Privacy-Aware Collaborative Learning for Skin Cancer Prediction
Abstract
1. Introduction
- We proposed an optimal classification for skin cancer detection.
- To improve the communication concern, we used asynchronous for skin cancer.
- We improved the convergence rate by using FL.
- We achieved more accuracy by using these methods.
2. Related Work
3. Materials and Methods
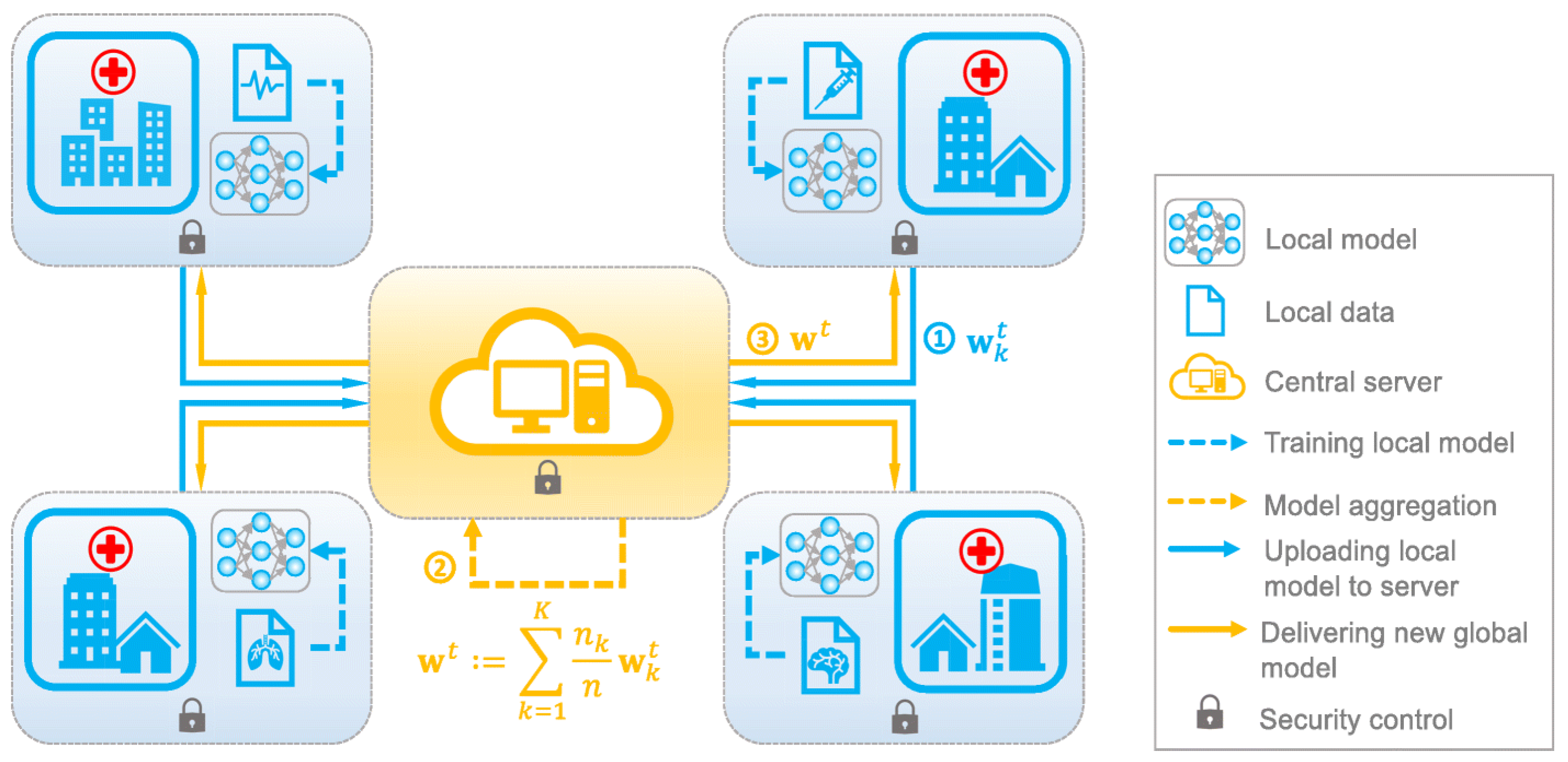
- Selection of Four Hospitals: The study involved the participation of four hospitals, chosen based on their capacity to contribute data to the project and their willingness to collaborate. The hospitals were chosen to ensure geographic diversity and representation of both public and private healthcare facilities.
- Local Training using SVM and CNN: The data collected from each hospital was preprocessed and used to train support vector machines (SVMs) and convolutional neural networks (CNNs) locally. This step ensured that each hospital’s data were used to create models that were specific to their patient population, thereby enhancing model accuracy.
- Conversion of Data into Weights: Following the completion of local training, the data were transformed into weight values that were representative of the learned patterns within the models. This process ensured that only model parameters were exchanged between hospitals, preserving data privacy and security.
- Transfer of Local Weights to Cloud for Training: The weight values obtained from the individual hospitals were then transferred to a cloud-based server for further training. The central server aggregated the weight values and used them to update a global model, which incorporated the latest learnings from all participating hospitals.
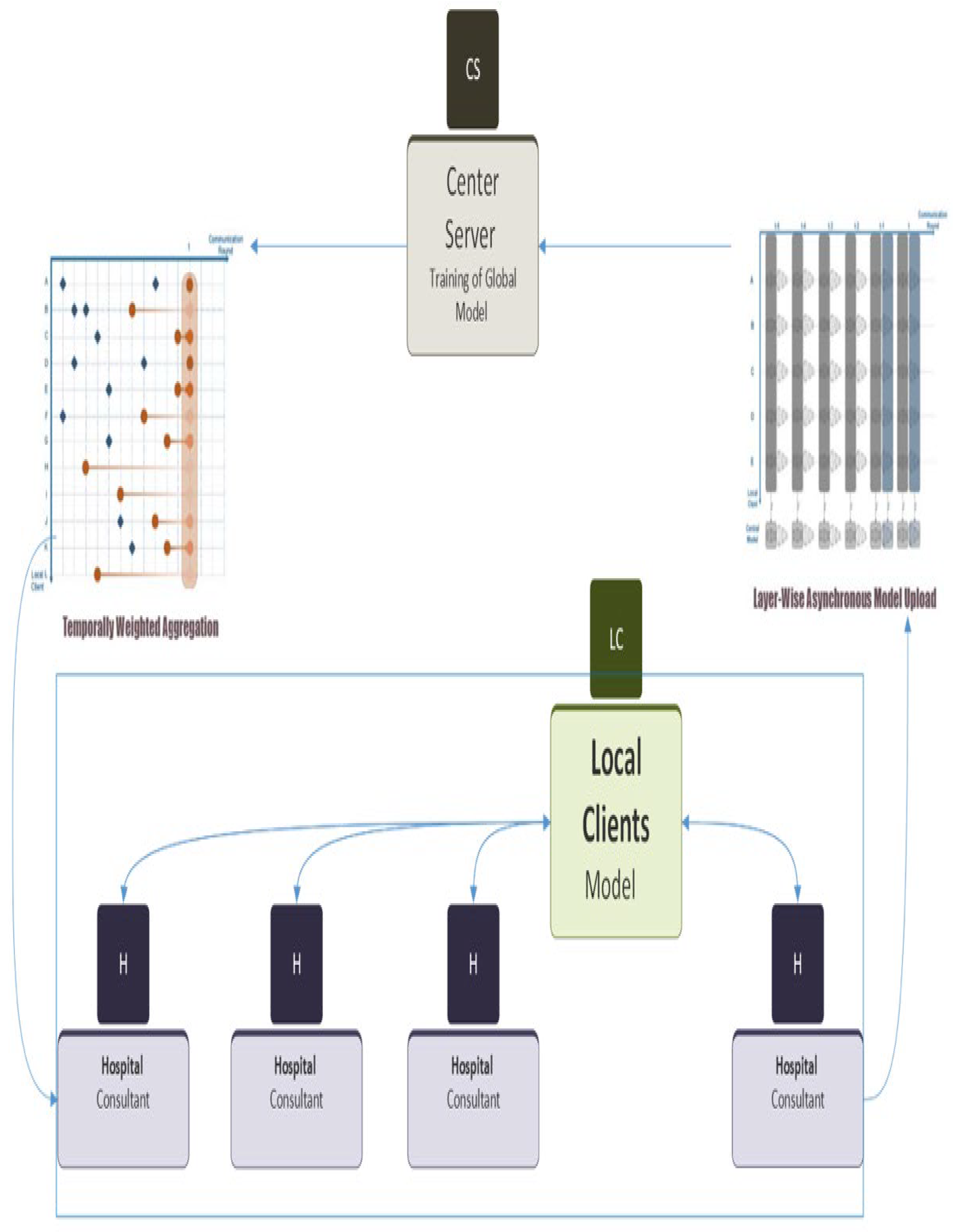
- Federated Learning using Asynchronous Method: The global model was then used to perform federated learning using an asynchronous method. This approach enabled the central server to train the model using the updated weights from each hospital, without the need for synchronous communication, which reduced the communication overhead and latency between clients and server.
- Distribution of Updated Weights to Clients: The updated weight values were sent back to each hospital according to their request. This step allowed each hospital to incorporate the latest learnings from the global model into their locally trained models, thereby improving model accuracy and generalization performance. The distribution of these weights is done asynchronously as depicted in Figure 3. The different colored weights shows weights at different time.
| Algorithm 1. Proposed Async-FL model for skin cancer prediction |
| Input: Skin Lesion data from various client users Output: Personalized models for skin cancer prediction //Working at the FL server end for round r = 1, 2, … do if r in every round_i n_loop ∈ Set ES then§ tag ← 1 else tag ← 0 max← maximum(A∗B.1) ts ← (random set of maxclients) every client j є ts in parallel do if tag then wj ← ClientUpdate(j, ws,tag) timeframekg ← s timeframeks ← s else wkg ← ClientUpdate(j, wk,s, tag) timeframekg ← s *fk(s,j)*wkg if tag then *ft (s,j)*wkg end function //Working at FL client client update (j, w, tag)//client j α ← (fragment bj into batches of size A) if tag then w ← w else wt ← w for local epoch j from 1 to f do for batch a є α do perform classification using radial basis SVM if tag then return w to server else return wt to server end function |
3.1. Rules for Skin Lesion Assessment
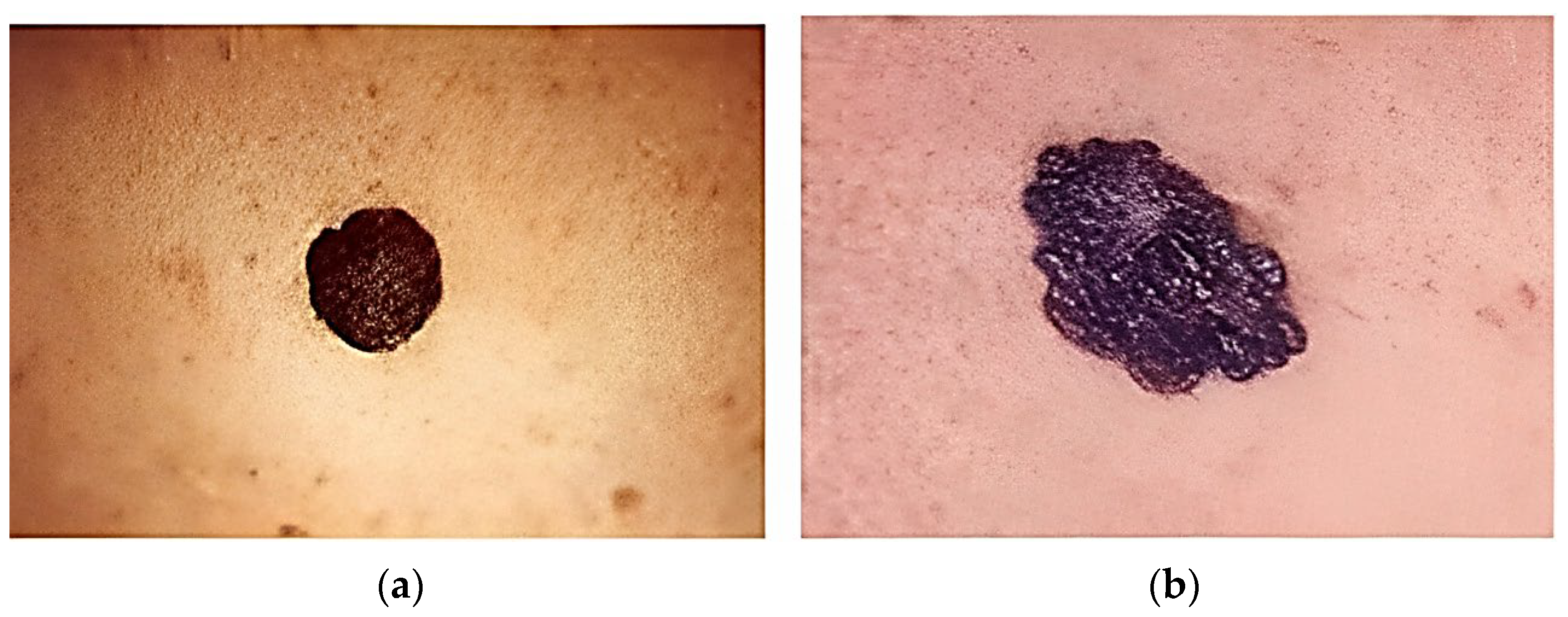
3.2. Experimentation Details and Dataset Utilized
4. Results and Discussion
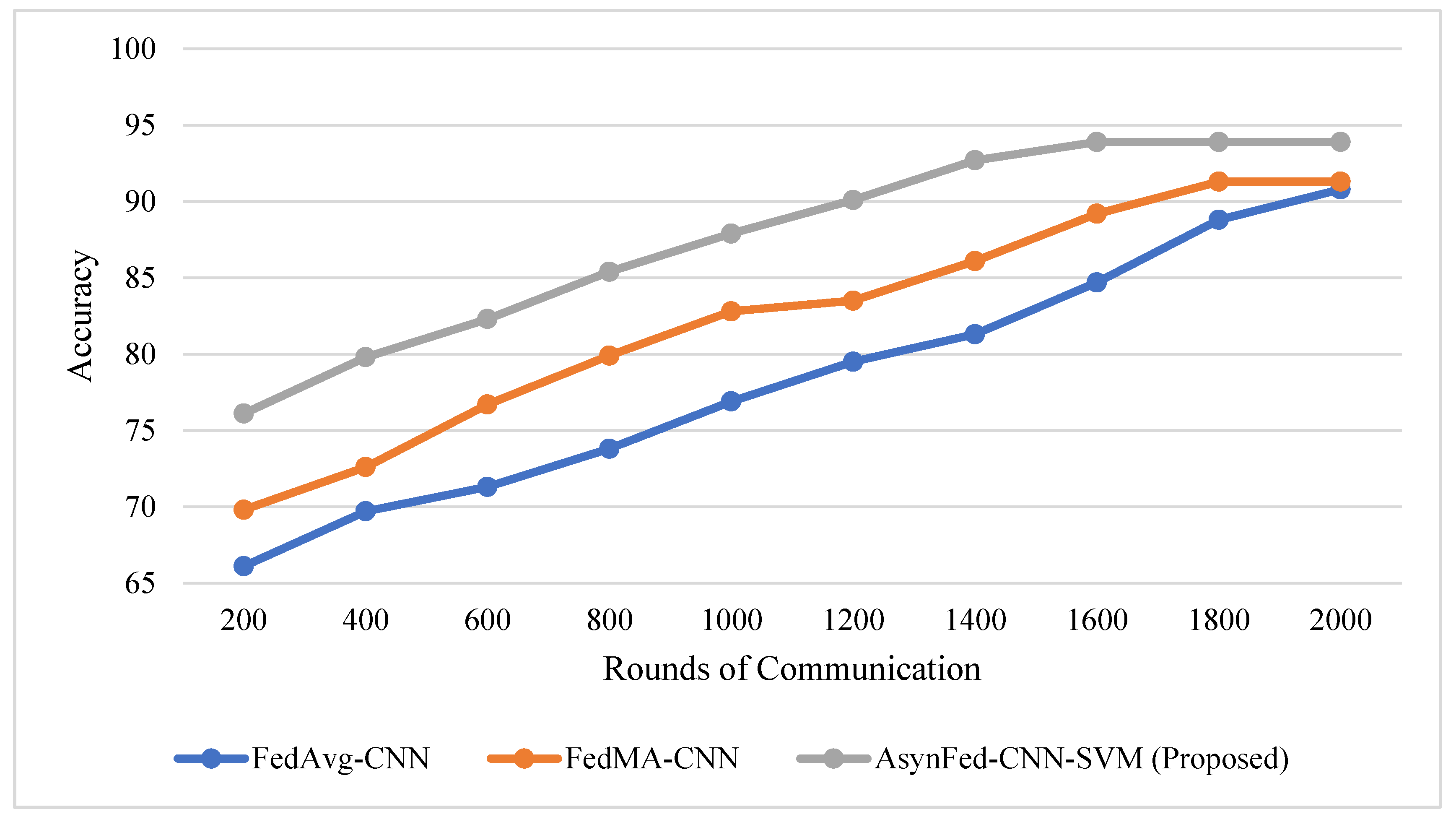
4.1. Experimental Results of the Proposed Model
4.2. Effect of Learning Rate in Training
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andre, G.P.; Renato, A.K. The impact of patient clinical information on automated skin cancer detection. Comput. Biol. Med. 2020, 116, 103545. [Google Scholar]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Voigt, P.; Von dem Bussche, A. Scope of Application of the GDPR. In The EU General Data Protection Regulation; Springer: Cham, Switzerland, 2017; pp. 9–30. [Google Scholar]
- Wagner, J. China’s Cybersecurity Law: What You Need to Know. The Diplomat, June 2017. Available online: https://thediplomat.com/2017/06/chinas-cybersecurity-law-what-you-need-to-know/ (accessed on 10 February 2023).
- De la Torre, L. A guide to the California Consumer Privacy Act of 2018. 2018. Available online: https://ssrn.com/abstract=3275571 (accessed on 12 February 2023).
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; y Areas, B.A. Communication-Efficient Learning of Deep Networks from Decentralized Data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Ft. Lauderdale, FL, USA, 20–22 April 2017; pp. 1273–1282. [Google Scholar]
- Bdair, T.M.; Navab, N.; Albarqouni, S. FedPerl: Semi-Supervised Federated Peer Learning for Skin Lesion Classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2021; Volume 12903. [Google Scholar]
- Murugan, A.; Nair, S.A.H.; Preethi, A.A.P.; Kumar, K.P.S. Diagnosis of skin cancer using machine learning techniques. Microprocess. Microsyst. 2020, 81, 103727. [Google Scholar] [CrossRef]
- Monika, M.K.; Vignesh, N.A.; Kumari, C.U.; Kumar, M.; Lydia, E.L. Skin cancer detection and classification using machine learning. Mater. Today Proc. 2020, 33, 4266–4270. [Google Scholar] [CrossRef]
- Jones, O.T.; Matin, R.N.; van der Schaar, M.; Bhayankaram, K.P.; Ranmuthu, C.K.I.; Islam, M.S.; Behiyat, D.; Boscott, R.; Calanzani, N.; Emery, J.; et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: A systematic review. Lancet Digit. Health 2022, 4, e466–e476. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Spasić, I.; Meyer, B.; Chapman, S.; Andres, F. Machine Learning on Mobile: An On-Device Inference App for Skin Cancer Detection. In Proceedings of the 2019 Fourth International Conference on Fog and Mobile Edge Computing (FMEC), Rome, Italy, 10–13 June 2019; pp. 301–305. [Google Scholar]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Daghrir, J.; Tlig, L.; Bouchouicha, M.; Sayadi, M. Melanoma Skin Cancer Detection Using Deep Learning and Classical Machine Learning Techniques: A Hybrid Approach. In Proceedings of the 2020 5th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Sousse, Tunisia, 2–5 September 2020; pp. 1–5. [Google Scholar]
- Md, S.; Jahurul, H.; Md, M.; Md, K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar]
- Nauta, M.; Walsh, R.; Dubowski, A.; Seifert, C. Uncovering and Correcting Shortcut Learning in Machine Learning Models for Skin Cancer Diagnosis. Diagnostics 2021, 12, 40. [Google Scholar] [CrossRef]
- Magalhaes, C.; Tavares, J.M.R.; Mendes, J.; Vardasca, R. Comparison of machine learning strategies for infrared thermography of skin cancer. Biomed. Signal Process. Control. 2021, 69, 1–6. [Google Scholar] [CrossRef]
- Putra, T.A.; Rufaida, S.I.; Leu, J.-S. Enhanced Skin Condition Prediction Through Machine Learning Using Dynamic Training and Testing Augmentation. IEEE Access 2020, 8, 40536–40546. [Google Scholar] [CrossRef]
- Ashraf, R.; Afzal, S.; Rehman, A.U.; Gul, S.; Baber, J.; Bakhtyar, M.; Mehmood, I.; Song, O.-Y.; Maqsood, M. Region-of-Interest Based Transfer Learning Assisted Framework for Skin Cancer Detection. IEEE Access 2020, 8, 147858–147871. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef]
- Das, K.; Cockerell, C.J.; Patil, A.; Pietkiewicz, P.; Giulini, M.; Grabbe, S.; Goldust, M. Machine Learning and Its Application in Skin Cancer. Int. J. Environ. Res. Public Health 2021, 18, 13409. [Google Scholar] [CrossRef]
- Wang, G.G.; Deb, S.; Cui, Z. Monarch butterfly optimization. Neural Comput. Appl. 2019, 31, 1995–2014. [Google Scholar] [CrossRef]
- Li, S.; Chen, H.; Wang, M.; Heidari, A.A.; Mirjalili, S. Slime mould algorithm: A new method for stochastic optimization. Future Gener. Comput. Syst. 2020, 111, 300–323. [Google Scholar] [CrossRef]
- Elaziz, M.A.; Xiong, S.; Jayasena, K.; Li, L. Task scheduling in cloud computing based on hybrid moth search algorithm and differential evolution. Knowl.-Based Syst. 2019, 169, 39–52. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, H.; Heidari, A.A.; Gandomi, A.H. Hunger games search: Visions, conception, implementation, deep analysis, perspectives, and towards performance shifts. Expert Syst. Appl. 2021, 177, 114864. [Google Scholar] [CrossRef]
- Ahmadianfar, I.; Heidari, A.A.; Gandomi, A.H.; Chu, X.; Chen, H. RUN beyond the metaphor: An efficient optimization algorithm based on Runge Kutta method. Expert Syst. Appl. 2021, 181, 115079. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris hawks optimization: Algorithm and applications. Futur. Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Govindaswamy, A.; Montague, E.; Raicu, D.; Furst, J. Cnn as a Feature Extractor in Gaze Recognition. In Proceedings of the 2020 3rd Artificial Intelligence and Cloud Computing Conference, Kyoto, Japan, 18–20 December 2020; pp. 31–37. [Google Scholar]
- Basly, H.; Ouarda, W.; Sayadi, F.; Ouni, B.; Alimi, A. Cnn-svm learning approach based human activity recognition. Image Signal Process. 2020, 12119, 271–281. [Google Scholar]
- Alzubaidi, L.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J.; Duan, Y. Deep Learning Models for Classification of Red Blood Cells in Microscopy Images to Aid in Sickle Cell Anemia Diagnosis. Electronics 2020, 9, 427. [Google Scholar] [CrossRef]
- Karungaru, S.; Dongyang, L.; Terada, K. Vehicle Detection and Type Classification Based on CNN-SVM. Int. J. Mach. Learn. Comput. 2021, 11, 304–310. [Google Scholar] [CrossRef]
- Yaqoob, M.M.; Alsulami, M.; Khan, M.A.; Alsadie, D.; Saudagar, A.K.J.; AlKhathami, M. Federated Machine Learning for Skin Lesion Diagnosis: An Asynchronous and Weighted Approach. Diagnostics 2023, 13, 1964. [Google Scholar] [CrossRef] [PubMed]
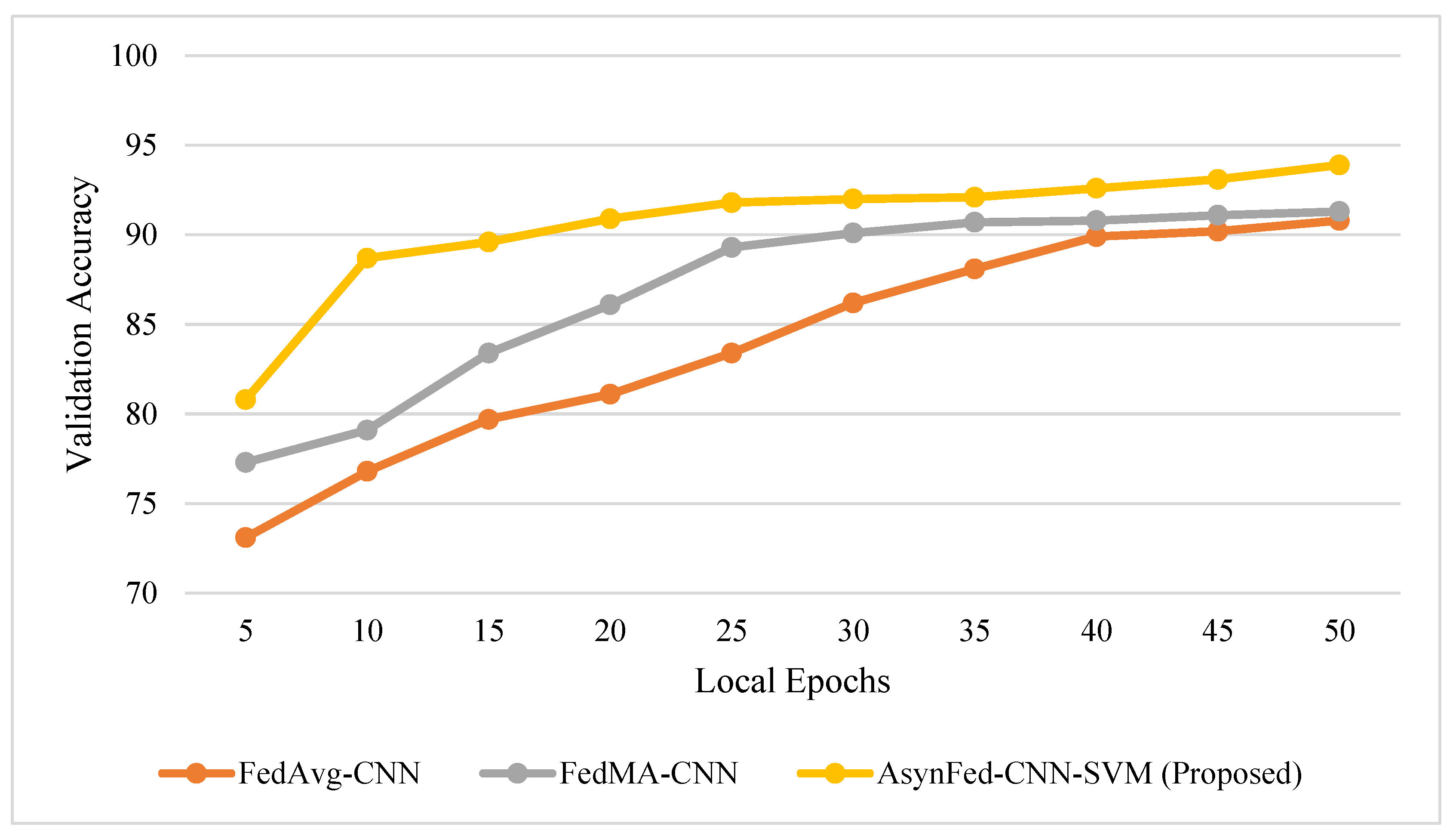
| No. of Epochs | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 |
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy training | 81.1 | 88.8 | 89.7 | 91.1 | 92.0 | 92.3 | 92.4 | 92.8 | 93.6 | 94.7 |
| Validation accuracy | 80.8 | 88.7 | 89.6 | 90.9 | 91.8 | 92.0 | 92.1 | 92.6 | 93.1 | 93.9 |
| Parameters | Name of the Classifier | ||
|---|---|---|---|
| SVM (Proposed) | KNN | Random Forest | |
| Sensitivity (%) | 90.1 | 76.9 | 77.1 |
| Specificity (%) | 89.1 | 72.7 | 79.9 |
| Accuracy (%) | 92.1 | 69.1 | 76.8 |
| Parameters | Name of Classifier | ||
|---|---|---|---|
| SVM (Proposed) | KNN | Random Forest | |
| Sensitivity | 86.1 | 65.8 | 74.9 |
| Specificity (%) | 87.1 | 68.7 | 77.2 |
| Accuracy (%) | 88.1 | 63.1 | 77.1 |
| Training | 94% |
| Validation | 94% |
| Testing | 93% |
| Training loss | 3% |
| Validation loss | 4% |
| Testing loss | 5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ain, Q.u.; Khan, M.A.; Yaqoob, M.M.; Khattak, U.F.; Sajid, Z.; Khan, M.I.; Al-Rasheed, A. Privacy-Aware Collaborative Learning for Skin Cancer Prediction. Diagnostics 2023, 13, 2264. https://doi.org/10.3390/diagnostics13132264
Ain Qu, Khan MA, Yaqoob MM, Khattak UF, Sajid Z, Khan MI, Al-Rasheed A. Privacy-Aware Collaborative Learning for Skin Cancer Prediction. Diagnostics. 2023; 13(13):2264. https://doi.org/10.3390/diagnostics13132264
Chicago/Turabian StyleAin, Qurat ul, Muhammad Amir Khan, Muhammad Mateen Yaqoob, Umar Farooq Khattak, Zohaib Sajid, Muhammad Ijaz Khan, and Amal Al-Rasheed. 2023. "Privacy-Aware Collaborative Learning for Skin Cancer Prediction" Diagnostics 13, no. 13: 2264. https://doi.org/10.3390/diagnostics13132264
APA StyleAin, Q. u., Khan, M. A., Yaqoob, M. M., Khattak, U. F., Sajid, Z., Khan, M. I., & Al-Rasheed, A. (2023). Privacy-Aware Collaborative Learning for Skin Cancer Prediction. Diagnostics, 13(13), 2264. https://doi.org/10.3390/diagnostics13132264






