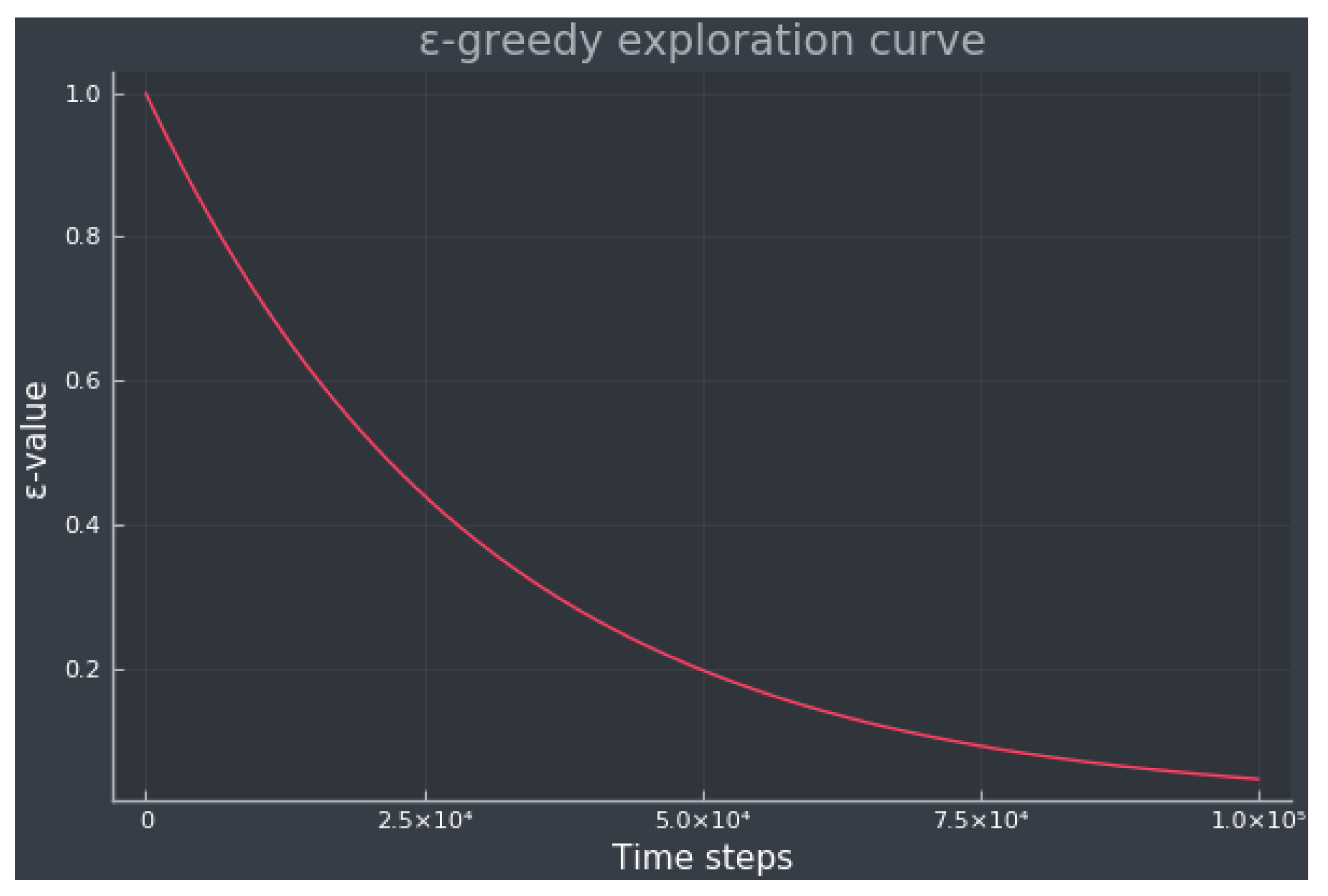
2.2.1. Q-Learning Extensions
- Deep Q-Learning (DQN): In DQN, the Q-value function is approximated using a neural network (NN). The input of the NN is the current state of the environment, whereas the output is the Q-value of all the possible actions the agent can take [38].
- Double Q-Learning (DDQN): This approach proposes two Q-value approximators represented by two NNs, one to estimate the target Q-values and the other one to estimate the predicted Q-values. DDQN reduces the overestimation of the Q-values, usually leading to a better performance compared to DQN [39].
- Dueling DQN and DDQN: This extension changes the standard DQN and DDQN architectures presenting two separate estimators, one for the state-value function and one for the advantage function. The advantage function is defined as the difference between the Q-value function and the state-value function, , and indicates the amount of reward that could have been obtained by the agent by taking the action a over any other action. This method increases the stability of the optimization [40].
- Prioritized Experience Replay (PR): DQN training is not efficient since transitions are randomly sampled to train the network parameters. PR gives priority measures to the transitions, sampling important transitions more frequently [41].
- Noisy DQN: Standard DQN uses a random policy resulting in an inefficient exploration, whereas noisy DQN adds parametric noise to the weights and biases for exploration [42].
- Categorical DQN: This algorithm learns a distribution of the Q-values instead of an estimation of the Q-value function, leading to a more stable and faster learning and usually outperforming standard DQN [43].
- Rainbow DQN: This algorithm integrates all the previous extensions introduced in this section, improving data efficiency and overall performance [44].
2.3. In Silico Simulation
2.3.1. Experiment Setup
- Breakfast: (40 + ) [g] of CHO at 8:00 ± 30 min.
- Lunch: (80 + ) [g] of CHO at 12:00 ± 30 min.
- Dinner: (60 + ) [g] of CHO at 18:00 ± 30 min.
- Supper: (30 + ) [g] of CHO at 22:00 ± 30 min.
- DQN and DDQN: A 4 layer fully connected network with 64 hidden units each. ReLU nonlinearity was used across all layers. The output layer has a linear output.
- Dueling DQN, dueling DDQN, PR DQN, and noisy DQN: A fully connected network consisting of two blocks, each individual block, is similar to the DQN network. Each output layer in the two blocks has linear outputs, representing the advantage and value streams. For the noisy DQN algorithm, we simply added noise to the linear layers and reset the noise parameters after every training batch.
- Categorical DQN: A 4 layer fully connected network with 64 hidden units each. The two first layers are without noise and the following are with noise. ReLU nonlinearity was used across all layers. The output layer has a linear output.
- Rainbow DQN: A fully connected network consisting of two blocks, each with a dense layer of 3 fully connected noisy layers. The input layer consists of an additional linear layer before splitting into the two streams. The amount of hidden units is 64, and ReLU nonlinearity was used across all layers. Each output layer is linear and represents the advantage and value streams as in the dueling DQN case.
3. Results
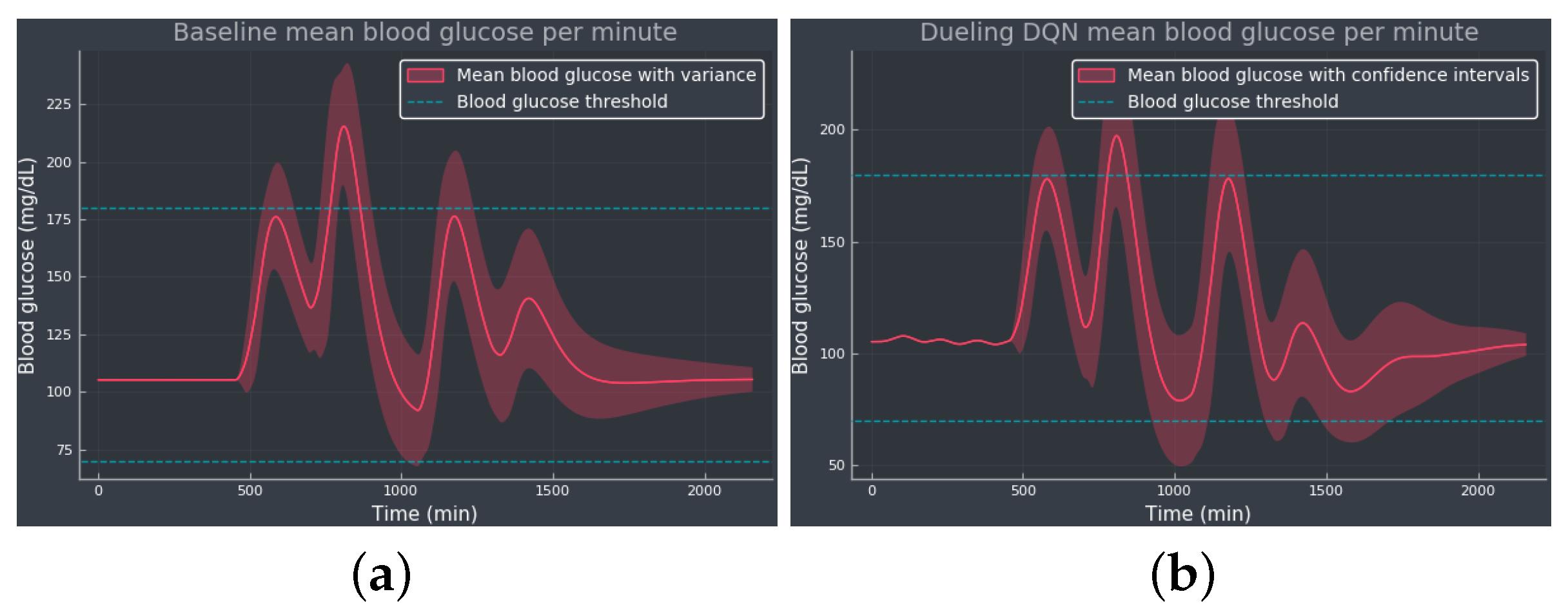
3.1. Experiment 1—Comparing Algorithms
3.2. Experiment 2—Expanded Action Space
3.3. Experiment 3—Meal Bolus Perturbation
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Association, A.D. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2010, 33, S62–S69. [Google Scholar] [CrossRef]
- WHO. Diabetes. 2023. Available online: https://www.who.int/health-topics/diabetes (accessed on 22 September 2023).
- Holt, R.I.; Cockram, C.; Flybjerg, A.; Goldstein, B.J.E. Textbook of Diabetes; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Tuomilehto, J. The emerging global epidemic of type 1 diabetes. Curr. Diabetes Rep. 2013, 6, 795–804. [Google Scholar] [CrossRef]
- El Fathi, A.; Smaoui, M.R.; Gingras, V.; Boulet, B.; Haidar, A. The Artificial Pancreas and Meal Control: An Overview of Postprandial Glucose Regulation in Type 1 Diabetes. IEEE Control Syst. Mag. 2018, 38, 67–85. [Google Scholar] [CrossRef]
- Faculty of Medicine for Doctors and Medical Students. All You Need to Know about the Glucose Tolerance Test. 2016. Available online: https://forum.facmedicine.com/threads/all-you-need-to-know-about-the-glucose-tolerance-test.25348/ (accessed on 22 September 2023).
- Misso, M.L.; Egberts, K.J.; Page, M.; O’Connor, D.; Shaw, J. Cochrane review: Continuous subcutaneous insulin infusion (CSII) versus multiple insulin injections for type 1 diabetes mellitus. Cochrane Database Syst. Rev. 2010, 5, 1726–1867. [Google Scholar] [CrossRef]
- Tamborlane, W.V.; Beck, R.W.; Bode, B.W.; Buckingham, B.; Chase, H.P.; Clemons, R.; Fiallo-Scharer, R.; Fox, L.A.; Gilliam, L.K.; Hirsch, I.B.; et al. Juvenile Diabetes Research Foundation Continuous Glucose Monitoring Study Group. Continuous glucose monitoring and intensive treatment of type 1 diabetes. N. Engl. J. Med. 2008, 359, 1464–1476. [Google Scholar]
- Hovorka, R. Closed-loop insulin delivery: From bench to clinical practice. Nat. Rev. Endocrinol. 2011, 7, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Cinar, A. Artificial pancreas systems: An introduction to the special issue. IEEE Control Syst. Mag. 2018, 38, 26–29. [Google Scholar]
- Bothe, M.K.; Dickens, L.; Reichel, K.; Tellmann, A.; Ellger, B.; Westphal, M.; Faisal, A.A. The use of reinforcement learning algorithms to meet the challenges of an artificial pancreas. Biomed. Signal Process. Control 2013, 10, 661–673. [Google Scholar] [CrossRef]
- Bastani, M. Model-Free Intelligent Diabetes Management Using Machine Learning. Master’s Thesis, University of Albertam, Edmonton, AB, Canada, 2014. [Google Scholar]
- Weiner, A.; Robinson, E.; Gandica, R. 1017-P: Effects of the T:slim X2 Insulin Pump with Basal-IQ Technology on Glycemic Control in a Pediatric Urban Academic Diabetes Practice. Diabetes 2020, 69, 1017-P. [Google Scholar] [CrossRef]
- Tauschmann, M.; Thabit, H.; Bally, L.; Allen, J.M.; Hartnell, S.; Wilinska, M.E.; Ruan, Y.; Sibayan, J.; Kollman, C.; Cheng, P.; et al. Closed-loop insulin delivery in suboptimally controlled type 1 diabetes: A multicentre, 12-week randomised trial. Lancet 2018, 392, 1321–1329. [Google Scholar] [CrossRef]
- Messer, L.H.; Forlenza, G.P.; Sherr, J.L.; Wadwa, R.P.; Buckingham, B.A.; Weinzimer, S.A.; Maahs, D.M.; Slover, R.H. Optimizing Hybrid Closed-Loop Therapy in Adolescents and Emerging Adults Using the MiniMed 670G System. Diabetes Care 2018, 41, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Leelarathna, L.; Choudhary, P.; Wilmot, E.G.; Lumb, A.; Street, T.; Kar, P.; Ng, S.M. Hybrid closed-loop therapy: Where are we in 2021? Diabetes Obes. Metab. 2020, 23, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Petruzelkova, L.; Soupal, J.; Plasova, V.; Jiranova, P.; Neuman, V.; Plachy, L.; Pruhova, S.; Sumnik, Z.; Obermannova, B. Excellent Glycemic Control Maintained by Open-Source Hybrid Closed-Loop AndroidAPS During and After Sustained Physical Activity. Diabetes Technol. Ther. 2018, 20, 744–750. [Google Scholar] [CrossRef]
- Chase, H.P.; Doyle, F.J.; Zisser, H.; Renard, E.; Nimri, R.; Cobelli, C.; Buckingham, B.A.; Maahs, D.M.; Anderson, S.; Magni, L.; et al. Multicenter Closed-Loop/Hybrid Meal Bolus Insulin Delivery with Type 1 Diabetes. Diabetes Technol. Ther. 2014, 16, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Steil, G.M.; Rebrin, K.; Darwin, C.; Hariri, F.; Saad, M.F. Feasibility of automating insulin delivery for the treatment of type 1 diabetes. Diabetes 2006, 55, 3344–3350. [Google Scholar] [CrossRef] [PubMed]
- Hovorka, R.; Canonico, V.; Chassin, L.J.; Haueter, U.; Massi-Benedetti, M.; Federici, M.O.; Pieber, T.R.; Schaller, H.C.; Schaupp, L.; Vering, T.; et al. Nonlinear model predictive control of glucose concentration in subjects with type 1 diabetes. Physiol. Meas. 2004, 25, 905. [Google Scholar] [CrossRef]
- Harvey, R.A.; Dassau, E.; Bevier, W.C.; Seborg, D.E.; Jovanovič, L.; Doyle III, F.J.; Zisser, H.C. Clinical evaluation of an automated artificial pancreas using zone-model predictive control and health monitoring system. Diabetes Technol. Ther. 2014, 16, 348–357. [Google Scholar] [CrossRef]
- Tejedor, M.; Woldaregay, A.Z.; Godtliebsen, F. Reinforcement learning application in diabetes blood glucose control: A systematic review. Artif. Intell. Med. 2020, 104, 101836. [Google Scholar] [CrossRef]
- Sun, Q.; Jankovic, M.V.; Mougiakakou, S.G. Reinforcement learning-based adaptive insulin advisor for individuals with type 1 diabetes patients under multiple daily injections therapy. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 3609–3612. [Google Scholar]
- Fox, I.; Wiens, J. Reinforcement Learning for Blood Glucose Control: Challenges and Opportunities. In Proceedings of the Reinforcement Learning for Real Life (RL4RealLife) Workshop in the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Lee, S.; Kim, J.; Park, S.W.; Jin, S.M.; Park, S.M. Toward a fully automated artificial pancreas system using a bioinspired reinforcement learning design: In silico validation. IEEE J. Biomed. Health Inform. 2020, 25, 536–546. [Google Scholar] [CrossRef]
- Zhu, T.; Li, K.; Herrero, P.; Georgiou, P. Basal Glucose Control in Type 1 Diabetes using Deep Reinforcement Learning: An In Silico Validation. arXiv 2020, arXiv:2005.09059. [Google Scholar] [CrossRef]
- Yamagata, T.; O’Kane, A.; Ayobi, A.; Katz, D.; Stawarz, K.; Marshall, P.; Flach, P.; Santos-Rodríguez, R. Model-Based Reinforcement Learning for Type 1 Diabetes Blood Glucose Control. arXiv 2020, arXiv:2010.06266. [Google Scholar]
- Emerson, H.; Guy, M.; McConville, R. Offline reinforcement learning for safer blood glucose control in people with type 1 diabetes. J. Biomed. Inform. 2023, 142, 104376. [Google Scholar] [CrossRef]
- Viroonluecha, P.; Egea-Lopez, E.; Santa, J. Evaluation of blood glucose level control in type 1 diabetic patients using deep reinforcement learning. PLoS ONE 2022, 17, e0274608. [Google Scholar] [CrossRef] [PubMed]
- Coronato, A.; Naeem, M.; De Pietro, G.; Paragliola, G. Reinforcement learning for intelligent healthcare applications: A survey. Artif. Intell. Med. 2020, 109, 101964. [Google Scholar] [CrossRef]
- Yu, C.; Liu, J.; Nemati, S.; Yin, G. Reinforcement Learning in Healthcare: A Survey. ACM Comput. Surv. 2021, 55, 1–36. [Google Scholar] [CrossRef]
- Choudhury, A.A.; Gupta, D. A Survey on Medical Diagnosis of Diabetes Using Machine Learning Techniques. In Recent Developments in Machine Learning and Data Analytics; Springer: Singapore, 2018; Volume 740. [Google Scholar]
- Khaleel, F.A.; Al-Bakry, A.M. Diagnosis of diabetes using machine learning algorithms. Mater. Today Proc. 2023, 80, 3200–3203. [Google Scholar] [CrossRef]
- Hjerde, S. Evaluating Deep Q-Learning Techniques for Controlling Type 1 Diabetes. Master’s Thesis, UiT The Arctic University of Norway, Tromsø, Norway, 2020. [Google Scholar]
- Sutton, R.S.; Barto, A.G. Reinforcement Learning: An Introduction; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Nachum, O.; Norouzi, M.; Xu, K.; Schuurmans, D. Bridging the gap between value and policy based reinforcement learning. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 2775–2785. [Google Scholar]
- Watkins, C.J.; Dayan, P. Q-learning. Mach. Learn. 1992, 8, 279–292. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Graves, A.; Antonoglou, I.; Wierstra, D.; Riedmiller, M. Playing Atari with Deep Reinforcement Learning. arXiv 2013, arXiv:1312.5602. [Google Scholar]
- van Hasselt, H.; Guez, A.; Silver, D. Deep Reinforcement Learning with Double Q-learning. arXiv 2015, arXiv:1509.06461. [Google Scholar] [CrossRef]
- Wang, Z.; Schaul, T.; Hessel, M.; van Hasselt, H.; Lanctot, M.; de Freitas, N. Dueling Network Architectures for Deep Reinforcement Learning. arXiv 2016, arXiv:1511.06581. [Google Scholar]
- Schaul, T.; Quan, J.; Antonoglou, I.; Silver, D. Prioritized Experience Replay. arXiv 2016, arXiv:1511.05952. [Google Scholar]
- Fortunato, M.; Azar, M.G.; Piot, B.; Menick, J.; Osband, I.; Graves, A.; Mnih, V.; Munos, R.; Hassabis, D.; Pietquin, O.; et al. Noisy Networks for Exploration. arXiv 2019, arXiv:1706.10295. [Google Scholar]
- Bellemare, M.G.; Dabney, W.; Munos, R. A Distributional Perspective on Reinforcement Learning. arXiv 2017, arXiv:1707.06887. [Google Scholar]
- Hessel, M.; Modayil, J.; van Hasselt, H.; Schaul, T.; Ostrovski, G.; Dabney, W.; Horgan, D.; Piot, B.; Azar, M.; Silver, D. Rainbow: Combining Improvements in Deep Reinforcement Learning. arXiv 2017, arXiv:1710.02298. [Google Scholar] [CrossRef]
- Bergman, R.N. Toward physiological understanding of glucose tolerance: Minimal-model approach. Diabetes 1989, 38, 1512–1527. [Google Scholar] [CrossRef] [PubMed]
- Dalla Man, C.; Rizza, R.A.; Cobelli, C. Meal simulation model of the glucose-insulin system. IEEE Trans. Biomed. Eng. 2007, 54, 1740–1749. [Google Scholar] [CrossRef]
- Kanderian, S.S.; Weinzimer, S.A.; Steil, G.M. The identifiable virtual patient model: Comparison of simulation and clinical closed-loop study results. J. Diabetes Sci. Technol. 2012, 6, 371–379. [Google Scholar] [CrossRef]
- Bergman, R.N. Minimal Model: Perspective from 2005. Horm. Res. Paediatr. 2005, 64, 8–15. [Google Scholar] [CrossRef]
- Wilinska, M.E.; Hovorka, R. Simulation models for in silico testing of closed-loop glucose controllers in type 1 diabetes. Drug Discov. Today Dis. Model. 2008, 5, 289–298. [Google Scholar] [CrossRef]
- Huyett, L.M.; Dassau, E.; Zisser, H.C.; Doyle III, F.J. Design and evaluation of a robust PID controller for a fully implantable artificial pancreas. Ind. Eng. Chem. Res. 2015, 54, 10311–10321. [Google Scholar] [CrossRef]
- Mosching, A. Reinforcement Learning Methods for Glucose Regulation in Type 1 Diabetes. Unpublished Master’s Thesis, Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland, 2016. [Google Scholar]
- diaTribe. Time-in-Range. 2023. Available online: https://diatribe.org/time-range (accessed on 22 September 2023).
- Fey, M.; Lenssen, J.E. Fast graph representation learning with PyTorch Geometric. arXiv 2019, arXiv:1903.02428. [Google Scholar]
- Brockman, G.; Cheung, V.; Pettersson, L.; Schneider, J.; Schulman, J.; Tang, J.; Zaremba, W. Openai gym. arXiv 2016, arXiv:1606.01540. [Google Scholar]


| Algorithm | TIR (%) | TAR (%) | TBR (%) | (mg/dL) | (mg/dL) |
|---|---|---|---|---|---|
| Baseline | 95.41 | 4.59 | 0.0 | 124.00 | 33.84 |
| DQN | 95.05 | 4.95 | 0.0 | 125.04 | 35.21 |
| DDQN | 92.82 | 1.90 | 5.28 | 111.67 | 33.31 |
| Dueling DQN | 93.33 | 0.45 | 6.25 | 124.32 | 32.24 |
| Dueling DDQN | 96.71 | 3.29 | 0.0 | 126.92 | 32.32 |
| PR DQN | 94.35 | 5.65 | 0.0 | 122.63 | 33.26 |
| Noisy DQN | 97.04 | 2.96 | 0.0 | 116.10 | 31.74 |
| Categorical DQN | 95.23 | 4.77 | 0.0 | 125.25 | 34.68 |
| Rainbow DQN | 94.35 | 0.0 | 5.65 | 100.66 | 32.20 |
| Algorithm | TIR (%) | TAR (%) | TBR (%) | (mg/dL) | (mg/dL) |
|---|---|---|---|---|---|
| Baseline | 95.41 | 4.59 | 0.0 | 124.00 | 33.84 |
| DQN | 95.23 | 4.77 | 0.0 | 124.46 | 34.88 |
| DDQN | 93.80 | 0.0 | 6.20 | 103.29 | 31.65 |
| Dueling DQN | 97.04 | 2.96 | 0.0 | 113.80 | 34.03 |
| Dueling DDQN | 94.91 | 5.09 | 0.0 | 119.61 | 31.60 |
| PR DQN | 93.75 | 0.65 | 5.60 | 107.04 | 33.70 |
| Noisy DQN | 94.17 | 0.0 | 5.83 | 109.54 | 32.98 |
| Categorical DQN | 93.89 | 0.0 | 6.11 | 106.28 | 32.28 |
| Rainbow DQN | 90.56 | 3.43 | 6.02 | 118.35 | 42.50 |
| Algorithm | TIR | TAR | TBR | (mg/dL) | (mg/dL) | (mU/min) |
|---|---|---|---|---|---|---|
| Baseline | 94.49% | 5.51% | 0.0% | 125.95 | 36.47 | 0.0 |
| DQN | 91.57% | 8.43% | 0.0% | 129.54 | 39.01 | |
| DDQN | 93.15% | 2.45% | 4.40% | 113.33 | 35.59 | 4.72 |
| Dueling DQN | 92.18% | 7.82% | 0.0% | 126.11 | 35.65 | 5.85 |
| Dueling DDQN | 96.16% | 3.84% | 0.0% | 123.41 | 34.43 | 9.06 |
| PR DQN | 93.61% | 6.39% | 0.0% | 123.50 | 34.41 | 6.92 |
| Noisy DQN | 96.20% | 3.8% | 0.0% | 117.73 | 34.97 | 4.50 |
| Categorical DQN | 94.49% | 5.51% | 0.0% | 125.95 | 36.47 | 8.88 |
| Rainbow DQN | 94.21% | 0.0% | 5.79% | 101.41 | 33.57 | 7.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tejedor, M.; Hjerde, S.N.; Myhre, J.N.; Godtliebsen, F. Evaluating Deep Q-Learning Algorithms for Controlling Blood Glucose in In Silico Type 1 Diabetes. Diagnostics 2023, 13, 3150. https://doi.org/10.3390/diagnostics13193150
Tejedor M, Hjerde SN, Myhre JN, Godtliebsen F. Evaluating Deep Q-Learning Algorithms for Controlling Blood Glucose in In Silico Type 1 Diabetes. Diagnostics. 2023; 13(19):3150. https://doi.org/10.3390/diagnostics13193150
Chicago/Turabian StyleTejedor, Miguel, Sigurd Nordtveit Hjerde, Jonas Nordhaug Myhre, and Fred Godtliebsen. 2023. "Evaluating Deep Q-Learning Algorithms for Controlling Blood Glucose in In Silico Type 1 Diabetes" Diagnostics 13, no. 19: 3150. https://doi.org/10.3390/diagnostics13193150
APA StyleTejedor, M., Hjerde, S. N., Myhre, J. N., & Godtliebsen, F. (2023). Evaluating Deep Q-Learning Algorithms for Controlling Blood Glucose in In Silico Type 1 Diabetes. Diagnostics, 13(19), 3150. https://doi.org/10.3390/diagnostics13193150






