Applying Deep Learning Model to Predict Diagnosis Code of Medical Records
Abstract
:1. Introduction
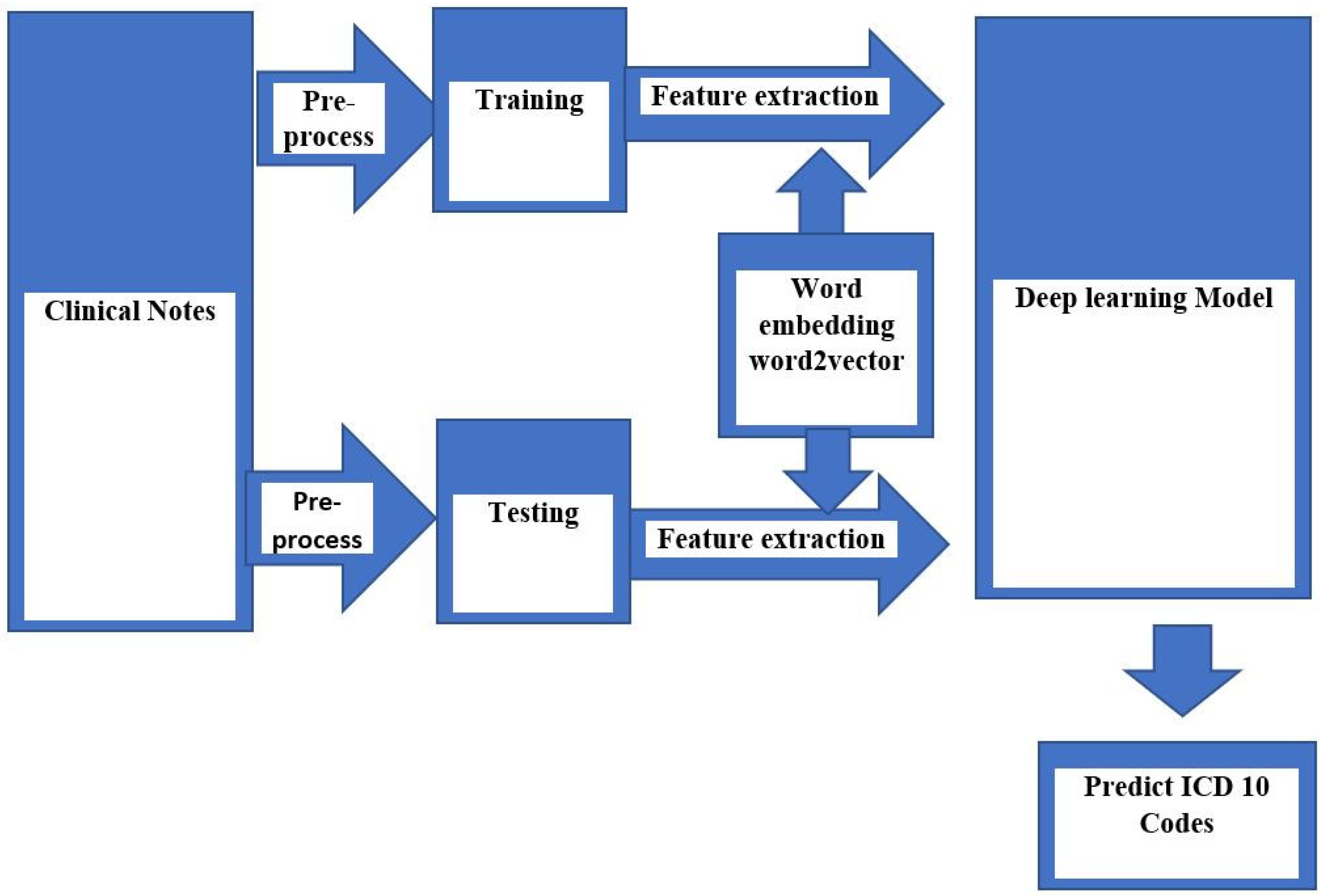
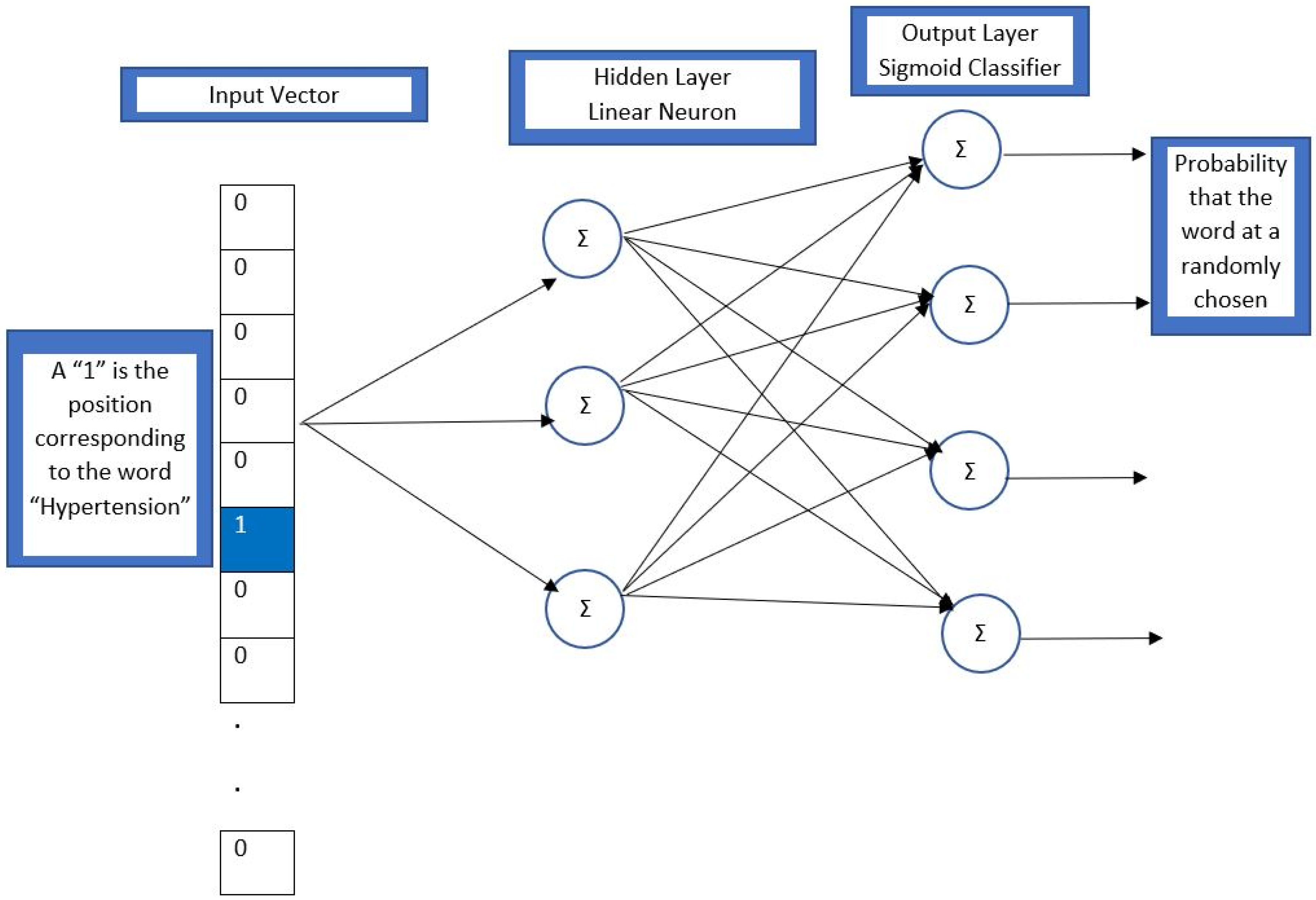
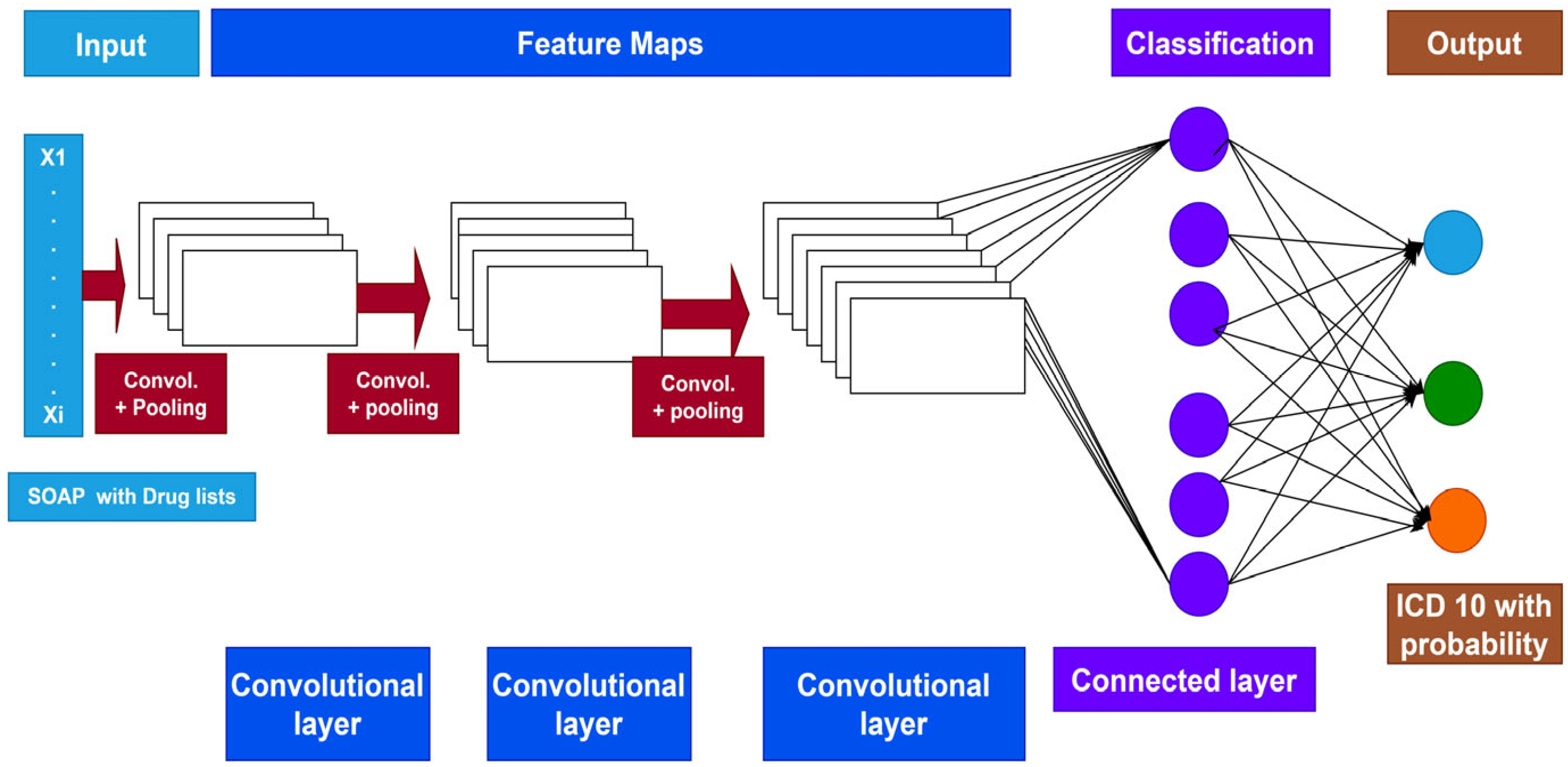
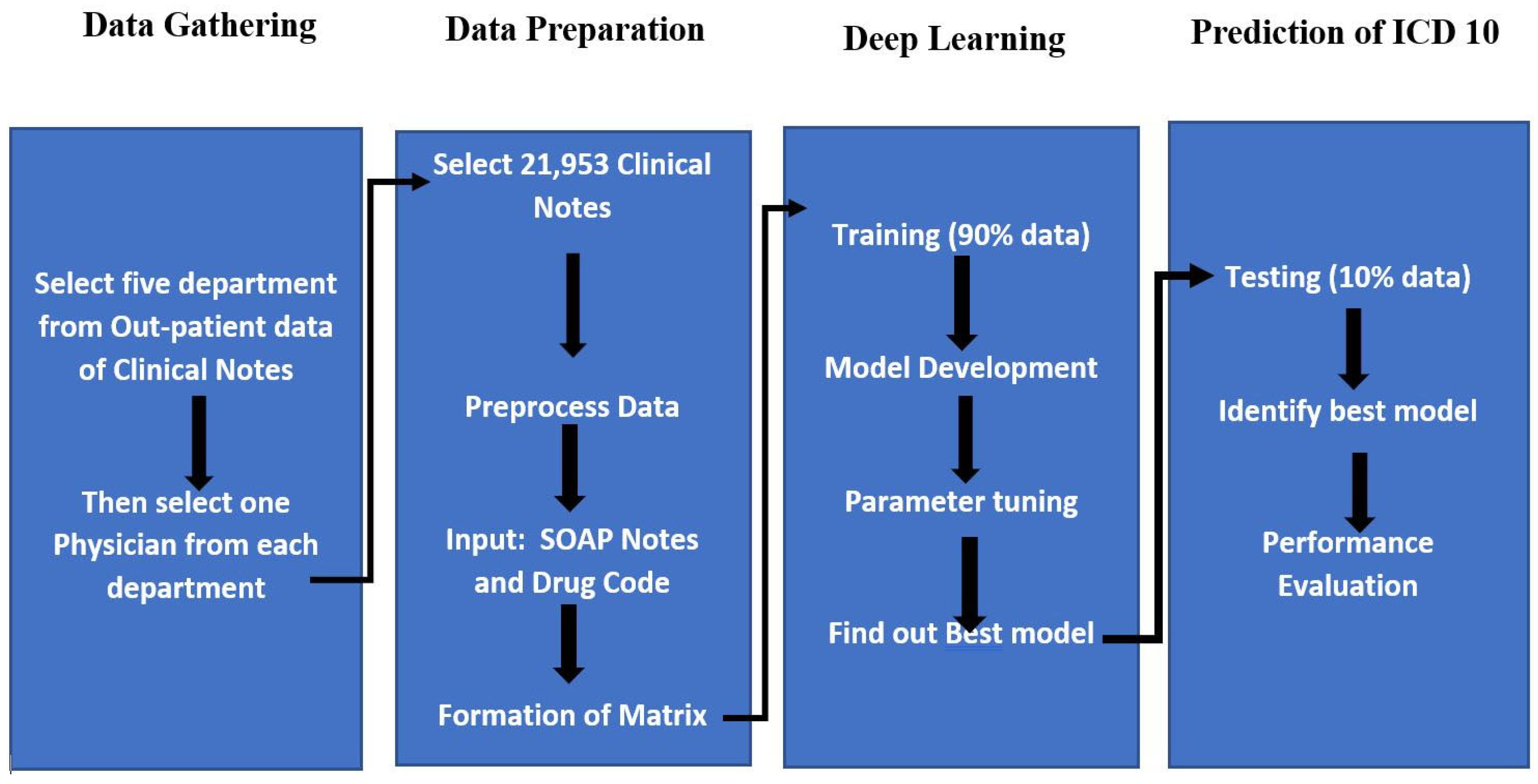
2. Methods
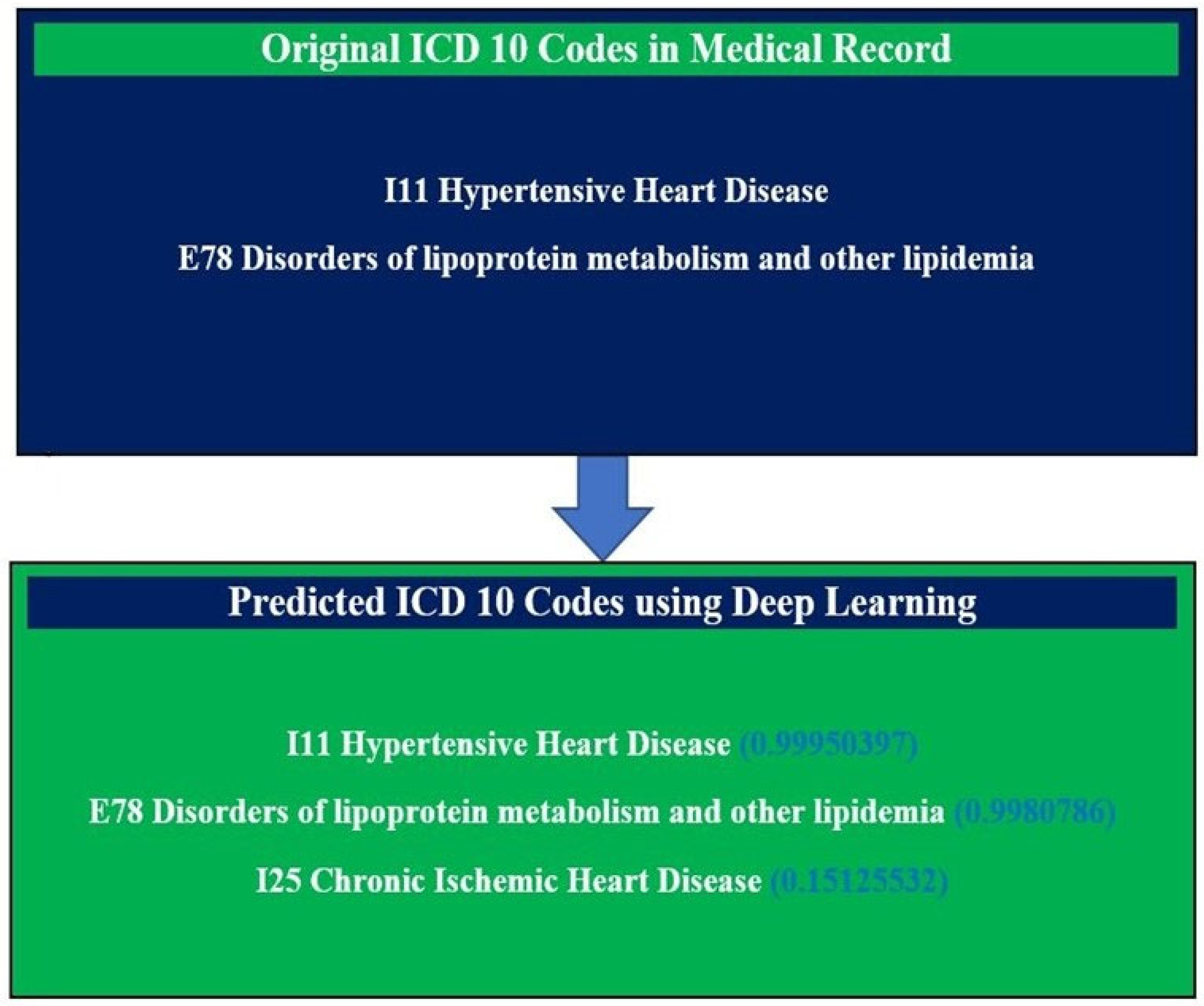
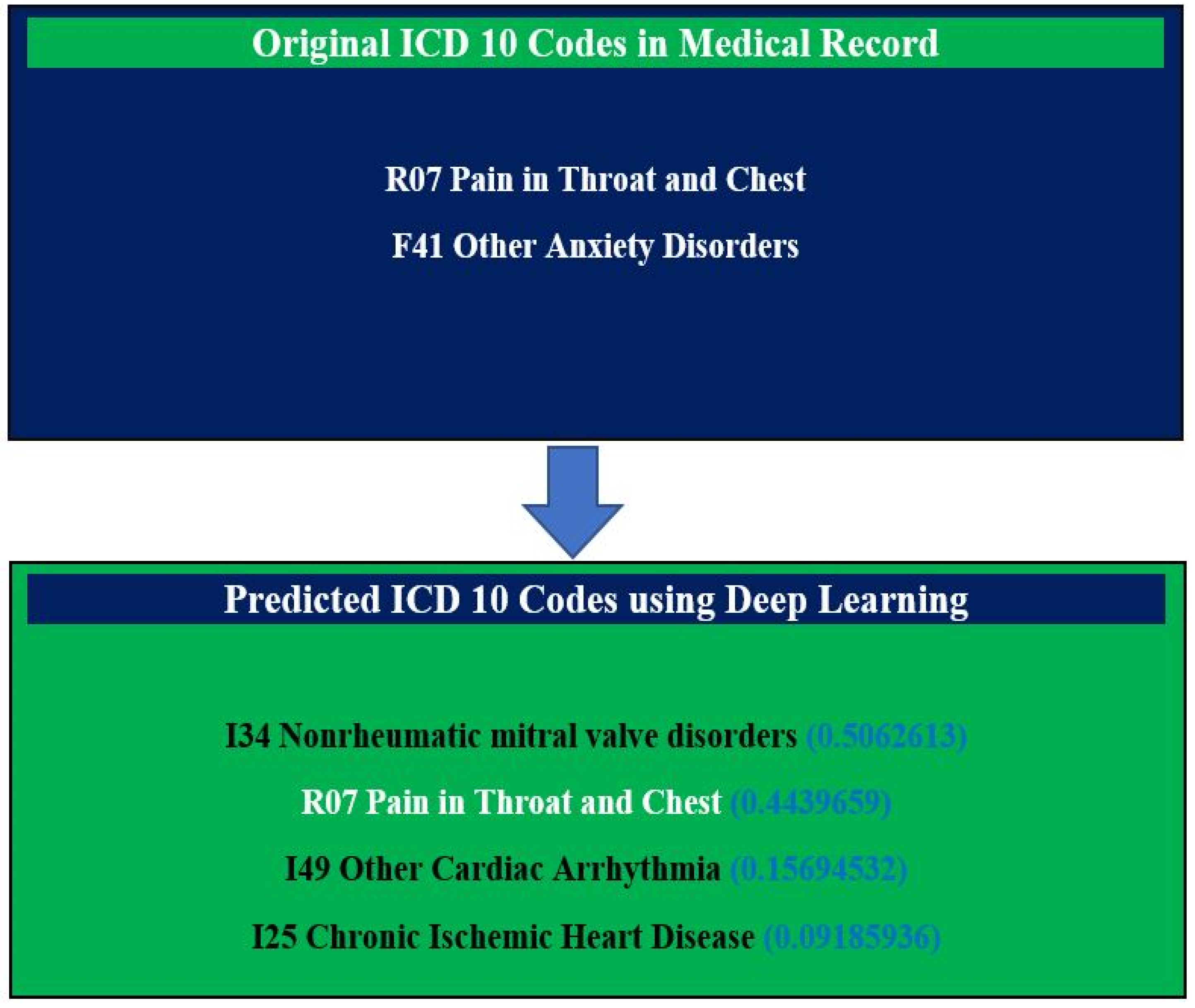
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. The International Classification of Diseases; 10th Revision; World Health Organization: Geneva, Switzerland, 2015; Available online: https://icd.who.int/browse10/2019/en (accessed on 3 March 2023).
- Rae, K.; Britt, H.; Orchard, J.; Finch, C. Classifying sports medicine diagnoses: A comparison of the International classification of diseases 10-Australian modification (ICD-10-AM) and the Orchard sports injury classification system (OSICS-8). Br. J. Sport. Med. 2005, 39, 907–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subotin, M.; Davis, A. A system for predicting ICD-10-PCS codes from electronic health records. In Proceedings of the BioNLP 2014, Baltimore, MD, USA, 26–27 June 2014; pp. 59–67. [Google Scholar]
- Nadathur, S.G. Maximising the value of hospital administrative datasets. Aust. Health Rev. 2010, 34, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Bottle, A.; Aylin, P. Intelligent information: A national system for monitoring clinical performance. Health Serv. Res. 2008, 43, 10–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, H.; Sundararajan, V.; Halfon, P.; Fong, A.; Burnand, B.; Luthi, J.C.; Saunders, L.D.; Beck, C.A.; Feasby, T.E.; Ghali, W.A. Coding algorithms for defining comorbidities in icd-9-cm and icd-10 administrative data. Med. Care 2005, 43, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Banerji, A.; Lai, K.H.; Li, Y.; Saff, R.R.; Camargo, C.A., Jr.; Blumenthal, K.G.; Zhou, L. Natural language processing combined with ICD-9-CM codes as a novel method to study the epidemiology of allergic drug reactions. J. Allergy Clin. Immunol. Pract. 2020, 8, 1032–1038.e1. [Google Scholar] [CrossRef]
- Adams, D.L.; Norman, H.; Burroughs, V.J. Addressing medical coding and billing part ii: A strategy for achieving compliance. a risk management approach for reducing coding and billing errors. J. Natl. Med. Assoc. 2002, 94, 430. [Google Scholar]
- Yang, L.; Kenny, E.M.; Ng, T.L.J.; Yang, Y.; Smyth, B.; Dong, R. Generating plausible counterfactual explanations for deep transformers in financial text classification. arXiv 2020, arXiv:2010.12512. [Google Scholar]
- Melville, P.; Gryc, W.; Lawrence, R.D. Sentiment analysis of blogs by combining lexical knowledge with text classification. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 1275–1284. [Google Scholar]
- Slater, L.T.; Karwath, A.; Williams, J.A.; Russell, S.; Makepeace, S.; Carberry, A.; Gkoutos, G.V. Towards similarity-based differential diagnostics for common diseases. Comput. Biol. Med. 2021, 133, 104360. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the NIPS 2012, Lake Tahoe, NV, USA, 3–8 December 2012. [Google Scholar]
- Zhang, Y.; Chen, R.; Tang, J.; Stewart, W.F.; Sun, J. LEAP: Learning to Prescribe Effective and Safe Treatment Combinations for Multimorbidity. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Halifax, NS, Canada, 13–17 August 2017; pp. 1315–1324. [Google Scholar] [CrossRef]
- Wang, S.M.; Chang, Y.H.; Kuo, L.C.; Lai, F.; Chen, Y.N.; Yu, F.Y.; Chen, C.H.; Lee, C.W.; Chung, Y. Using Deep Learning for Automatic Icd-10 Classification from FreeText Data. Eur. J. Biomed. Inform. 2020, 16, 1–10. [Google Scholar] [CrossRef]
- Chen, P.F.; Wang, S.M.; Liao, W.C.; Kuo, L.C.; Chen, K.C.; Lin, Y.C.; Yang, C.Y.; Chiu, C.H.; Chang, S.C.; Lai, F. Automatic ICD-10 Coding and Training System: Deep Neural Network Based on Supervised Learning. JMIR Med. Inform. 2021, 9, e23230. [Google Scholar] [CrossRef]
- Wang, S.M.; Lai, F.; Sung, C.S.; Chen, Y. ICD-10 Auto-coding System Using Deep Learning. In Proceedings of the 10th International Workshop on Computer Science and Engineering (WCSE 2020), Yangon, Myanmar, 26–28 February 2020; pp. 46–51. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural Language Processing (almost) from Scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Kalchbrenner, N.; Grefenstette, E.; Blunsom, P. A Convolutional Neural Network for Modelling Sentences. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Baltimore, MD, USA, 23–24 June 2014; Association for Computational Linguistics: Baltimore, MD, USA, 2014; pp. 655–665. [Google Scholar]
- Kim, Y. Convolutional Neural Networks for Sentence Classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; Association for Computational Linguistics: Doha, Qatar, 2014; pp. 1746–1751. [Google Scholar]
- Gangavarapu, T.; Jayasimha, A.; Krishnan, G.S.; Kamath, S. Predicting ICD-9 Code Groups with Fuzzy Similarity Based Supervised Multi-label Classification of Unstructured Clinical Nursing Notes. Knowl.-Based Syst. 2020, 190, 105321. [Google Scholar] [CrossRef]
- Chen, M.; Hao, Y.; Hwang, K.; Wang, L.; Wang, L. Disease Prediction by Machine Learning over Big Data from Healthcare Communities. IEEE Access 2017, 5, 8869–8879. [Google Scholar] [CrossRef]
- Moons, E.; Khanna, A.; Akkasi, A.; Moens, M.F. A comparison of deep learning methods for ICD coding of clinical records. Appl. Sci. 2020, 10, 5262. [Google Scholar] [CrossRef]
- Krishnan, G.S.; Kamath, S.S. Evaluating the Quality of Word Representation Models for Unstructured Clinical Text Based ICU Mortality Prediction. In Proceedings of the 20th International Conference on Distributed Computing and Networking, ICDCN’19, Bangalore, India, 4–7 January 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 480–485. [Google Scholar]
- Mauch, J.T.; Rios-Diaz, A.J.; Kozak, G.M.; Zhitomirsky, A.; Broach, R.B.; Fischer, J.P. How to Develop a Risk Prediction Smartphone App. Surg. Innov. 2020, 28, 438–448. [Google Scholar] [CrossRef]
- Kavuluru, R.; Rios, A.; Lu, Y. An empirical evaluation of supervised learning approaches in assigning diagnosis codes to electronic medical records. Artif. Intell. Med. 2015, 65, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, X.; Ramanathan, M.; Zhang, A. Prediction and Informative Risk Factor Selection of Bone Diseases. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 12, 79–91. [Google Scholar] [CrossRef]
- Jin, B.; Che, C.; Liu, Z.; Zhang, S.; Yin, X.; Wei, X. Predicting the Risk of Heart Failure With EHR Sequential Data Modeling. IEEE Access 2018, 6, 9256–9261. [Google Scholar] [CrossRef]
- Arnaud, É.; Elbattah, M.; Gignon, M.; Dequen, G. NLP-Based Prediction of Medical Specialties at Hospital Admission Using Triage Notes. In Proceedings of the 2021 IEEE 9th International Conference on Healthcare Informatics (ICHI), Victoria, BC, Canada, 9–12 August 2021; pp. 548–553. [Google Scholar] [CrossRef]
- Vinod, P.; Safar, S.; Mathew, D.; Venugopal, P.; Joly, L.M.; George, J. Fine-tuning the BERTSUMEXT model for Clinical Report Summarization. In Proceedings of the 2020 International Conference for Emerging Technology (INCET), Belgaum, India, 5–7 June 2020; pp. 1–7. [Google Scholar]
- Teng, F.; Ma, Z.; Chen, J.; Xiao, M.; Huang, L. Automatic medical code assignment via deep learning approach for intelligent healthcare. IEEE J. Biomed. Health Inform. 2020, 24, 2506–2515. [Google Scholar] [CrossRef]
- Bird, S. NLTK: The natural language toolkit. In Proceedings of the COLING/ACL 2006 Interactive Presentation Sessions, Sydney, Australia, 17–18 July 2006; pp. 69–72. [Google Scholar]
- Rehurek, R.; Sojka, P. Gensim–Python Framework for Vector Space Modelling; NLP Centre, Faculty of Informatics, Masaryk University: Brno, Czech Republic, 2011; Volume 3. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Chollet, F. Keras. Internet. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 11 April 2023).
- Hu, S.; Teng, F.; Huang, L.; Yan, J.; Zhang, H. An explainable CNN approach for medical codes prediction from clinical text. BMC Med. Inform. Decis. Mak. 2021, 21, 256. [Google Scholar] [CrossRef]
- Suo, Q.; Ma, F.; Yuan, Y.; Huai, M.; Zhong, W.; Zhang, A.; Gao, J. Personalized Disease Prediction using a CNN-based Similarity Learning Method. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 811–816. [Google Scholar]
- Cheng, Y.; Wang, F.; Zhang, P.; Hu, J. Risk Prediction with Electronic Health Records: A Deep Learning Approach. In Proceedings of the 2016 SIAM International Conference on Data Mining, Miami, FL, USA, 5–7 May 2016; pp. 432–440. [Google Scholar]
- Diao, X.; Huo, Y.; Zhao, S.; Yuan, J.; Cui, M.; Wang, Y.; Lian, X.; Zhao, W. Automated ICD coding for primary diagnosis via clinically interpretable machine learning. Int. J. Med. Inform. 2021, 153, 104543. [Google Scholar] [CrossRef] [PubMed]
- Rashidian, S.; Hajagos, J.; Moffitt, R.A.; Wang, F.; Noel, K.M.; Gupta, R.R.; Tharakan, M.A.; Saltz, J.H.; Saltz, M.M. Deep Learning on Electronic Health Records to Improve Disease Coding Accuracy. AMIA Jt. Summits Transl. Sci. Proc. 2019, 2019, 620–629. [Google Scholar] [PubMed]
- Li, M.; Fei, Z.; Zeng, M.; Wu, F.X.; Li, Y.; Pan, Y.; Wang, J. Automated ICD-9 Coding via A Deep Learning Approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 1193–1202. [Google Scholar] [CrossRef]
- Choi, E.; Bahadori, M.T.; Schuetz, A.; Stewart, W.F.; Sun, J. Doctor AI: Predicting Clinical Events via Recurrent Neural Networks. JMLR Workshop Conf. Proc. 2016, 56, 301–318. [Google Scholar]
- Hsu, C.C.; Chang, P.C.; Chang, A. Multi-Label Classification of ICD Coding Using Deep Learning. In Proceedings of the International Symposium on Community-Centric Systems (CcS), Tokyo, Japan, 23–26 September 2020; pp. 1–6. [Google Scholar]
- Gangavarapu, T.; Krishnan, G.S.; Kamath, S.; Jeganathan, J. FarSight: Long-Term Disease Prediction Using Unstructured Clinical Nursing Notes. IEEE Trans. Emerg. Top. Comput. 2020, 9, 1151–1169. [Google Scholar] [CrossRef]
- Samonte, M.J.C.; Gerardo, B.D.; Fajardo, A.C.; Medina, R.P. ICD-9 tagging of clinical notes using topical word embedding. In Proceedings of the 2018 International Conference on Internet and e-Business, Taipei, Taiwan, 16–18 May 2018; pp. 118–123. [Google Scholar]
- Obeid, J.S.; Dahne, J.; Christensen, S.; Howard, S.; Crawford, T.; Frey, L.J.; Stecker, T.; Bunnell, B.E. Identifying and Predicting intentional self-harm in electronic health record clinical notes: Deep learning approach. JMIR Med. Inform. 2020, 8, e17784. [Google Scholar] [CrossRef]
- Hsu, J.L.; Hsu, T.J.; Hsieh, C.H.; Singaravelan, A. Applying Convolutional Neural Networks to Predict the ICD-9 Codes of Medical Records. Sensors 2020, 20, 7116. [Google Scholar] [CrossRef]
- Xie, P.; Xing, E. A Neural Architecture for Automated ICD Coding. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia, 15–20 July 2018; Association for Computational Linguistics: Melbourne, Australia, 2018; pp. 1066–1076. [Google Scholar]
- Singaravelan, A.; Hsieh, C.-H.; Liao, Y.-K.; Hsu, J.L. Predicting ICD-9 Codes Using Self-Report of Patients. Appl. Sci. 2021, 11, 10046. [Google Scholar] [CrossRef]
- Zeng, M.; Li, M.; Fei, Z.; Yu, Y.; Pan, Y.; Wang, J. Automatic ICD-9 coding via deep transfer learning. Neurocomputing 2019, 324, 43–50. [Google Scholar] [CrossRef]
- Huang, J.; Osorio, C.; Sy, L.W. An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Comput. Methods Programs Biomed. 2019, 177, 141–153. [Google Scholar] [CrossRef] [Green Version]
| Characteristics | Frequency (n) |
|---|---|
| Total number of clinical notes from Doctors of five Departments | 21,953 |
| Doctors’ notes from Neurology | 6027 |
| Doctors’ notes from Psychiatry | 5789 |
| Doctors’ notes from Nephrology | 3707 |
| Doctors’ notes from Cardiology | 3668 |
| Doctors’ notes from Metabolism | 2762 |
| Number of ICD 10 codes | 1131 |
| Number of Drugs | 807 |
| Name of Department | Test Cases | Number of ICD-10 Codes | Number of Drugs | Precision | Recall | F-Score | Accuracy |
|---|---|---|---|---|---|---|---|
| Doctors’ notes from Cardiology | 284 | 148 | 145 | 0.96 | 0.99 | 0.98 | 0.99 |
| Doctors’ notes from Metabolism | 307 | 155 | 136 | 0.78 | 0.97 | 0.86 | 0.97 |
| Doctors’ notes from Psychiatry | 475 | 193 | 128 | 0.64 | 0.91 | 0.75 | 0.91 |
| Doctors’ notes from Neurology | 282 | 358 | 177 | 0.60 | 0.85 | 0.71 | 0.85 |
| Doctors’ notes from Nephrology | 432 | 277 | 221 | 0.52 | 0.88 | 0.65 | 0.88 |
| Batch | Accuracy | Precision | Recall | F-Score | Loss | Error Rate | Computational Time (Minutes) |
|---|---|---|---|---|---|---|---|
| 64 | 0.94 | 0.66 | 0.94 | 0.77 | 0.03 | 0.06 | 14 |
| 128 | 0.89 | 0.45 | 0.89 | 0.60 | 0.05 | 0.11 | 14 |
| 512 | 0.88 | 0.44 | 0.88 | 0.59 | 0.06 | 0.12 | 14 |
| 1000 | 0.84 | 0.40 | 0.84 | 0.54 | 0.07 | 0.16 | 14 |
| 1024 | 0.84 | 0.39 | 0.84 | 0.53 | 0.07 | 0.16 | 14 |
| 2000 | 0.81 | 0.38 | 0.81 | 0.52 | 0.08 | 0.19 | 14 |
| 2048 | 0.81 | 0.36 | 0.81 | 0.50 | 0.08 | 0.19 | 14 |
| Training Accuracy | Testing Accuracy |
|---|---|
| 0.94 | 0.94 |
| Work | Data | Method | Target Variable | Performance Measure |
|---|---|---|---|---|
| Hsu et al. [42] | Discharge summary | Deep learning | (i) 19 distinct ICD-9 chapter codes, (ii) top 50 ICD-9 codes, (iii) top 100 ICD-9 codes | (i) Micro F1 score of 0.76, (ii) Micro F1 score of 0.57, (iii) Micro F1 score of 0.51 |
| Gangavarapu et al. [43] | Nursing notes | Deep learning | 19 distinct ICD-9 chapter codes | Accuracy of 0.833 |
| Samonte et al. [44] | Discharge summary | Deep learning | 10 distinct ICD-9 codes | Precision of 0.780, Recall of 0.620, F1 score of 0.678 |
| Obeid et al. [45] | Clinical notes | Deep learning | ICD-9 code from E950-E959 | Area under the ROC curve score of 0.882, F-score of 0.769 |
| Hsu et al. [46] | Subjective component | Deep learning | (i) 17 distinct ICD-9 chapter codes, (ii) 2017 distinct ICD-9 codes | (i) Accuracy of 0.580, (ii) Accuracy of 0.409 |
| Xie et al. [47] | Diagnosis description | Deep learning | 2833 ICD-9 codes | Sensitivity score of 0.29, Specificity score of 0.33 |
| Singaravelan et al. [48] | Subjective component | Deep learning | 1871 ICD-9 codes | Recall score for chapter code, 0.57; Recall score for block, 0.49; Recall score for three-digit code, 0.43; Recall score for full code, 0.45 |
| Zeng et al. [49] | Discharge summary | Deep learning | 6984 ICD-9 codes | F1 score of 0.42 |
| Huang et al. [50] | Discharge summary | Deep learning | (i) 10 ICD-9 codes, (ii) 10 blocks | (i) F1 score of 0.69, (ii) F1 score of 0.72 |
| Our study | Clinical notes | Deep learning | 1131 ICD-10 codes | Precision of 0.96, Recall of 0.99, F-score of 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masud, J.H.B.; Kuo, C.-C.; Yeh, C.-Y.; Yang, H.-C.; Lin, M.-C. Applying Deep Learning Model to Predict Diagnosis Code of Medical Records. Diagnostics 2023, 13, 2297. https://doi.org/10.3390/diagnostics13132297
Masud JHB, Kuo C-C, Yeh C-Y, Yang H-C, Lin M-C. Applying Deep Learning Model to Predict Diagnosis Code of Medical Records. Diagnostics. 2023; 13(13):2297. https://doi.org/10.3390/diagnostics13132297
Chicago/Turabian StyleMasud, Jakir Hossain Bhuiyan, Chen-Cheng Kuo, Chih-Yang Yeh, Hsuan-Chia Yang, and Ming-Chin Lin. 2023. "Applying Deep Learning Model to Predict Diagnosis Code of Medical Records" Diagnostics 13, no. 13: 2297. https://doi.org/10.3390/diagnostics13132297
APA StyleMasud, J. H. B., Kuo, C.-C., Yeh, C.-Y., Yang, H.-C., & Lin, M.-C. (2023). Applying Deep Learning Model to Predict Diagnosis Code of Medical Records. Diagnostics, 13(13), 2297. https://doi.org/10.3390/diagnostics13132297






