Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning
Abstract
:1. Introduction
- -
- astrocytoma (isocitrate dehydrogenase (IDH)-mutant), which can be graded as 2, 3, or 4, depending on increasing mitotic activity and the presence of necrosis, microvascular proliferation, and/or homozygous deletion of CDKN2A/CDKN2B (in grade 4). Most of these tumors show a loss of alpha thalassemia retardation syndrome X-linked gene (ATRX) [6]
- -
- oligodendroglioma (IDH-mutant and 1p/19q-codeleted), which can be graded in 2 or 3, the latter showing high cellularity with atypia, microvascular proliferation, and increased mitotic activity. ATRX-staining is mostly retained [6]
- -
- and glioblastoma IDH-wildtype, historically defined as high grade, with hypercellularity, microvascular proliferation, and necrosis [6]
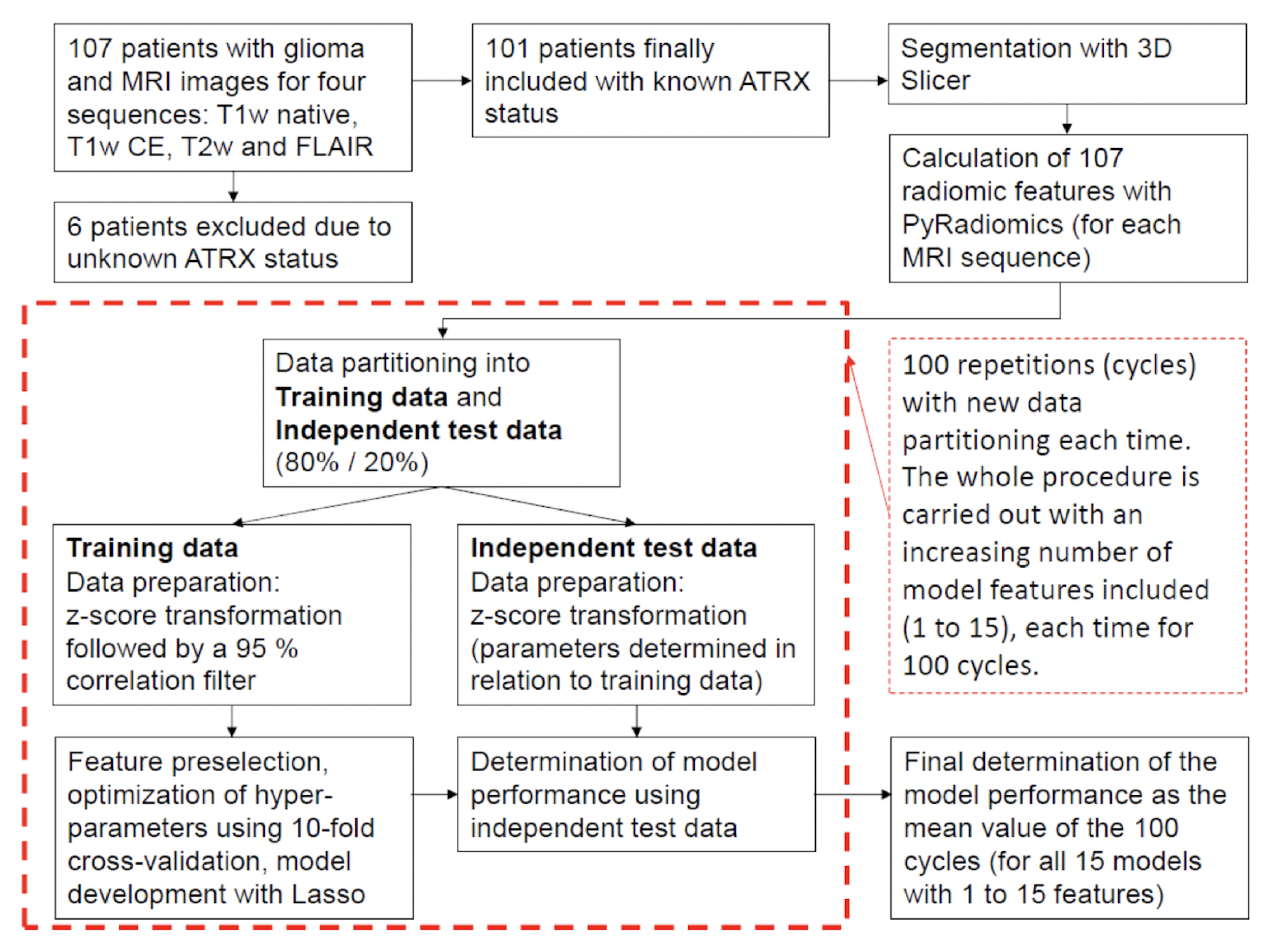
2. Materials and Methods
2.1. Radiomics
2.2. Statistical Analysis
3. Results
3.1. Results of Comparing MRI Sequences to Predict ATRX Status
3.2. Results for the Final Models: Fixed Feature Model Approach
4. Literature Review
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNS | Central Nervous System |
| WHO | World Health Organisation |
| MRI | Magnetic Resonance Imaging |
| FAIR | Fluid attenuated inversion recovery |
| ATRX | alpha thalassemia retardation syndrome X-linked |
| TMZ | Temozolomid |
References
- Aiman, W.; Gasalberti, D.P.; Rayi, A. Low-Grade Gliomas. 2023 May 6. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Nicholson, J.G.; Fine, H.A. Diffuse Glioma Heterogeneity and Its Therapeutic Implications. Cancer Discov. 2021, 11, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.; Hoggard, N.; McKevitt, F.M. Imaging in low-grade glioma: A guide for neurologists. Pract. Neurol. 2018, 18, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Mesfin, F.B.; Al-Dhahir, M.A. Gliomas. 2023 May 20. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System tumours: A practical update on what neurosurgeons need to know-a minireview. Acta Neurochir. (Wien) 2022, 164, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.L. 2021 updates to the World Health Organization classification of adult-type and pediatric-type diffuse gliomas: A clinical practice review. Chin. Clin. Oncol. 2023, 12, 7. [Google Scholar] [CrossRef]
- Cevik, L.; Landrove, M.V.; Aslan, M.T.; Khammad, V.; Garagorry Guerra, F.J.; Cabello-Izquierdo, Y.; Wang, W.; Zhao, J.; Becker, A.P.; Czeisler, C.; et al. Information theory approaches to improve glioma diagnostic workflows in surgical neuropathology. Brain Pathol. 2022, 32, e13050. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- An, N.; Husain, N.; Varshney, R.; Malhotra, K.P.; Kaif, M. Molecular classification and stratification of adult diffuse gliomas: A tertiary care center study. J. Carcinog. 2021, 20, 20. [Google Scholar]
- Bai, J.; Varghese, J.; Jain, R. Adult Glioma WHO Classification Update, Genomics, and Imaging: What the Radiologists Need to Know. Top. Magn. Reson. Imaging 2020, 29, 71–82. [Google Scholar] [CrossRef]
- Tsiouris, S.; Bougias, C.; Fotopoulos, A. Principles and current trends in the correlative evaluation of glioma with advanced MRI techniques and PET. Hell. J. Nucl. Med. 2019, 22, 206–219. [Google Scholar]
- Habib, A.; Jovanovich, N.; Hoppe, M.; Ak, M.; Mamindla, P.; RColen, R.; Zinn, P.O. MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. J. Clin. Med. 2021, 10, 1411. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Riche, M.; Amelot, A.; Peyre, M.; Capelle, L.; Carpentier, A.; Mathon, B. Complications after frame-based stereotactic brain biopsy: A systematic review. Neurosurg. Rev. 2021, 44, 301–307. [Google Scholar] [CrossRef]
- Abrigo, J.M.; Fountain, D.M.; Provenzale, J.M.; Law, E.K.; Kwong, J.S.; Hart, M.G.; Tam, W.W.S. Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation. Cochrane Database Syst. Rev. 2018, 1, CD011551. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Chintalapudi, N.; Angeloni, U.; Battineni, G.; di Canio, M.; Marotta, C.; Rezza, G.; Sagaro, G.G.; Silenzi, A.; Amenta, F. LASSO Regression Modeling on Prediction of Medical Terms among Seafarers’ Health Documents Using Tidy Text Mining. Bioengineering 2022, 9, 124. [Google Scholar] [CrossRef]
- Musigmann, M.; Akkurt, B.H.; Krähling, H.; Nacul, N.G.; Remonda, L.; Sartoretti, T.; Henssen, D.; Brokinkel, B.; Stummer, W.; Heindel, W.; et al. Testing the applicability and performance of Auto ML for potential applications in diagnostic neuroradiology. Sci. Rep. 2022, 12, 13648. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Chang, K.; Bai, H.X.; Xiao, B.; Su, C.; Bi, W.L.; Zhang, P.J.; Senders, J.T.; Vallières, M.; Kavouridis, V.K.; et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neurooncol. 2019, 142, 299–307. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, X.; Rui, W.; Pang, H.; Qiu, T.; Wang, J.; Xie, Q.; Jin, T.; Zhang, H.; Chen, H.; et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J. Magn. Reson. Imaging 2019, 49, 808–817. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, R.; Fa, L.; Zhang, L.; Wang, L.; Shao, G. ATRX status in patients with gliomas: Radiomics analysis. Medicine (Baltimore) 2022, 101, e30189. [Google Scholar] [CrossRef]
- Wick, W.; Bendszus, M.; Goldbrunner, R.; Grosu, A.; Hattingen, E.; Hau, P.; Herrlinger, U.; Kessler, T.; Platten, M.; Pukrop, T.; et al. Gliome, S2k-Leitlinie, 2021. In Deutsche Gesellschaft für Neurologie (Hrsg.), Leitlinien für Diagnostik und Therapie in der Neurologie. Available online: www.dgn.org/leitlinien (accessed on 25 January 2023).
- Han, B.; Meng, X.; Wu, P.; Li, Z.; Li, S.; Zhang, Y.; Zha, C.; Ye, Q.; Jiang, C.; Cai, J.; et al. ATRX/EZH2 complex epigenetically regulates FADD/PARP1 axis, contributing to TMZ resistance in glioma. Theranostics 2020, 10, 3351–3365. [Google Scholar] [CrossRef] [PubMed]
- Haase, S.; Garcia-Fabiani, M.B.; Carney, S.; Altshuler, D.; Núñez, F.J.; Méndez, F.M.; Núñez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Pladevall-Morera, D.; Castejón-Griñán, M.; Aguilera, P.; Gaardahl, K.; Ingham, A.; Brosnan-Cashman, J.A.; Meeker, A.K.; Lopez-Contreras, A.J. ATRX-Deficient High-Grade Glioma Cells Exhibit Increased Sensitivity to RTK and PDGFR Inhibitors. Cancers 2022, 14, 1790. [Google Scholar] [CrossRef] [PubMed]
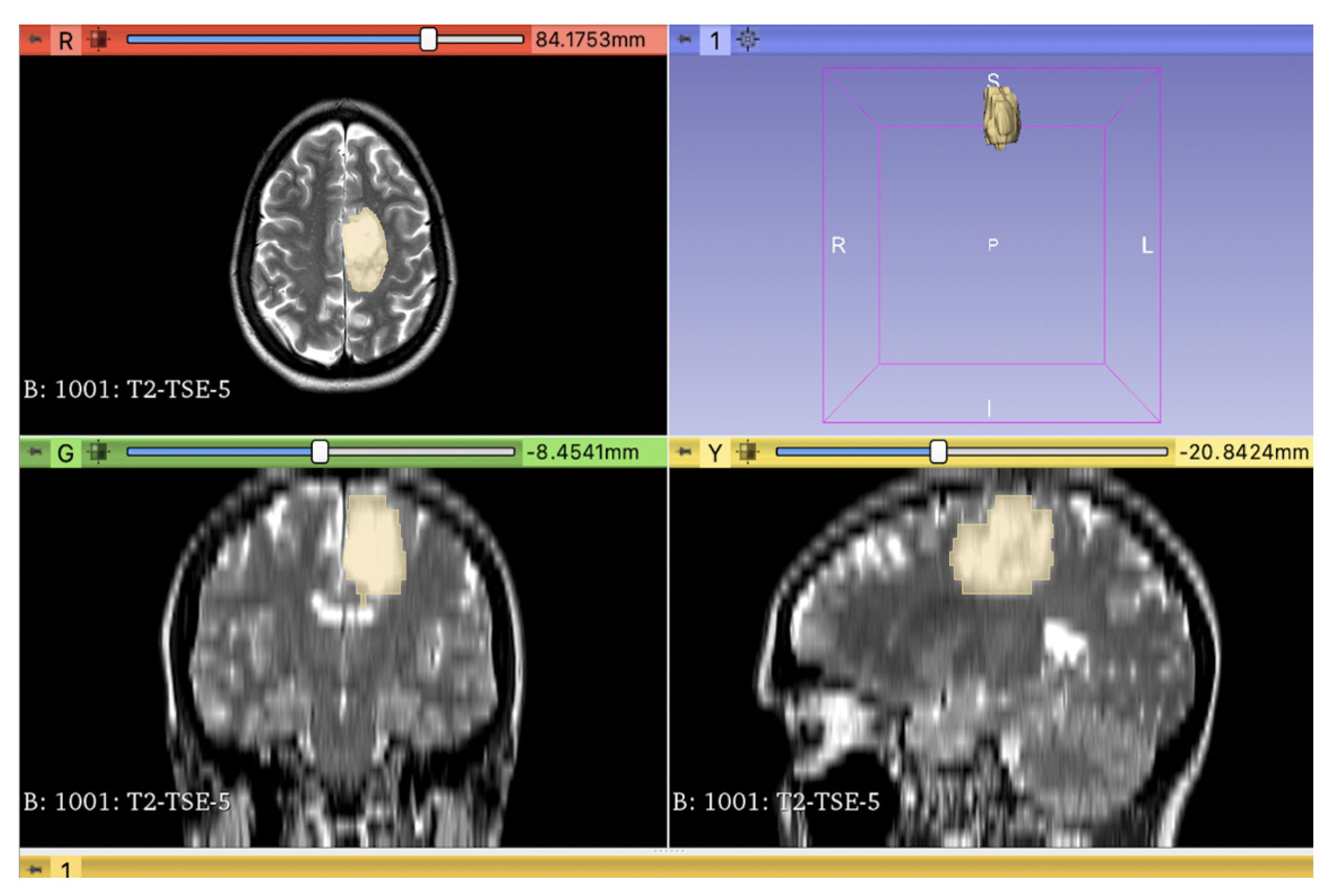

| Training Data | Independent Test Data | Total Data | |
|---|---|---|---|
| Number of Patients | 81 | 20 | 101 |
| Gender in percentage | |||
| Male | 53.44 | 53.55 | 53.47 |
| Female | 46.56 | 46.45 | 46.53 |
| Mean Age in years | 42.98 | 43.25 | 43.04 |
| ATRX Status in percentage | |||
| Retained, not mutated | 62.96 | 65.00 | 63.37 |
| Lost, mutated | 37.04 | 35.00 | 36.63 |
| Level of Importance | Feature Name | Number of Runs Included |
|---|---|---|
| 1 | original shape Flatness | 100 |
| 2 | Age at dignose | 99 |
| 3 | original firstorder Skewness | 98 |
| 4 | original gldm DependenceVariance | 97 |
| 5 | original shape LeastAxisLength | 92 |
| 6 | original glszm SmallAreaEmphasis | 91 |
| 7 | original glszm GrayLevelNonUniformity | 82 |
| 8 | original glszm SizeZoneNonUniformityNormalized | 79 |
| 9 | original gldm DependenceNonUniformityNormalized | 67 |
| 10 | original shape SurfaceVolumeRatio | 59 |
| 11 | original gldm LargeDependenceHighGrayLevelEmphasis | 47 |
| 12 | original firstorder Kurtosis | 36 |
| 13 | original glszm GrayLevelNonUniformityNormalized | 27 |
| 14 | original shape Elongation | 25 |
| 15 | original glrlm ShortRunLowGrayLevelEmphasis | 24 |
| Models with 7 Features | Models with 7 Features | Models with 12 Features | Models with 12 Features | |
|---|---|---|---|---|
| Different Features | Fixed Features | Different Features | Fixed Features | |
| AUC | 0.781 [0.546:0.949] | 0.831 [0.637:0.966] | 0.792 [0.556:0.945] | 0.829 [0.611:0.961] |
| Accuracy | 0.714 [0.453:0.874] | 0.746 [0.550:0.900] | 0.734 [0.503:0.874] | 0.752 [0.576:0.950] |
| Sensitivity | 0.756 [0.425:0.923] | 0.772 [0.502:0.923] | 0.768 [0.538:0.960] | 0.778 [0.538:1.000] |
| Specificity | 0.636 [0.286:0.925] | 0.697 [0.286:1.000] | 0.670 [0.361:1.000] | 0.703 [0.429:1.000] |
| PPV | 0.798 [0.582:0.960] | 0.832 [0.655:1.000] | 0.817 [0.641:1.000] | 0.835 [0.704:1.000] |
| NPV | 0.598 [0.256:0.857] | 0.636 [0.349:0.857] | 0.627 [0.329:0.925] | 0.652 [0.423:1.000] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nacul Mora, N.G.; Akkurt, B.H.; Kasap, D.; Blömer, D.; Heindel, W.; Mannil, M.; Musigmann, M. Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning. Diagnostics 2023, 13, 2216. https://doi.org/10.3390/diagnostics13132216
Nacul Mora NG, Akkurt BH, Kasap D, Blömer D, Heindel W, Mannil M, Musigmann M. Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning. Diagnostics. 2023; 13(13):2216. https://doi.org/10.3390/diagnostics13132216
Chicago/Turabian StyleNacul Mora, Nabila Gala, Burak Han Akkurt, Dilek Kasap, David Blömer, Walter Heindel, Manoj Mannil, and Manfred Musigmann. 2023. "Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning" Diagnostics 13, no. 13: 2216. https://doi.org/10.3390/diagnostics13132216
APA StyleNacul Mora, N. G., Akkurt, B. H., Kasap, D., Blömer, D., Heindel, W., Mannil, M., & Musigmann, M. (2023). Comparison of MRI Sequences to Predict ATRX Status Using Radiomics-Based Machine Learning. Diagnostics, 13(13), 2216. https://doi.org/10.3390/diagnostics13132216






