Development and Implementation of an Internal Quality Control Sample to Standardize Oligomer-Based Diagnostics of Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Monomerization Aβ1–42 Peptide Stock and Aggregation Protocol
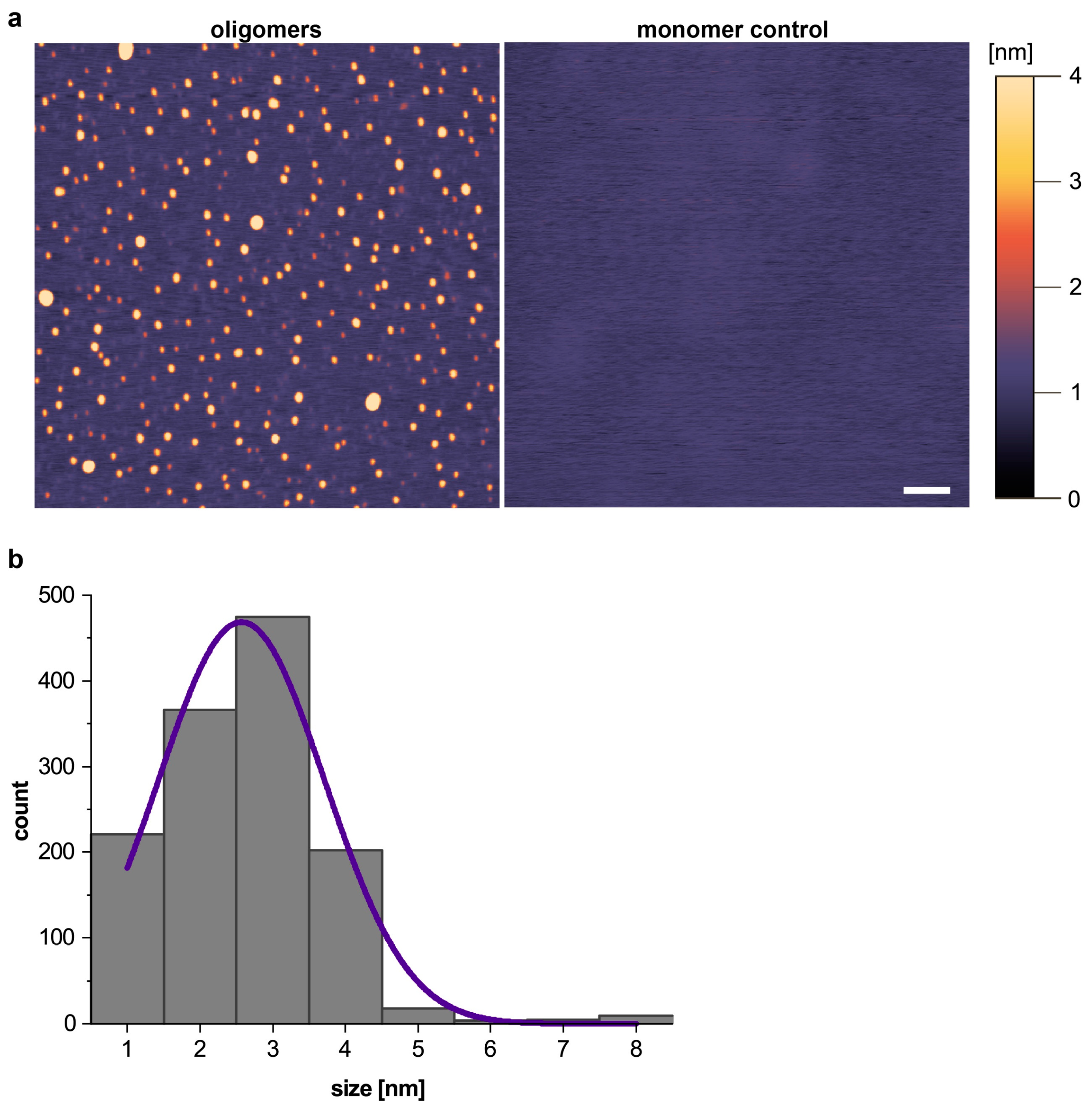
2.2. Atomic Force Microscopy
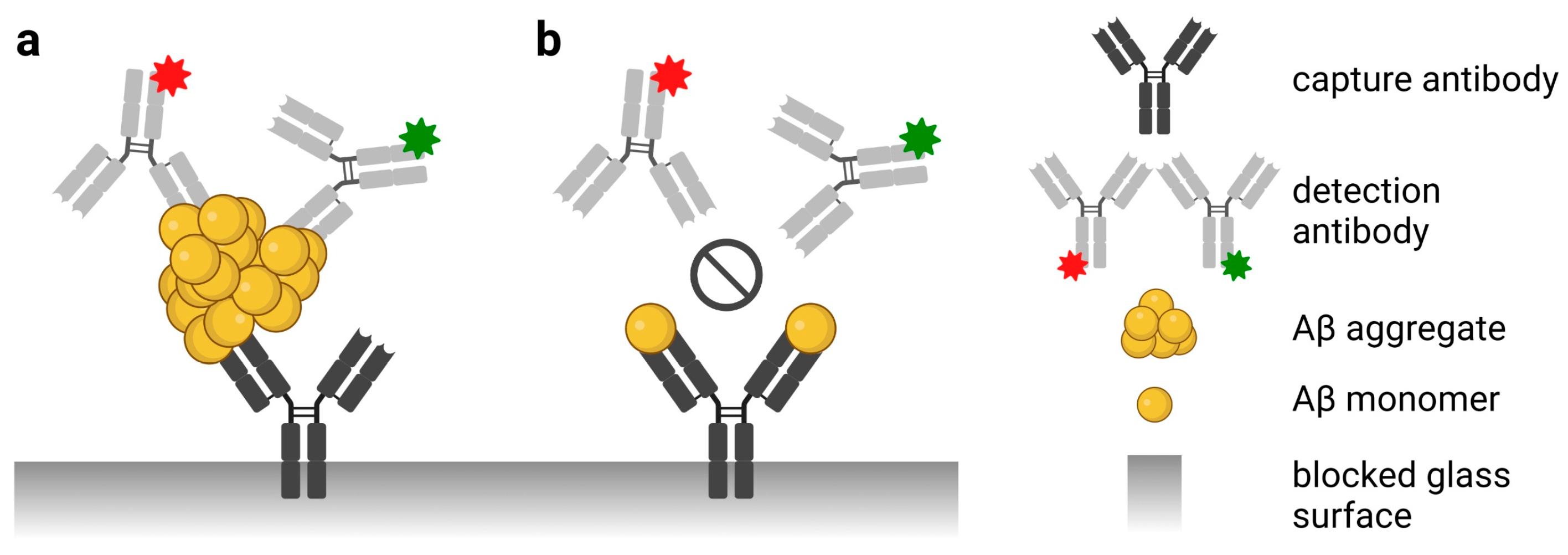
2.3. sFIDA
2.3.1. Synthesis of SiNaPs Coated with Aβ1–15
2.3.2. Labeling of Antibodies
2.3.3. Assay Protocol
2.3.4. Image Data Acquisition
2.4. Statistics
2.4.1. Analysis of Image Data
2.4.2. Calibration
2.4.3. Analytical Validation: Detection and Quantification Limits
2.4.4. Analytical Validation: Analytical Selectivity
2.4.5. Establishment of QC-Tool
3. Results
3.1. Aβ Oligomer-Based IQC Sample Displays High Homogeneity
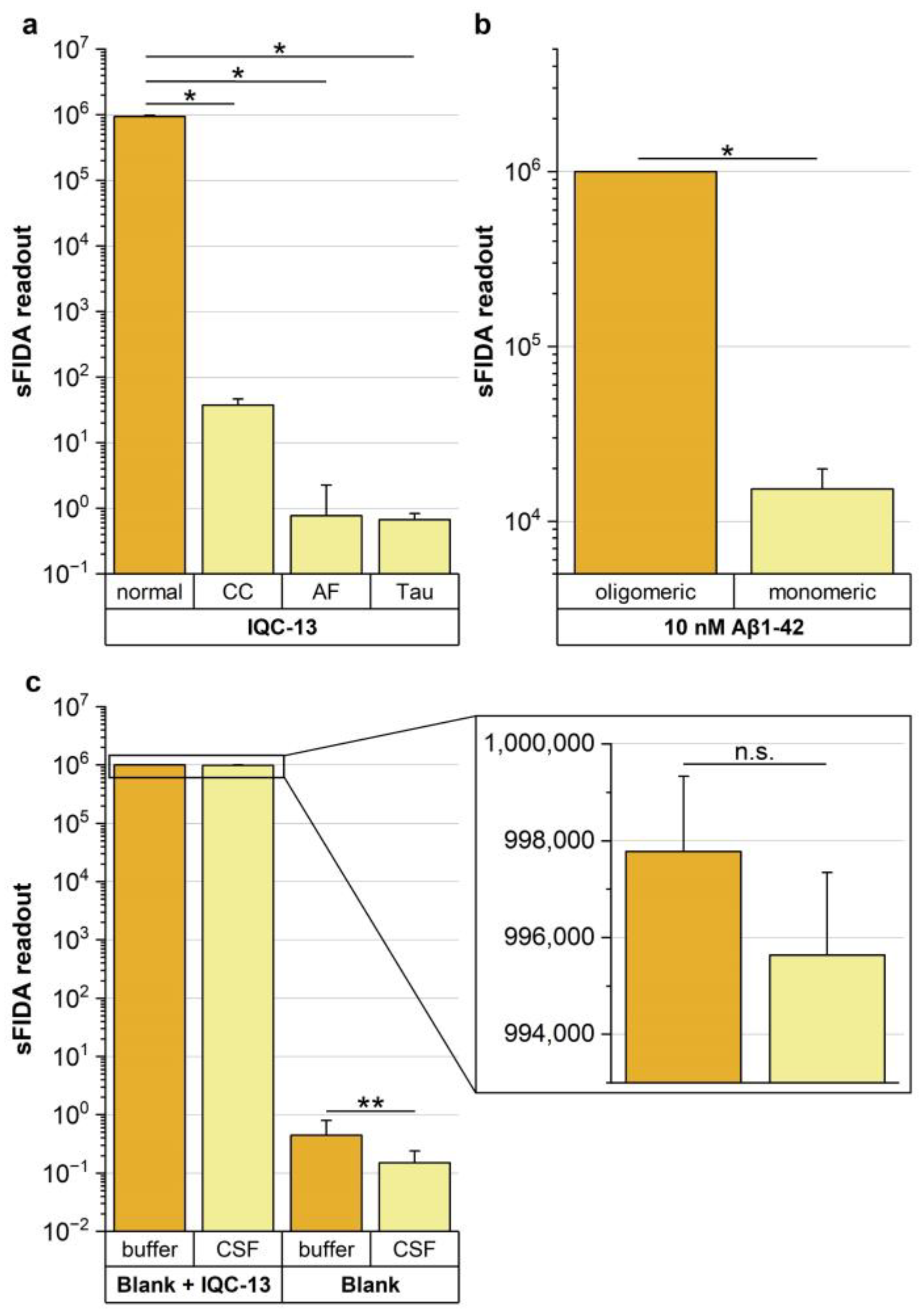
3.2. Successful Application of the Aβ Oligomer-Based IQC Sample in the sFIDA Assay
3.3. sFIDA Features High Selectivity for the Aβ Oligomer-Based IQC Sample
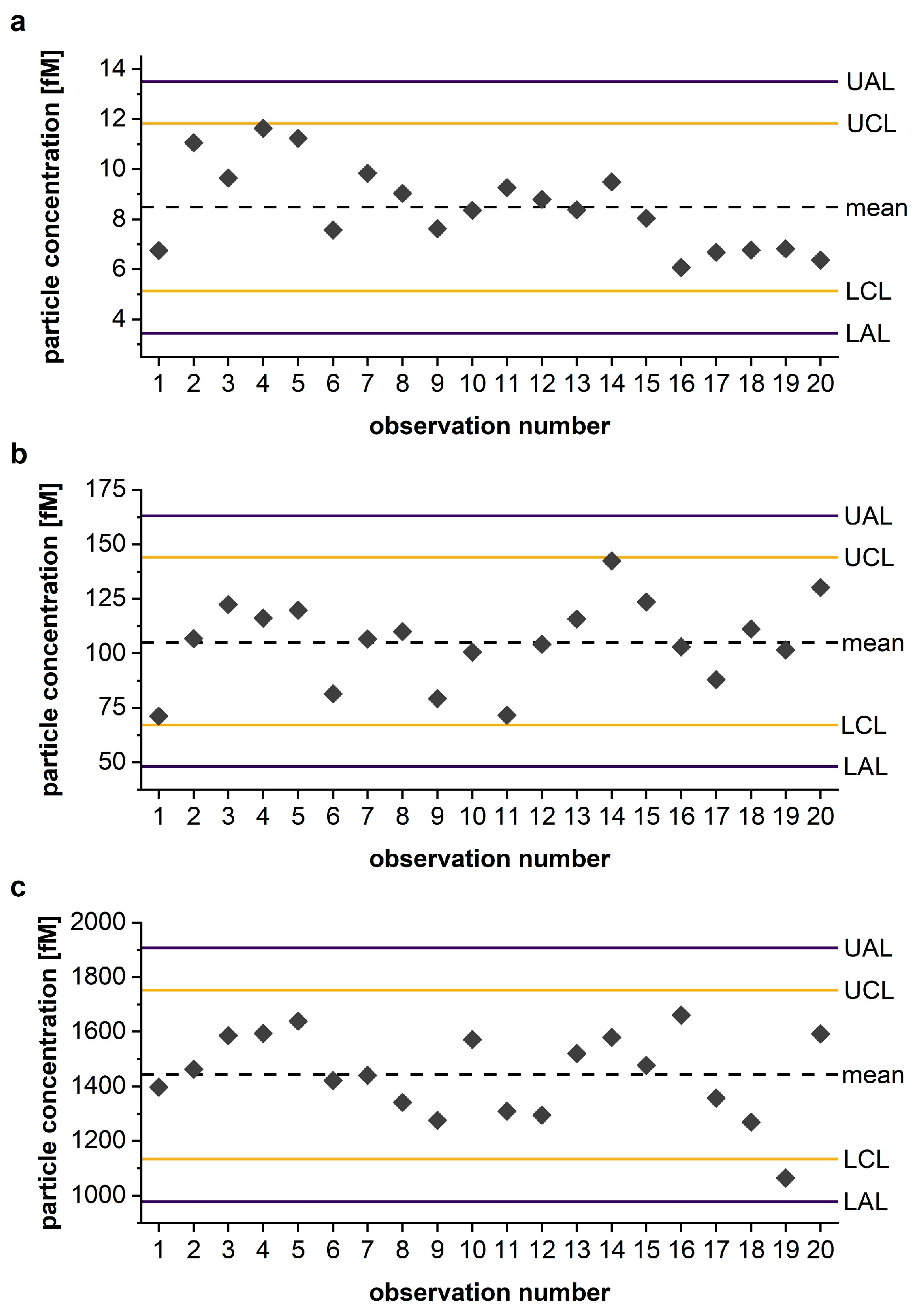
3.4. Shewhart Chart as a Reliable QC-Tool for Monitoring IQC Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brettschneider, J.; Del Tredici, K.; Lee, V.M.; Trojanowski, J.Q. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 2015, 16, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Dulewicz, M.; Kulczyńska-Przybik, A.; Mroczko, P.; Kornhuber, J.; Lewczuk, P.; Mroczko, B. Biomarkers for the Diagnosis of Alzheimer’s Disease in Clinical Practice: The Role of CSF Biomarkers during the Evolution of Diagnostic Criteria. Int. J. Mol. Sci. 2022, 23, 8598. [Google Scholar] [CrossRef] [PubMed]
- Tolar, M.; Abushakra, S.; Sabbagh, M. The path forward in Alzheimer’s disease therapeutics: Reevaluating the amyloid cascade hypothesis. Alzheimers Dement. 2020, 16, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Lotz, G.P.; Legleiter, J. The role of amyloidogenic protein oligomerization in neurodegenerative disease. J. Mol. Med. 2013, 91, 653–664. [Google Scholar] [CrossRef]
- Kulenkampff, K.; Wolf Perez, A.-M.; Sormanni, P.; Habchi, J.; Vendruscolo, M. Quantifying misfolded protein oligomers as drug targets and biomarkers in Alzheimer and Parkinson diseases. Nat. Rev. Chem. 2021, 5, 277–294. [Google Scholar] [CrossRef]
- Mroczko, B.; Groblewska, M.; Litman-Zawadzka, A.; Kornhuber, J.; Lewczuk, P. Amyloid β oligomers (AβOs) in Alzheimer’s disease. J. Neural. Transm. 2018, 125, 177–191. [Google Scholar] [CrossRef]
- Herrmann, Y.; Kulawik, A.; Kühbach, K.; Hülsemann, M.; Peters, L.; Bujnicki, T.; Kravchenko, K.; Linnartz, C.; Willbold, J.; Zafiu, C.; et al. sFIDA automation yields sub-femtomolar limit of detection for Aβ aggregates in body fluids. Clin. Biochem. 2017, 50, 244–247. [Google Scholar] [CrossRef]
- Blömeke, L.; Pils, M.; Kraemer-Schulien, V.; Dybala, A.; Schaffrath, A.; Kulawik, A.; Rehn, F.; Cousin, A.; Nischwitz, V.; Willbold, J.; et al. Quantitative detection of α-Synuclein and Tau oligomers and other aggregates by digital single particle counting. NPJ Parkinsons Dis. 2022, 8, 68. [Google Scholar] [CrossRef]
- Kass, B.; Schemmert, S.; Zafiu, C.; Pils, M.; Bannach, O.; Kutzsche, J.; Bujnicki, T.; Willbold, D. Aβ oligomer concentration in mouse and human brain and its drug-induced reduction ex vivo. Cell. Rep. Med. 2022, 3, 100630. [Google Scholar] [CrossRef]
- Wang-Dietrich, L.; Funke, S.A.; Kühbach, K.; Wang, K.; Besmehn, A.; Willbold, S.; Cinar, Y.; Bannach, O.; Birkmann, E.; Willbold, D. The amyloid-β oligomer count in cerebrospinal fluid is a biomarker for Alzheimer’s disease. J. Alzheimers Dis. 2013, 34, 985–994. [Google Scholar] [CrossRef]
- Kulawik, A.; Heise, H.; Zafiu, C.; Willbold, D.; Bannach, O. Advancements of the sFIDA method for oligomer-based diagnostics of neurodegenerative diseases. FEBS Lett. 2018, 592, 516–534. [Google Scholar] [CrossRef] [PubMed]
- Schaffrath, A.; Schleyken, S.; Seger, A.; Jergas, H.; Özdüzenciler, P.; Pils, M.; Blömeke, L.; Cousin, A.; Willbold, J.; Bujnicki, T.; et al. Patients with isolated REM-sleep behavior disorder have elevated levels of alpha-synuclein aggregates in stool. NPJ Parkinsons Dis. 2023, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Hülsemann, M.; Zafiu, C.; Kühbach, K.; Lühmann, N.; Herrmann, Y.; Peters, L.; Linnartz, C.; Willbold, J.; Kravchenko, K.; Kulawik, A.; et al. Biofunctionalized Silica Nanoparticles: Standards in Amyloid-β Oligomer-Based Diagnosis of Alzheimer’s Disease. J. Alzheimers Dis. 2016, 54, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, M.; Röder, C.; Schröder, G.F. Challenges in sample preparation and structure determination of amyloids by cryo-EM. J. Biol. Chem. 2021, 297, 100938. [Google Scholar] [CrossRef] [PubMed]
- Vanderstichele, H.; Stoops, E.; Vanmechelen, E.; Jeromin, A. Potential sources of interference on Abeta immunoassays in biological samples. Alzheimers Res. Ther. 2012, 4, 39. [Google Scholar] [CrossRef]
- Blacksell, S.D.; Cameron, A.R.; Chamnanpood, C.; Chamnanpood, P.; Tatong, D.; Monpolsiri, M.; Westbury, H.A. Implementation of internal laboratory quality control procedures for the monitoring of ELISA performance at a regional veterinary laboratory. Vet. Microbiol. 1996, 51, 1–9. [Google Scholar] [CrossRef]
- Sturgeon, C.M. Chapter 6.2—Quality Assurance. In The Immunoassay Handbook, 4th ed.; Wild, D., Ed.; Elsevier: Oxford, UK, 2013; pp. 441–454. [Google Scholar]
- Lock, R.J. My approach to internal quality control in a clinical immunology laboratory. J. Clin. Pathol. 2006, 59, 681–684. [Google Scholar] [CrossRef]
- Ryan, D.A.; Narrow, W.C.; Federoff, H.J.; Bowers, W.J. An improved method for generating consistent soluble amyloid-beta oligomer preparations for in vitro neurotoxicity studies. J. Neurosci. Methods 2010, 190, 171–179. [Google Scholar] [CrossRef]
- Kotler, S.A.; Ramamoorthy, A. Preparation of Stable Amyloid-β Oligomers Without Perturbative Methods. Methods Mol. Biol. 2018, 1777, 331–338. [Google Scholar] [CrossRef]
- Stine, W.B.; Jungbauer, L.; Yu, C.; LaDu, M.J. Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 2011, 670, 13–32. [Google Scholar] [CrossRef]
- Brener, O.; Dunkelmann, T.; Gremer, L.; van Groen, T.; Mirecka, E.A.; Kadish, I.; Willuweit, A.; Kutzsche, J.; Jürgens, D.; Rudolph, S.; et al. QIAD assay for quantitating a compound’s efficacy in elimination of toxic Aβ oligomers. Sci. Rep. 2015, 5, 13222. [Google Scholar] [CrossRef]
- Novo, M.; Freire, S.; Al-Soufi, W. Critical aggregation concentration for the formation of early Amyloid-β(1–42) oligomers. Sci. Rep. 2018, 8, 1783. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S49–S52. [Google Scholar] [PubMed]
- Reichenbächer, M.; Einax, J.W. Challenges in Analytical Quality Assurance; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Michaels, T.C.T.; Šarić, A.; Curk, S.; Bernfur, K.; Arosio, P.; Meisl, G.; Dear, A.J.; Cohen, S.I.A.; Dobson, C.M.; Vendruscolo, M.; et al. Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nat. Chem. 2020, 12, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Lewczuk, P.; Łukaszewicz-Zając, M.; Mroczko, P.; Kornhuber, J. Clinical significance of fluid biomarkers in Alzheimer’s Disease. Pharmacol. Rep. 2020, 72, 528–542. [Google Scholar] [CrossRef]
- Shea, D.; Colasurdo, E.; Smith, A.; Paschall, C.; Jayadev, S.; Keene, C.D.; Galasko, D.; Ko, A.; Li, G.; Peskind, E.; et al. SOBA: Development and testing of a soluble oligomer binding assay for detection of amyloidogenic toxic oligomers. Proc. Natl. Acad. Sci. USA 2022, 119, e2213157119. [Google Scholar] [CrossRef]
- Savage, M.J.; Kalinina, J.; Wolfe, A.; Tugusheva, K.; Korn, R.; Cash-Mason, T.; Maxwell, J.W.; Hatcher, N.G.; Haugabook, S.J.; Wu, G.; et al. A sensitive aβ oligomer assay discriminates Alzheimer’s and aged control cerebrospinal fluid. J. Neurosci. 2014, 34, 2884–2897. [Google Scholar] [CrossRef]
- IBL-International. Human Amyloid β Toxic Oligomer Assay Kit—IBL. Instruction for Use Code No. 27709. Available online: https://www.ibl-japan.co.jp/files/topics/4751_ext_02_en_0.pdf (accessed on 29 March 2023).
- Hölttä, M.; Hansson, O.; Andreasson, U.; Hertze, J.; Minthon, L.; Nägga, K.; Andreasen, N.; Zetterberg, H.; Blennow, K. Evaluating amyloid-β oligomers in cerebrospinal fluid as a biomarker for Alzheimer’s disease. PLoS ONE 2013, 8, e66381. [Google Scholar] [CrossRef]
- Kasai, T.; Tokuda, T.; Taylor, M.; Nakagawa, M.; Allsop, D. Utilization of a multiple antigenic peptide as a calibration standard in the BAN50 single antibody sandwich ELISA for Aβ oligomers. Biochem. Biophys Res. Commun. 2012, 422, 375–380. [Google Scholar] [CrossRef]
- Esparza, T.J.; Zhao, H.; Cirrito, J.R.; Cairns, N.J.; Bateman, R.J.; Holtzman, D.M.; Brody, D.L. Amyloid-β oligomerization in Alzheimer dementia versus high-pathology controls. Ann. Neurol. 2013, 73, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, U.; Perret-Liaudet, A.; van Waalwijk van Doorn, L.J.; Blennow, K.; Chiasserini, D.; Engelborghs, S.; Fladby, T.; Genc, S.; Kruse, N.; Kuiperij, H.B.; et al. A Practical Guide to Immunoassay Method Validation. Front. Neurol. 2015, 6, 179. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.M.; Kokjohn, T.A.; Kalback, W.; Luehrs, D.; Galasko, D.R.; Chevallier, N.; Koo, E.H.; Emmerling, M.R.; Roher, A.E. Amyloid-beta peptides interact with plasma proteins and erythrocytes: Implications for their quantitation in plasma. Biochem. Biophys Res. Commun. 2000, 268, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Sehlin, D.; Söllvander, S.; Paulie, S.; Brundin, R.; Ingelsson, M.; Lannfelt, L.; Pettersson, F.E.; Englund, H. Interference from heterophilic antibodies in amyloid-β oligomer ELISAs. J. Alzheimers Dis. 2010, 21, 1295–1301. [Google Scholar] [CrossRef] [PubMed]
- Vanderstichele, H.M.; Janelidze, S.; Demeyer, L.; Coart, E.; Stoops, E.; Herbst, V.; Mauroo, K.; Brix, B.; Hansson, O. Optimized Standard Operating Procedures for the Analysis of Cerebrospinal Fluid Aβ42 and the Ratios of Aβ Isoforms Using Low Protein Binding Tubes. J. Alzheimers Dis. 2016, 53, 1121–1132. [Google Scholar] [CrossRef]
- Schoonenboom, N.S.; Mulder, C.; Vanderstichele, H.; Van Elk, E.J.; Kok, A.; Van Kamp, G.J.; Scheltens, P.; Blankenstein, M.A. Effects of processing and storage conditions on amyloid β(1–42) and tau concentrations in cerebrospinal fluid: Implications for use in clinical practice. Clin. Chem. 2005, 51, 189–195. [Google Scholar] [CrossRef]
- van de Merbel, N.; Savoie, N.; Yadav, M.; Ohtsu, Y.; White, J.; Riccio, M.F.; Dong, K.; de Vries, R.; Diancin, J. Stability: Recommendation for best practices and harmonization from the Global Bioanalysis Consortium Harmonization Team. Aaps J. 2014, 16, 392–399. [Google Scholar] [CrossRef]

| Reference | Assay Setup | Calibration Standard | Sensitivity [fM] |
|---|---|---|---|
| sFIDA | Single-particle analysis with fluorescence microscopy, overlapping epitopes capture: Nab228 aa1–11 detection: IC16 aa2–8 + Nab228 aa1–11 | Aβ1–15 SiNaPs | LoD = 0.28 |
| Savage et al. [30] | Single particle analysis with bead-based assay capture: 19.3 oligomer-specific detection: 82E1 aa1–16 | Aβ1–42 oligomers (MW 1117 kDa) | LoD = 0.08 |
| IBL [31] | ELISA capture: 82E1 aa1–16 detection: 24B3 oligomer-specific | E22P–Aβ40 Dimer | 31.4 (N/A) |
| Hölttä et al. [32] | ELISA, overlapping epitopes capture and detection: 82E1 aa1–16 | Dimer Aβ1–11 | LLoQ = 90.9 |
| Kasai et al. [33] | ELISA, overlapping epitopes capture and detection: Ban50 aa1–10 | MAP 16-mer (lysine core) | LoD = 190 |
| Esparza et al. [34] | Single particle analysis with bead-based assay overlapping epitopes capture and detection: HJ3.4 aa1–13 | Aβ1–40Ser26Cys dimer | LLoQ = 720 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pils, M.; Dybala, A.; Rehn, F.; Blömeke, L.; Bujnicki, T.; Kraemer-Schulien, V.; Hoyer, W.; Riesner, D.; Willbold, D.; Bannach, O. Development and Implementation of an Internal Quality Control Sample to Standardize Oligomer-Based Diagnostics of Alzheimer’s Disease. Diagnostics 2023, 13, 1702. https://doi.org/10.3390/diagnostics13101702
Pils M, Dybala A, Rehn F, Blömeke L, Bujnicki T, Kraemer-Schulien V, Hoyer W, Riesner D, Willbold D, Bannach O. Development and Implementation of an Internal Quality Control Sample to Standardize Oligomer-Based Diagnostics of Alzheimer’s Disease. Diagnostics. 2023; 13(10):1702. https://doi.org/10.3390/diagnostics13101702
Chicago/Turabian StylePils, Marlene, Alexandra Dybala, Fabian Rehn, Lara Blömeke, Tuyen Bujnicki, Victoria Kraemer-Schulien, Wolfgang Hoyer, Detlev Riesner, Dieter Willbold, and Oliver Bannach. 2023. "Development and Implementation of an Internal Quality Control Sample to Standardize Oligomer-Based Diagnostics of Alzheimer’s Disease" Diagnostics 13, no. 10: 1702. https://doi.org/10.3390/diagnostics13101702
APA StylePils, M., Dybala, A., Rehn, F., Blömeke, L., Bujnicki, T., Kraemer-Schulien, V., Hoyer, W., Riesner, D., Willbold, D., & Bannach, O. (2023). Development and Implementation of an Internal Quality Control Sample to Standardize Oligomer-Based Diagnostics of Alzheimer’s Disease. Diagnostics, 13(10), 1702. https://doi.org/10.3390/diagnostics13101702






