Multi-Classification of Breast Cancer Lesions in Histopathological Images Using DEEP_Pachi: Multiple Self-Attention Head
Abstract
:1. Introduction
1.1. Diagnostic Medical Methods Used in the Investigation of BC
1.2. Related Studies
- ❖
- This research reviews several Medical BC imaging techniques, their robustness and limitation, and associated public dataset.
- ❖
- This paper proposed a fine-tuned approach termed “DEEP_Pachi,” an end-to-end deep learning model incorporating multiple self-attention network heads and Multilayer Perceptron for the multiclassification of Breast cancer diseases using histopathological images.
- ❖
- According to the comprehensive study via transfer learning experiment, the suggested feature extractor discriminates remarkably between benign breast tumors such as Adenosis, Fibroadenoma, Phyllodes_tumor and Tubular_adenoma malignant breast tumors Ductal_carcinoma, Lobular_Carcinoma, Mucinous_Cancinoma, and Papillary_carcinoma to help medical diagnosis even when professional radiologists are not accessible.
- ❖
- We reported a well robust deep learning method in Accuracy, Specificity, Sensitivity, Precision, F1 Score, Confusion matrix, and AUC using receiver operating characteristics (ROC) for the multiclassification of Breast cancer diseases using histopathological images based on the detailed experimental evaluation of the proposed model and comparison with state-of-the-art results.
- ❖
- Finally, this research suggests that the proposed model “DEEP_Pachi” can also be used to increase ensemble deep learning models’ detection and classification accuracies.
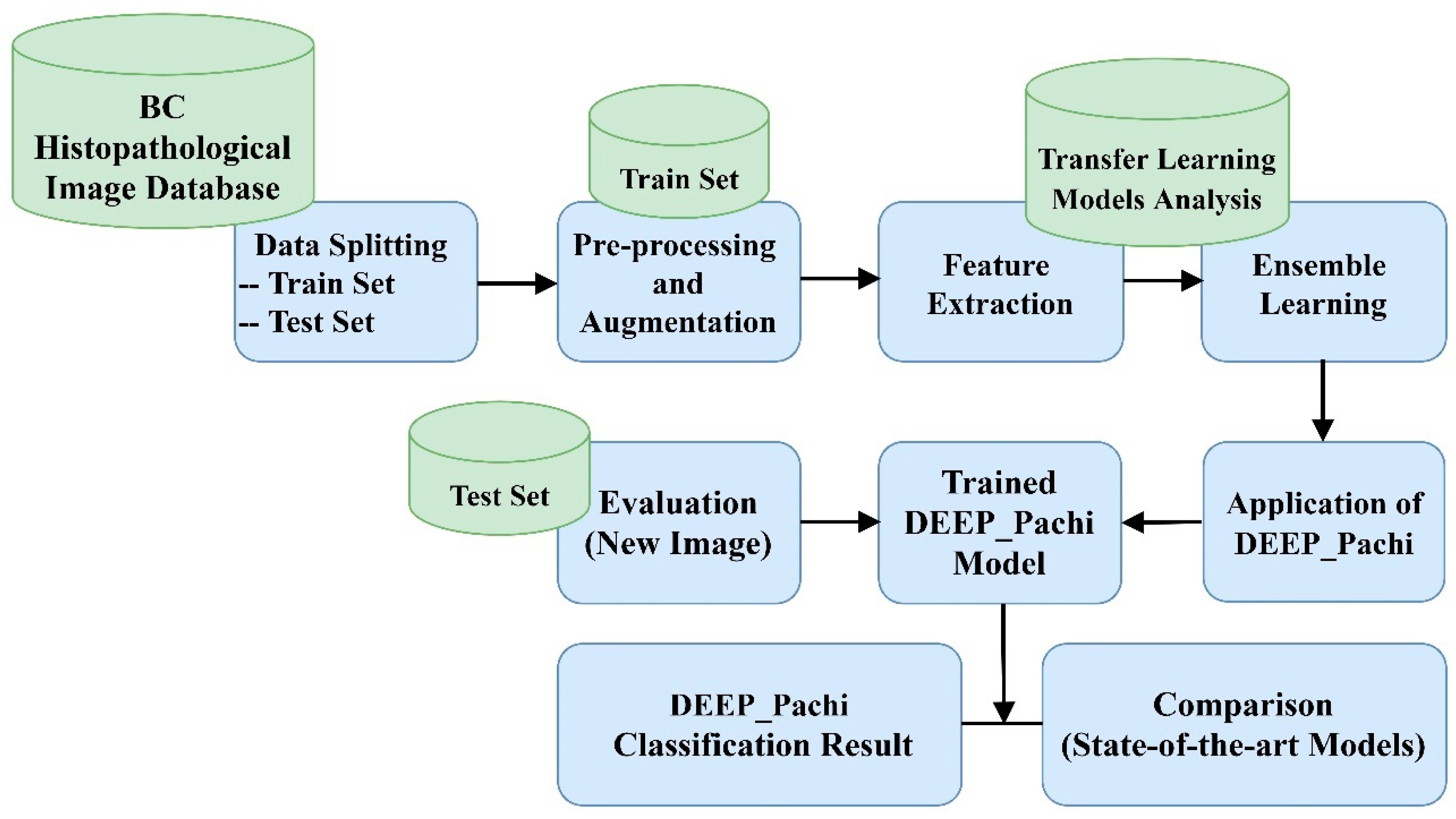
2. Materials and Methods
- ❖
- Step 1: Data collection, splitting, and data preprocessing
- ❖
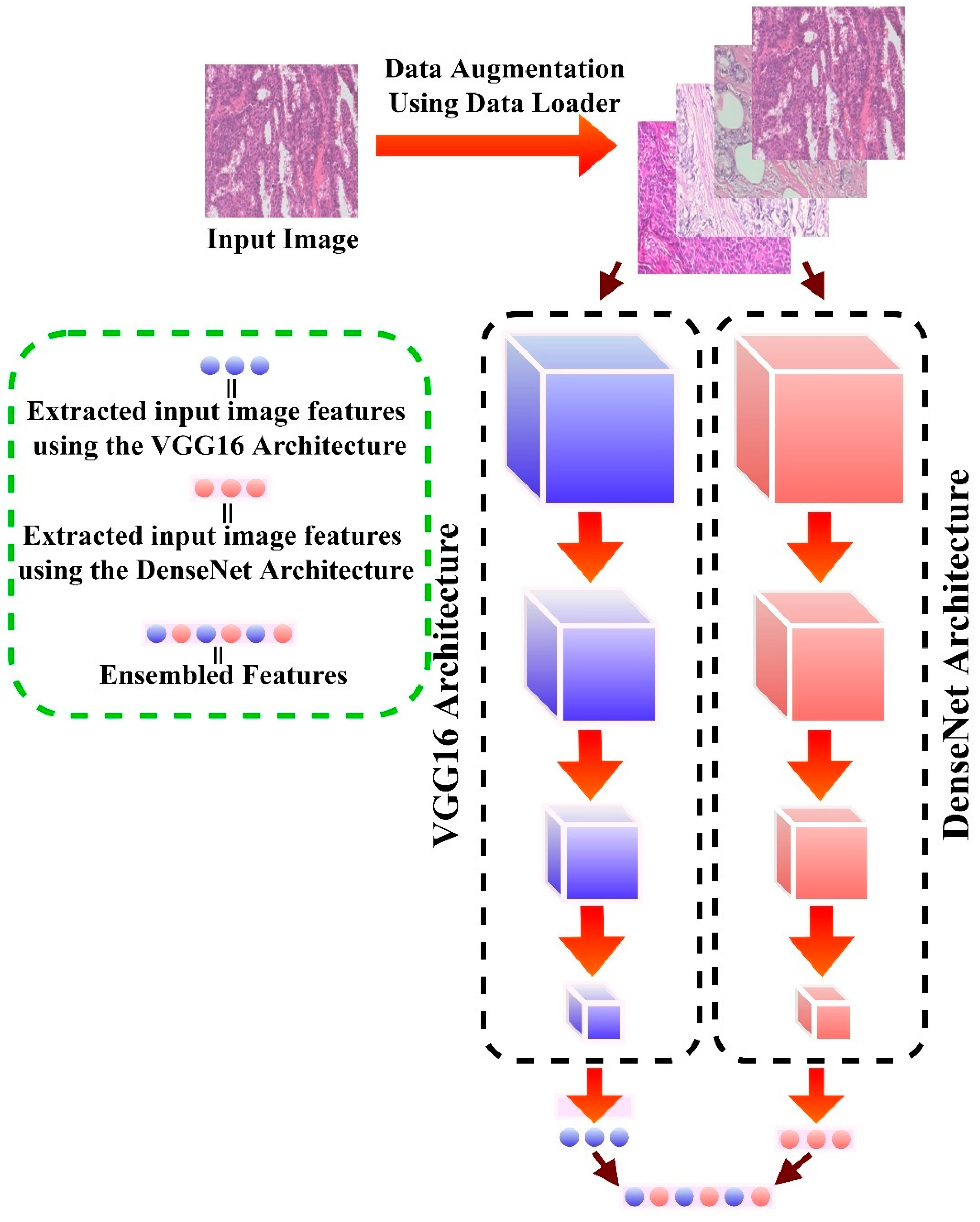
- Step 2: Backbone selection and Ensembling for more robust and generalized features. The examined models were DenseNet201, VGG16, Xception, and InceptionResNetV3 architecture.
- ❖
- Step 3: Feeding the extracted features from the ensemble model into DEEP_Pach architecture.
- ❖
- Step 4: This is the last stage of the proposed model: the identification and classification stage. The learned features are passed into the classification layer for the final result prediction.
- ❖
- Step 5: Then, evaluation with the test set is performed after training.
2.1. Dataset
2.2. Data Pre-Processing/Augmentation
2.3. Network Backbone
- ❖
- VGG16 [96]: VGG16 consists of 16 layers. Following preprocessing, the captured values are fed into a stacked Convolutional layer with 3 × 3 receptive-field filters and a fixed stride of 1. Following that, five max-pooling convolutional layers are used to perform spatial pooling. A 2 × 2 filter’s max-pooling layer is run with a stride of 2. To finalize the design, two fully connected layers (FC) and SoftMax (for the output) are added at the end of the final convolution.
- ❖
- DenseNet201 [97]: This architecture assures information flow across network levels by linking each layer to each layer in a feed-forward fashion (with equal feature-map size). It concatenates (.) the previous layer’s output with the output of the next layer. The transition layers consist of a 1 × 1 convolution followed by a 2 × 2 average pooling. Global pooling is utilized after the last dense block before applying SoftMax.
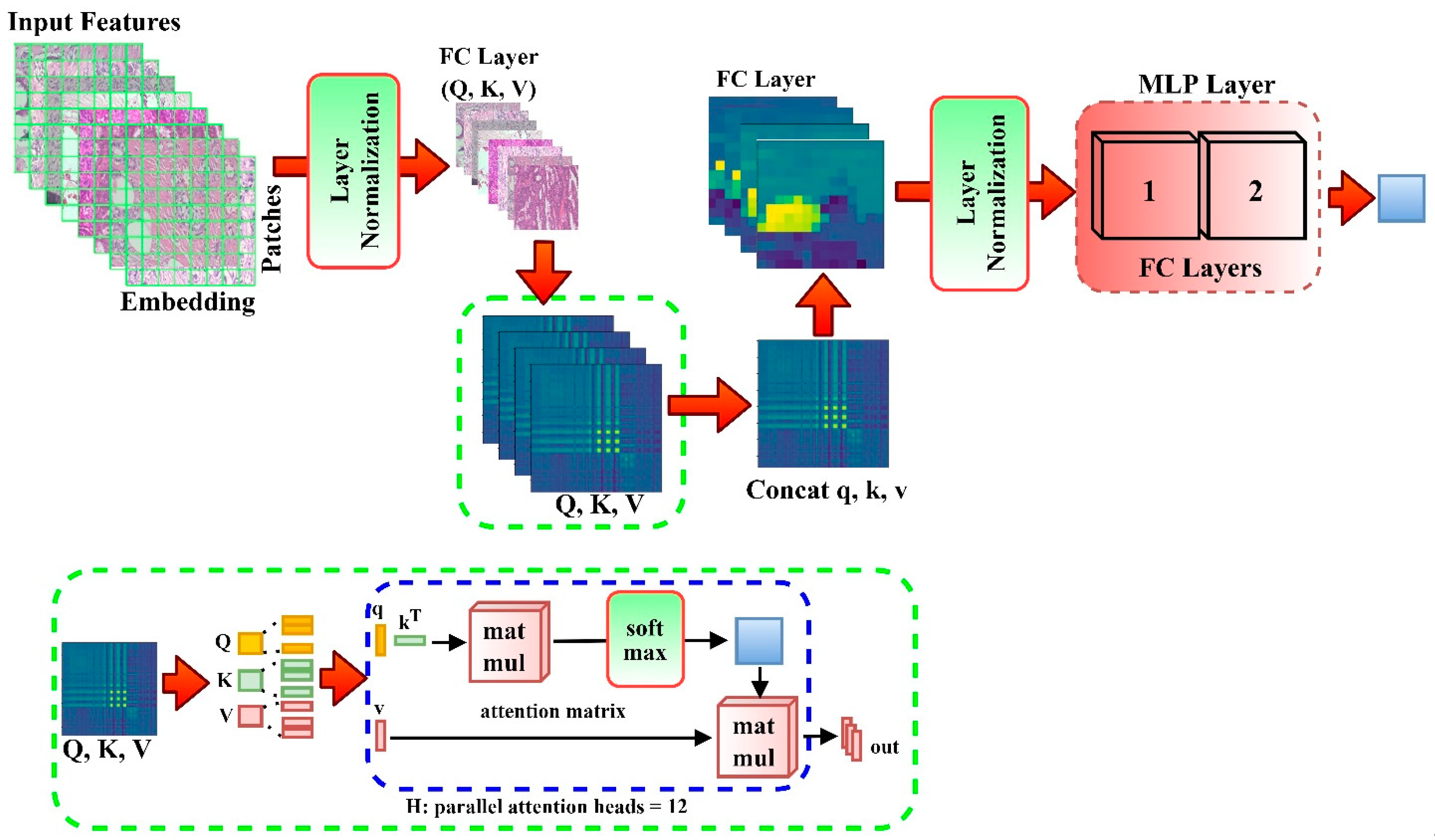
2.4. DEEP_Pachi Architecture
2.5. Experimental Setup
2.6. Evaluation
3. Results
3.1. Parameter Sensitivity Analysis of the Proposed Method
3.2. Transfer Learning Experiment for Backbone Network Selection
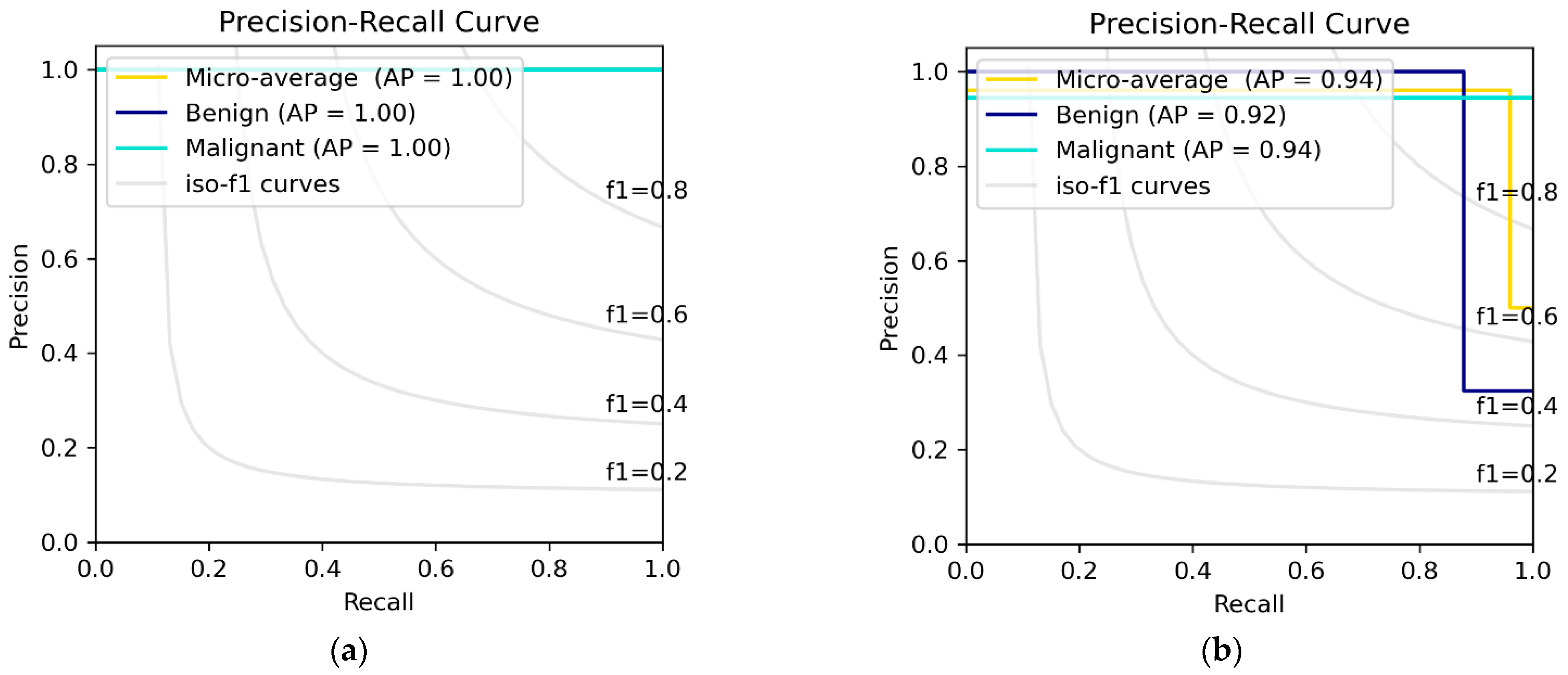
3.3. DEEP_Pachi Architecture Classification Result
4. Discussion
4.1. Visualization the Influence of DEEP_Pachi Framework
4.2. Comparison with the State-of-the-Art Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anastasiadi, Z.; Lianos, G.D.; Ignatiadou, E.; Harissis, H.V.; Mitsis, M. Breast Cancer in Young Women: An Overview. Updates Surg. 2017, 69, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Franch-Expósito, S.; Li, L.; Xiang, T.; Wu, J.; Ren, G. 34P Comprehensive clinical and molecular portraits of grade 3 ER+ HER- breast cancer. Ann. Oncol. 2020, 31, S27. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Man, R.; Yang, P.; Xu, B. Classification of Breast Cancer Histopathological Images Using Discriminative Patches Screened by Generative Adversarial Networks. IEEE Access 2020, 8, 155362–155377. [Google Scholar] [CrossRef]
- Mambou, S.; Maresova, P.; Krejcar, O.; Selamat, A.; Kuca, K. Breast Cancer Detection Using Infrared Thermal Imaging and a Deep Learning Model. Sensors 2018, 18, 2799. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, T.; Li, J.; Pei, Y.; Akhtar, F.; Imran, A.; Rehman, K.U. A Brief Survey on Breast Cancer Diagnostic with Deep Learning Schemes Using Multi-Image Modalities. IEEE Access 2020, 8, 165779–165809. [Google Scholar] [CrossRef]
- Chiao, J.Y.; Chen, K.-Y.; Liao, K.Y.-K.; Hsieh, P.-H.; Zhang, G.; Huang, T.-C. Detection and Classification the Breast Tumors Using Mask R-CNN On Sonograms. Medicine 2019, 98, e15200. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.C.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and Reproducible Invasive Breast Cancer Detection in Whole-Slide Images: A Deep Learning Approach for Quantifying Tumor Extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef] [Green Version]
- Talbert, P.Y.; Frazier, M.D. Inflammatory Breast Cancer Disease: A Literature Review. Cancer Stud. 2019, 2. [Google Scholar] [CrossRef]
- Saha, M.; Chakraborty, C.; Racoceanu, D. Efficient Deep Learning Model for Mitosis Detection Using Breast Histopathology Images. Comput. Med. Imaging Graph. 2018, 64, 29–40. [Google Scholar]
- Domingues, I.; Pereira, G.; Martins, P.; Duarte, H.; Santos, J.; Abreu, P.H. Using Deep Learning Techniques in Medical Imaging: A Systematic Review of Applications on CT And PET. Artif. Intell. Rev. 2019, 53, 4093–4160. [Google Scholar] [CrossRef]
- Murtaza, G.; Shuib, L.; Wahab, A.W.A.; Mujtaba, G.; Mujtaba, G.; Nweke, H.F.; Al-garadi, M.A.; Zulfiqar, F.; Raza, G.; Azmi, N.A. Deep Learning-Based Breast Cancer Classification Through Medical Imaging Modalities: State of The Art and Research Challenges. Artif. Intell. Rev. 2019, 53, 1655–1720. [Google Scholar] [CrossRef]
- Pavithra, S.; Vanithamani, R.; Justin, J. Computer-aided breast cancer detection using ultrasound images. Mater. Today Proc. 2020, 33, 4802–4807. [Google Scholar] [CrossRef]
- Moghbel, M.; Ooi, C.Y.; Ismail, N.; Hau, Y.W.; Memari, N. A Review of Breast Boundary and Pectoral Muscle Segmentation Methods in Computer-Aided Detection/Diagnosis of Breast Mammography. Artif. Intell. Rev. 2019, 53, 1873–1918. [Google Scholar] [CrossRef]
- Prabha, S. Thermal Imaging Techniques for Breast Screening—A Survey. Curr. Med. Imaging 2020, 16, 855–862. [Google Scholar]
- Hadadi, I.; Rae, W.; Clarke, J.; McEntee, M.; Ekpo, E. Diagnostic Performance of Adjunctive Imaging Modalities Compared to Mammography Alone in Women with Non-Dense and Dense Breasts: A Systematic Review and Meta-Analysis. Clin. Breast Cancer 2021, 21, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Nahid, A.A.; Ali, F.B.; Kong, Y. Histopathological Breast-Image Classification with Image Enhancement by Convolutional Neural Network. In Proceedings of the 2017 20th International Conference of Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 22–24 December 2017. [Google Scholar]
- Bardou, D.; Zhang, K.; Ahmad, S.M. Classification of Breast Cancer Based on Histology Images Using Convolutional Neural Networks. IEEE Access 2018, 6, 24680–24693. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of Breast Cancer Histology Images Using Convolutional Neural Networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef]
- Roy, K.; Banik, D.; Bhattacharjee, D.; Nasipuri, M. Patch-Based System for Classification of Breast Histology Images Using Deep Learning. Comput. Med. Imaging Graph. 2019, 71, 90–103. [Google Scholar] [CrossRef]
- Kausar, T.; Wang, M.; Malik, M.S.S. Cancer Detection in Breast Histopathology with Convolution Neural Network Based Approach. In Proceedings of the 2019 IEEE/ACS 16th International Conference on Computer Systems and Applications (AICCSA), Abu Dhabi, United Arab Emirates, 3–7 November 2019. [Google Scholar]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of A Convolutional Neural Network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017. [Google Scholar]
- Perumal, V.; Narayanan, V.; Rajasekar, S.J.S. Detection of Brain Tumor with Magnetic Resonance Imaging using Deep Learning Techniques. In Brain Tumor MRI Image Segmentation Using Deep Learning Techniques; Elsevier: Amsterdam, The Netherlands, 2022; pp. 183–196. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-And-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Huang, Z.; Wang, X.; Huang, L.; Huang, C.; Wei, Y.; Liu, W. CCNET: Criss-Cross Attention for Semantic Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 603–612. [Google Scholar]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual Attention Network for Image Classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3156–3164. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-Local Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Sarikaya, I. Breast Cancer and Pet Imaging. Nucl. Med. Rev. Cent. East. Eur. 2021, 24, 16–26. [Google Scholar] [CrossRef]
- Vaishnavi, J.; Devi, M.A.; Punitha, S.; Ravi, S. Computer-aided mammography techniques for detection and classification of microcalcifications in digital mammograms. Int. J. Image Min. 2018, 3, 48. [Google Scholar] [CrossRef]
- Loizidou, K.; Skouroumouni, G.; Nikolaou, C.; Pitris, C. An Automated Breast Micro-Calcification Detection and Classification Technique Using Temporal Subtraction of Mammograms. IEEE Access 2020, 8, 52785–52795. [Google Scholar] [CrossRef]
- Suh, Y.J.; Jung, J.; Cho, B.-J. Automated Breast Cancer Detection in Digital Mammograms of Various Densities Via Deep Learning. J. Pers. Med. 2020, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Berg, W.A.; Peng, H.; Luo, Y.; Jankowitz, R.C.; Wu, S. A Deep Learning Method for Classifying Mammographic Breast Density Categories. Med. Phys. 2018, 45, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, M.; Ayub, E.; Ahmad, F.; Alruwaili, M.; Alrowaili, Z.A.; Alanazi, S.; Humayun, M.; Rizwan, M.; Naseem, S.; Alyas, T. Machine Learning Enabled Early Detection of Breast Cancer by Structural Analysis of Mammograms. Comput. Mater. Contin. 2021, 67, 641–657. [Google Scholar] [CrossRef]
- Fiorica, J.V. Breast Cancer Screening, Mammography, And Other Modalities. Clin. Obstet. Gynecol. 2017, 59, 688–709. [Google Scholar] [CrossRef]
- Li, Q.; Shi, W.; Yang, H.; Zhang, H.; Li, G.; Chen, T.; Mori, K.; Jiang, Z. Computer-aided diagnosis of mammographic masses using geometric verification-based image retrieval. Med. Imaging 2017 Comput. Aided Diagn. 2017, 10134, 746–753. [Google Scholar]
- Kaur, P.; Singh, G.; Kaur, P. Intellectual detection and validation of automated mammogram breast cancer images by multi-class SVM using deep learning classification. Inform. Med. Unlocked 2019, 16, 100151. [Google Scholar] [CrossRef]
- Tran, T.S.H.; Nguyen, H.M.T. Application of 2D Ultrasound, Elastography Arfi and Mammography for Diagnosis of solid tumors in breast. J. Med. Pharm. 2019, 58–65. [Google Scholar] [CrossRef]
- Han, J.; Li, F.; Peng, C.; Huang, Y.; Lin, Q.; Liu, Y.; Cao, L.; Zhou, J. Reducing Unnecessary Biopsy of Breast Lesions: Preliminary Results with Combination of Strain and Shear-Wave Elastography. Ultrasound Med. Biol. 2019, 45, 2317–2327. [Google Scholar] [CrossRef]
- Ucar, H.; Kacar, E.; Karaca, R. The Contribution of a Solid Breast Mass Gray-Scale Histographic Analysis in Ascertaining a Benign-Malignant Differentiation. J. Diagn. Med. Sonogr. 2022, 875647932210782. [Google Scholar] [CrossRef]
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE J. Biomed. Health Inf. 2017, 22, 1218–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Early Initiation of MRI-Based Breast Cancer Screening Predicted to Halve Breast Cancer Deaths in Childhood Cancer Survivor. Default Digital Object Group 2019. [CrossRef]
- Sriussadaporn, S.; Sriussadaporn, S.; Pak-art, R.; Kritayakirana, K.; Prichayudh, S.; Samorn, P. Ultrasonography increases sensitivity of mammography for diagnosis of multifocal, multicentric breast cancer using 356 whole breast histopathology as a gold standard. Surg. Pract. 2022. [Google Scholar] [CrossRef]
- Pujara, A.C.; Kim, E.; Axelrod, D.; Melsaether, A.N. PET/MRI in Breast Cancer. J. Magn. Reson. Imaging 2018, 49, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Athanasiou, A.; Baltzer, P.A.T.; Camps-Herrero, J.; Clauser, P.; Fallenberg, E.M.; Forrai, G.; Fuchsjäger, M.H.; Helbich, T.H.; Killburn-Toppin, F.; et al. Breast cancer screening in women with extremely dense breasts recommendations of the European Society of Breast Imaging (EUSOBI). Eur. Radiol. 2022, 1–10. [Google Scholar] [CrossRef]
- Houssami, N.; Cho, N. Screening Women with A Personal History of Breast Cancer: Overview of The Evidence on Breast Imaging Surveillance. Ultrasonography 2018, 37, 277. [Google Scholar] [CrossRef]
- Greenwood, H.I. Abbreviated Protocol Breast MRI: The Past, Present, And Future. Clin. Imaging 2019, 53, 169–173. [Google Scholar] [CrossRef]
- Zelst, J.C.V.; Vreemann, S.; Witt, H.-J.; Gubern-Merida, A.; Dorrius, M.D.; Duvivier, K.; Lardenoije-Broker, S.; Lobbes, M.B.; Loo, C.; Veldhuis, W.; et al. Multireader Study on The Diagnostic Accuracy of Ultrafast Breast Magnetic Resonance Imaging for Breast Cancer Screening. Investig. Radiol. 2018, 53, 579–586. [Google Scholar] [CrossRef]
- Heller, S.L.; Moy, L. MRI Breast Screening Revisited. J. Magn. Reson. Imaging 2019, 49, 1212–1221. [Google Scholar] [CrossRef]
- Aswathy, M.; Jagannath, M. Detection of Breast Cancer On Digital Histopathology Images: Present Status And Future Possibilities. Inf. Med. Unlocked 2017, 8, 74–79. [Google Scholar] [CrossRef] [Green Version]
- Tellez, D.; Balkenhol, M.; Karssemeijer, N.; Litjens, G.; Laak, J.V.D.; Ciompi, F. H and E Stain Augmentation Improves Generalization of Convolutional Networks for Histopathological Mitosis Detection. In Medical Imaging 2018: Digital Pathology; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; p. 105810Z. [Google Scholar]
- Jaglan, P.; Dass, R.; Duhan, M. Breast Cancer Detection Techniques: Issues and Challenges. J. Inst. Eng. Ser. B 2019, 100, 379–386. [Google Scholar] [CrossRef]
- Posso, M.; Puig, T.; Carles, M.; Ru’e, M.; Canelo-Aybar, C.; Bonfill, X. Effectiveness and Cost-Effectiveness of Double Reading in Digital Mammography Screening: A Systematic Review and Meta-Analysis. Eur. J. Radiol. 2017, 96, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, L.; Thomas, V.; Sharma, N. Microcalcification On Mammography: Approaches to Interpretation and Biopsy. Br. J. Radiol. 2017, 90, 20160594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houssami, N. Evidence on Synthesized Two-dimensional Mammography Versus Digital Mammography When Using Tomosynthesis (Three-dimensional Mammography) for Population Breast Cancer Screening. Clin. Breast Cancer 2018, 18, 255–260.e1. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Tamura, T.; Kawasaki, Y. The relationship between compressed breast thickness in mammography and other factors that influence breast cancer. J. Jpn. Assoc. Breast Cancer Screen. 2021, 30, 177–181. [Google Scholar] [CrossRef]
- Rapelyea, J.A.; Marks, C.G. Breast Ultrasound Past, Present, And Future. In Breast Imaging; Intech Open: Rijeka, Croatia, 2018; pp. 21–48. [Google Scholar]
- Sood, R.; Rositch, A.F.; Shakoor, D.; Ambinder, E.; Pool, K.-L.; Pollack, E.; Mollura, D.J.; Mullen, L.A.; Harvey, S.C. Ultrasound for Breast Cancer Detection Globally: A Systematic Review and Meta-Analysis. J. Global. Oncol. 2019, 5, 1–17. [Google Scholar] [CrossRef]
- Youk, J.H.; Gweon, H.M.; Son, E.J. Shear-Wave Elastography in Breast Ultrasonography: The State of the Art. Ultrasonography 2017, 36, 300. [Google Scholar] [CrossRef] [Green Version]
- Radiological Society of North America. Ultrasound Images. Available online: https://www.radiologyinfo.org/en/info/genus (accessed on 22 March 2022).
- García, E.; Diez, Y.; Diaz, O.; Llado, X.; Martí, R.; Martí, J.; Oliver, A. A Step-By-Step Review on Patient-Specific Biomechanical Finite Element Models for Breast M.R.I. To X-Ray Mammography Registration. Med. Phys. 2018, 45, e6–e31. [Google Scholar] [CrossRef] [Green Version]
- Kalantarova, A.; Zembol, N.J.; Kufel-Grabowska, J. Pregnancy-Associated Breast Cancer as A Screening and Diagnostic Challenge: A Case Report. Nowotwory 2021, 71, 162–164. [Google Scholar] [CrossRef]
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine Learning in Breast MRI. J. Magn. Reson. Imaging 2020, 52, 998–1018. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep Feature Learning for Histopathological Image Classification of Canine Mammary Tumors and Human Breast Cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Beevi, K.S.; Nair, M.S.; Bindu, G.R. Automatic Mitosis Detection In Breast Histopathology Images Using Convolutional Neural Network Based Deep Transfer Learning. Biocybern. Biomed. Eng. 2019, 39, 214–223. [Google Scholar] [CrossRef]
- Dodballapur, V.; Song, Y.; Huang, H.; Chen, M.; Chrzanowski, W.; Cai, W. Mask-Driven Mitosis Detection in Histopathology Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar]
- Wang, Y.; Lei, B.; Elazab, A.; Tan, E.-L.; Wang, W.; Huang, F.; Gong, X.; Wang, T. Breast Cancer Image Classification via Multi-Network Features and Dual-Network Orthogonal Low-Rank Learning. IEEE Access 2020, 8, 27779–27792. [Google Scholar] [CrossRef]
- Das, A.; Nair, M.S.; Peter, S.D. Sparse Representation Over Learned Dictionaries on the Riemannian Manifold for Automated Grading of Nuclear Pleomorphism in Breast Cancer. IEEE Trans. Image Process. 2019, 28, 1248–1260. [Google Scholar] [CrossRef]
- Dimitropoulos, K.; Barmpoutis, P.; Zioga, C.; Kamas, A.; Patsiaoura, K.; Grammalidis, N. Grading of Invasive Breast Carcinoma Through Grassmannian VLAD Encoding. PLoS ONE 2017, 12, e0185110. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Ma, Y.; Shi, H.; Zhao, Y. Histopathological Whole Slide Image Analysis Using Context-Based CBIR. IEEE Trans. Med. Imaging 2018, 37, 1641–1652. [Google Scholar] [CrossRef]
- Biswas, R.; Roy, S.; Biswas, A. Mammogram Classification using Curvelet Coefficients and Gray Level Co-Occurrence Matrix for Detection of Breast Cancer. Int. J. Innov. Technol. Explor. Eng. 2019, 8, 4819–4824. [Google Scholar]
- Reis, S.; Gazinska, P.; Hipwell, J.H.; Mertzanidou, T.; Naidoo, K.; Williams, N.; Pinder, S.; Hawkes, D.J. Automated Classification of Breast Cancer Stroma Maturity from Histological Images. IEEE Trans. Biomed. Eng. 2017, 64, 2344–2352. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Mari, K.; Delphina, A.A. An Intelligent Framework for Detection and Classification of MRI Brain Tumour using SIFT-SURF Features and K-nearest Neighbour Approach. Strad Res. 2020, 7, 1–10. [Google Scholar]
- Krystel-Whittemore, M.; Wen, H.Y. Update on HER2 expression in breast cancer. Diagn. Histopathol. 2022, 28, 170–175. [Google Scholar] [CrossRef]
- Nateghi, R.; Danyali, H.; Helfroush, M.S. Maximized Inter-Class Weighted Mean for Fast and Accurate Mitosis Cells Detection in Breast Cancer Histopathology Images. J. Med. Syst. 2017, 41, 146. [Google Scholar] [CrossRef] [PubMed]
- Burçak, K.C.; Baykan, Ö.K.; Uğuz, H. A New Deep Convolutional Neural Network Model for Classifying Breast Cancer Histopathological Images and The Hyperparameter Optimization of The Proposed Model. J. Supercomput. 2020, 77, 973–989. [Google Scholar] [CrossRef]
- Li, L.; Pan, X.; Yang, H.; Liu, Z.; He, Y.; Li, Z.; Fan, Y.; Cao, Z.; Zhang, L. Multi-Task Deep Learning for Fine-Grained Classification and Grading in Breast Cancer Histopathological Images. Multimed. Tools Appl. 2018, 79, 14509–14528. [Google Scholar] [CrossRef]
- Gour, M.; Jain, S.; Kumar, T.S. Residual Learning-Based CNN For Breast Cancer Histopathological Image Classification. Int. J. Imaging Syst. Technol. 2020, 30, 621–635. [Google Scholar] [CrossRef]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast Cancer Histopathological Image Classification Using A Hybrid Deep Neural Network. Methods 2020, 173, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Breast Cancer Histopathological Image Classification Using Convolutional Neural Networks with Small SE-Resnet Module. PLoS ONE 2019, 14, e0214587. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Zhang, X.; Zhou, X.; Liu, S. Parallel Structure Deep Neural Network Using CNN and RNN with an Attention Mechanism for Breast Cancer Histology Image Classification. Cancers 2019, 11, 1901. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.P.C. A Novel Deep Learning-Based Framework for The Detection and Classification of Breast Cancer Using Transfer Learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, R.; Zargari, A.; Thai, T.C.; Gunderson, C.C.; Moxley, K.M.; Liu, H.; Zheng, B.; Qiu, Y. Classification of Tumor Epithelium and Stroma by Exploiting Image Features Learned by Deep Convolutional Neural Networks. Ann. Biomed. Eng. 2018, 46, 1988–1999. [Google Scholar] [CrossRef]
- Wang, P.; Song, Q.; Li, Y.; Lv, S.; Wang, J.; Li, L.; Zhang, H. Cross-Task Extreme Learning Machine for Breast Cancer Image Classification with Deep Convolutional Features. Biomed. Signal Process. Control 2020, 57, 101789. [Google Scholar] [CrossRef]
- Xie, J.; Liu, R.; Luttrell, J.; Zhang, C. Deep Learning-Based Analysis of Histopathological Images of Breast Cancer. Front. Genet. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Kausar, T.; Wang, M.; Idrees, M.; Lu, Y. HWDCNN: Multi-Class Recognition in Breast Histopathology with HAAR Wavelet Decomposed Image-Based Convolution Neural Network. Biocybern. Biomed. Eng. 2019, 39, 967–982. [Google Scholar] [CrossRef]
- Yang, H.; Kim, J.-Y.; Kim, H.; Adhikari, S.P. Guided Soft Attention Network for Classification of Breast Cancer Histopathology Images. IEEE Trans. Med. Imaging 2020, 39, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 4172. [Google Scholar]
- Nazeri, K.; Aminpour, A.; Ebrahimi, M. Two-Stage Convolutional Neural Network for Breast Cancer Histology Image Classification. Image Anal. Recognit. 2018, 10882, 717–726. [Google Scholar]
- Xu, B.; Liu, J.; Hou, X.; Liu, B.; Garibaldi, J.; Ellis, I.O.; Green, A.; Shen, L.; Qiu, G. Attention by Selection: A Deep Selective Attention Approach to Breast Cancer Classification. IEEE Trans. Med. Imaging 2020, 39, 1930–1941. [Google Scholar] [CrossRef]
- Shallu; Mehra, R. Breast Cancer Histology Images Classification: Training from Scratch or Transfer Learning? ICT Express 2018, 4, 247–254. [Google Scholar] [CrossRef]
- Wan, T.; Cao, J.; Chen, J.; Qin, Z. Automated Grading of Breast Cancer Histopathology Using Cascaded Ensemble with Combination of Multi-Level Image Features. Neurocomputing 2017, 229, 34–44. [Google Scholar] [CrossRef]
- Saxena, S.; Shukla, S.; Gyanchandani, M. Pre-Trained Convolutional Neural Networks as Feature Extractors for Diagnosis of Breast Cancer Using Histopathology. Int. J. Imaging Syst. Technol. 2020, 30, 577–591. [Google Scholar] [CrossRef]
- Sharma, S.; Mehra, R. Conventional Machine Learning and Deep Learning Approach for Multi-Classification of Breast Cancer Histopathology Images—A Comparative Insight. J. Digit. Imaging 2020, 33, 632–654. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Song, F.; Wang, Y.; Dong, H.; Guo, Y.; Liu, J. Breast Cancer Histopathology Image Classification Through Assembling Multiple Compact CNNS. BMC Med. Inform. Decis. Mak. 2019, 19, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karen, S.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. BACH: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- George, K.; Faziludeen, S.; Sankaran, P. Breast cancer detection from biopsy images using nucleus guided transfer learning and belief-based fusion. Comput. Biol. Med. 2020, 124, 103954. [Google Scholar] [CrossRef]
- Kwok, S. Multiclass Classification of Breast Cancer in Whole-Slide Images. Image Anal. Recognit. 2018, 10882, 931–940. [Google Scholar]
- Wong, W.S.; Amer, M.; Maul, T.; Liao, I.Y.; Ahmed, A. Conditional Generative Adversarial Networks for Data Augmentation in Breast Cancer Classification. Recent Adv. Soft Comput. Data Min. 2019, 978, 392–402. [Google Scholar]
- Thuy, M.B.H.; Hoang, V.T. Fusing of Deep Learning, Transfer Learning and GAN for Breast Cancer Histopathological Image Classification. Adv. Intell. Syst. Comput. 2019, 1121, 255–266. [Google Scholar]
- Li, G.; Li, C.; Wu, G.; Ji, D.; Zhang, H. Multi-View Attention-Guided Multiple Instance Detection Network for Interpretable Breast Cancer Histopathological Image Diagnosis. IEEE Access 2021, 9, 79671–79684. [Google Scholar] [CrossRef]
- Boumaraf, S.; Liu, X.; Zheng, Z.; Ma, X.; Ferkous, C. A new transfer learning-based approach to magnification dependent and independent classification of breast cancer in histopathological images. Biomed. Signal Process. Control 2021, 63, 102192. [Google Scholar] [CrossRef]
- Gupta, K.; Chawla, N. Analysis of Histopathological Images for Prediction of Breast Cancer Using Traditional Classifiers with Pre-Trained CNN. Procedia Comput. Sci. 2020, 167, 878–889. [Google Scholar] [CrossRef]
- Budak, Ü.; Güzel, A.B. Automatic Grading System for Diagnosis of Breast Cancer Exploiting Co-occurrence Shearlet Transform and Histogram Features. IRBM 2020, 41, 106–114. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, J.; Huang, S.; Liu, B. Breast cancer histopathological image classification using attention high-order deep network. Int. J. Imaging Syst. Technol. 2021, 32, 266–279. [Google Scholar] [CrossRef]
- Ibraheem, A.M.; Rahouma, K.H.; Hamed, H.F.A. 3PCNNB-Net: Three Parallel CNN Branches for Breast Cancer Classification Through Histopathological Images. J. Med. Biol. Eng. 2021, 41, 494–503. [Google Scholar] [CrossRef]
- Liu, W.; Juhas, M.; Zhang, Y. Fine-Grained Breast Cancer Classification with Bilinear Convolutional Neural Networks (BCNNs). Front. Genet. 2020, 11, 1061. [Google Scholar] [CrossRef]
- Kashyap, R. Evolution of histopathological breast cancer images classification using stochastic dilated residual ghost model. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2758–2779. [Google Scholar] [CrossRef]
- Nahid, A.A.; Mehrabi, M.A.; Kong, Y. Histopathological Breast Cancer Image Classification by Deep Neural Network Techniques Guided by Local Clustering. BioMed Res. Int. 2018, 2018, 2362108. [Google Scholar] [CrossRef]
- Nawaz, M.; Sewissy, A.A.; Hassan, T. Multi-Class Breast Cancer Classification using Deep Learning Convolutional Neural Network. Int. J. Adv. Comput. Sci. Appl. 2018, 9, 316–332. [Google Scholar] [CrossRef]
- Sanchez-Morillo, D.; González, J.; García-Rojo, M.; Ortega, J. Classification of Breast Cancer Histopathological Images Using KAZE Features. Lect. Notes Comput. Sci. 2018, 10814, 276–286. [Google Scholar]
- Zhang, X.; Zhang, Y.; Qian, B.; Liu, X.; Li, X.; Wang, X.; Yin, C.; Lv, X.; Song, L.; Wang, L. Classifying Breast Cancer Histopathological Images Using a Robust Artificial Neural Network Architecture. Lect. Notes Comput. Sci. 2019, 11465, 204–215. [Google Scholar]
- Alom, M.Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast Cancer Histopathological Image Classification using Deep Convolutional Neural Network. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Mewada, H.K.; Patel, A.V.; Hassaballah, M.; Alkinani, M.H.; Mahant, K. Spectral–Spatial Features Integrated Convolution Neural Network for Breast Cancer Classification. Sensors 2020, 20, 4747. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Shukla, S.; Gyanchandani, M. Breast cancer histopathology image classification using kernelized weighted extreme learning machine. Int. J. Imaging Syst. Technol. 2020, 31, 168–179. [Google Scholar] [CrossRef]
- Sharma, S.; Mehra, R.; Kumar, S. Optimised CNN in conjunction with efficient pooling strategy for the multi-classification of breast cancer. IET Image Process. 2020, 15, 936–946. [Google Scholar] [CrossRef]
- Hao, Y.; Qiao, S.; Zhang, L.; Xu, T.; Bai, Y.; Hu, H.; Zhang, W.; Zhang, G. Breast Cancer Histopathological Images Recognition Based on Low Dimensional Three-Channel Features. Front. Oncol. 2021, 11, 657560. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, R.; Prasad, K.; Udupa, C.B.K. BCHisto-Net: Breast histopathological image classification by global and local feature aggregation. Artif. Intell. Med. 2021, 121, 102191. [Google Scholar]
- Carvalho, E.D.; Filho, A.O.C.; Silva, R.R.V.; Araújo, F.H.D.; Diniz, J.O.B.; Silva, A.C.; Paiva, A.C.; Gattass, M. Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif. Intell. Med. 2020, 105, 101845. [Google Scholar] [CrossRef]
- Pimkin, A.; Makarchuk, G.; Kondratenko, V.; Pisov, M.; Krivov, E.; Belyaev, M. Ensembling Neural Networks for Digital Pathology Images Classification and Segmentation. Image Anal. Recognit. 2018, 10882, 877–886. [Google Scholar]
- Yang, Z.; Ran, L.; Zhang, S.; Xia, Y.; Zhang, Y. EMS-Net: Ensemble of Multiscale Convolutional Neural Networks for Classification of Breast Cancer Histology Images. Neurocomputing 2019, 366, 46–53. [Google Scholar] [CrossRef]
- Sitaula, C.; Aryal, S. Fusion of whole and part features for the classification of histopathological image of breast tissue. Health Inf. Sci. Syst. 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Piao, Y.; Zhang, G. Dilated and soft attention-guided convolutional neural network for breast cancer histology images classification. Microsc. Res. Tech. 2022, 85, 1248–1257. [Google Scholar] [CrossRef] [PubMed]






| Imaging Techniques | Robustness | Constraints | Public Datasets |
|---|---|---|---|
| MG | 1. Reliable and premium approach for capturing, storing, and processing images of breast tissue [52,53] 2. Unlike HP images, they do not need a comprehensive experience or professional understanding to analyze and classify. | 1. Due to their microscopic dimensions and scattered form features, they have restricted abilities in acquiring segments and sub in the human breast [54]. 2. Unsuitable for detecting breast cancer in thick breasts due to the absence of malignant tissues [55]. 3. Not reliable in identifying BC; hence more screening may be necessary for accurate assessments [56]. | -BCDR -CBIS-DDSM -MIAS -Mini-MIAS -DDSM -InBreast |
| US | 1. Does not make patients vulnerable to dangerous rays and is thus regarded exceedingly safe, particularly for expectant mothers [57]. 2. These are specifically convenient imaging techniques for identifying BC in thick breasts, where MGs fail [58]. 3. Allows for viewing a breast tumor from multiple viewpoints and configurations, lowering the possibility of a negative result assessment. | 1. Often yield false diagnoses if the scanner probe is not moved or pushed appropriately [59]. 2. They cannot correctly portray the tumor outline in the breast due to its signal weakness to the human muscles [60]. 3. US images are of low quality compared to the images of MGs; thus, obtaining ROI for more advanced analysis is challenging with US imaging. | -BCDR -BUSI |
| MRI | 1. MRI can detect questionable spots, which can be explored further with autopsy (MRI-assisted biopsy). 2. MRI, just like US, does not make patients vulnerable to any dangerous radioactive materials. 3. MRI gives a thorough description of soft breast internal tissues as well as the ability to record | 1. To improve MRI images, supplement chemicals are frequently administered, which might cause sensitivities or other issues and are thus not suggested for patients, particularly renal patients [61]. 2. MRI is typically not suggested throughout pregnancy [62] and is primarily advised as a follow-up test only after an MGs-based examination has been performed. 3. MRI is a pricey procedure relative to MGs or US; hence, it is not often used for BC diagnosis. MRI offers highly accurate data about the interior breast tissues, but it can overlook some malignant areas that MGs can identify [63]. | Duke-Breast-Cancer RIDER Breast MRI |
| HP | 1. Images of HP are RGB images that are very efficient in diagnosing many types of malignancies and provide a greater efficacy for an early phase of BC. 2. An in-depth study of breast tissues is feasible with HP images, resulting in a more reliable examination of BC than other imaging alternatives. 3. Multi ROI images may be produced from full flip HP images, increasing the likelihood of detecting cancer tissues and lowering the number of false positives. | 1. HP images are obtained by mammogram, which is an expensive approach with significant potential complications, necessitating special attention from pathologists as comparable to other imaging alternatives 2. HP images are easy to misinterpret, and the conventional examination of HP images takes a long time [64]. As a result, experts are needed for correct interpretation. 3. Extreme caution is required during histopathology specimen preparation (From the extraction of a tissue sample from the breast to the application of microscope to the extracted tissue sample, the adjustment/control of the color disparities caused by different staining processes) to reduce the possibility of a mistaken diagnosis. | UCI (Wisconsin) BICBH BreakHis |
| Identified Public site for BC Dataset | http://peipa.essex.ac.uk/info/mias.html, http://marathon.csee.usf.edu/Mammography/Database.html, https://biokeanos.com/source/INBreast, https://bcdr.ceta-ciemat.es/information/about https://wiki.cancerimagingarchive.net/display/Public/, https://www.repository.cam.ac.uk/handle/1810/250394, accessed on 20 March 2022. | ||
| Ref | Year | Image Type | Techniques | Task | Recorded Result |
|---|---|---|---|---|---|
| [8] | 2017 | - | ConvNet classifier | Detection | 75.86% Dice coefficient 71.62% positive prediction 96.77% negative prediction (pixel-by-pixel evaluation) |
| [12] | 2017 | - | Multiscale Basic Image Features, Local Binary Patterns, Random Decision Trees Classifier | Classification | 84% Accuracy |
| [32] | 2017 | BreaKHis Augmented BreaKHis | CSDCNN model | Multi-Classification | 93.2% accuracy |
| [37] | 2017 | - | Hybrid Contour Model-Based Segmentation with SVM Classifier | Binary Classification Multi-Classification | 88% AUC. |
| [36] | 2018 | BreaKHis | VGG16, VGG19, and ResNet50 with Logistic Regression | Binary Classification | 92.60% accuracy, 95.65% AUC, 95.95% precision score |
| [33] | 2018 | BACH (ICIAR 2018) | Two-Stage CNN | Multi-Classification | 95% accuracy |
| [4] | 2018 | BreaKHis | DL model with handcrafted features | Mitosis detection | 92% Precision 88% Recall 90% F-Score |
| [5] | 2018 | BreaKHis | Transfer Learning based CNN | Mitosis detection | 15% F1-Score improvement |
| [27] | 2018 | TMAD, OUHSC | Transfer Learning. | Binary Classification | 90.2% Accuracy with GoogleNet |
| [23] | 2019 | BACH (ICIAR 2018) | Hybrid CNN + Deep RNN | Multi-Classification | 91.3% Accuracy |
| [24] | 2019 | BreaKHis | Small SE-ResNet | Binary Classification Multi-Classification | 98.87–99.34% Binary Classification Accuracy 90.66–93.81% Multi-Classification Accuracy |
| [25] | 2019 | BACH (ICIAR 2018) Bioimaging2015 Extended Bioimaging2015 | CNN + RNN + Attention Mechanism | Multi-Classification | - |
| [6] | 2019 | BreaKHis | Mask R-CNN network, with features obtained from Handcrafted and DCNN | Mitosis detection | - |
| [26] | 2019 | BreaKHis L.R.H. hospital Peshawar Data | Transfer Learning. GoogleNet, VGGNet, ResNet | Binary Classification | 97.53% Accuracy |
| [28] | 2019 | BreaKHis | D2TL and ICELM | Binary Classification | Classification Accuracy 96.67%, 96.96%, 98.18% |
| [29] | 2019 | BreaKHis | Inception_V3 Inception_ResNet_V2 | Multi-Classification | - |
| [30] | 2019 | BreaKHis BACH (ICIAR 2018) | Deep CNN with Wavelet decomposed mages | Binary Classification Multi-Classification | 96.85% Accuracy 98.2% Accuracy |
| [34] | 2019 | deep selective attention | Classification | 98% accuracy | |
| [21] | 2020 | B.H.I.s BreaKHis | Modified Inception Network/Transfer Learning | Classification multiclass | - |
| [22] | 2020 | BreaKHis | ResHist model (Residual Learning CNN) | Classification | 84.34% Accuracy 90.49% F1-Score 92.52% Accuracy (DA) 93.45% F1-score (DA) |
| [31] | 2020 | BACH (ICIAR 2018) | Attention Guided CNN | Detection and Classification | 90.25 ± Accuracy 0.98425 AUC Single 88% Accuracy Ensemble 93% Accuracy |
| [35] | 2020 | BreaKHis BACH (ICIAR 2018) | CNN and multi-resolution Spatial Features wavelet transform | Binary Classification Multi-Classification | 97.58% Accuracy 97.45% Accuracy |
| [38] | 2020 | BreaKHis | CNN With Several Classifiers | Binary Classification | |
| [39] | 2020 | VGG16, VGG19, and ResNet50 with SVM | |||
| [19] | 2021 | BHIs | DCNN with several Optimizers | Classification | 99.05% accuracy |
| Class | Sub_Class | Magnification | Total | Nos_Patients | |||
|---|---|---|---|---|---|---|---|
| 40× | 100× | 200× | 400× | ||||
| Benign | Adenosis | 114 | 113 | 111 | 106 | 444 | 24 |
| Fibroadenoma | 253 | 260 | 264 | 237 | 1014 | ||
| Phyllodes_tumor | 109 | 121 | 108 | 115 | 453 | ||
| Tubular_adenoma | 149 | 150 | 140 | 130 | 569 | ||
| Malignant | Ductal_carcinoma | 864 | 903 | 896 | 788 | 3451 | 58 |
| Lobular_carcinoma | 156 | 170 | 163 | 137 | 626 | ||
| Mucinous_carcinoma | 205 | 222 | 196 | 169 | 792 | ||
| Papillary_carcinoma | 145 | 142 | 135 | 138 | 560 | ||
| Total | 1995 | 2081 | 2013 | 1820 | 7090 | 82 | |
| Import Augmentor |
|---|
| def upsample(dir, num_samples): |
| p = Augmentor.Pipeline(dir) |
| p.rotate(probability = 1, max_left_rotation = 5, max_right_rotation = 5) |
| p.zoom(probability = 0.2, min_factor = 1.1, max_factor = 1.2) |
| p.skew(probability = 0.2) |
| p.shear(probability = 0.2, max_shear_left = 2, max_shear_right = 2) |
| p.crop_random(probability = 0.5, percentage_area = 0.8) |
| p.flip_random(probability = 0.2) |
| p.sample(num_samples) |
| p.random_distortion(probability = 1, grid_width = 4, grid_height = 4, magnitude = 8) |
| p.flip_left_right(probability = 0.8) |
| p.flip_top_bottom(probability = 0.3) |
| p.rotate90(probability = 0.5) |
| p.rotate270(probability = 0.5) |
| src_dir = ‘D:/Pachigo/Breast_Cancer/Train/Benign/40 |
| src_dir = ‘D:/Pachigo/Breast_Cancer/Train/Benign/100 |
| src_dir = ‘D:/Pachigo/Breast_Cancer/Train/Benign/200 |
| src_dir = ‘D:/Pachigo/Breast_Cancer/Train/Benign/400 |
| upsample(src_dir, 1500) |
| Models | Learning Rate | Loss Function | Trainable Parameter | Non-Trainable Parameter | Total Parameter | Optimizers | Nos. of Epochs |
|---|---|---|---|---|---|---|---|
| DenseNet201 | 0.001 | Categorical smooth loss | 1,106,179 | 18,321,984 | 19,428,163 | Adam | Early stop |
| VGG16 | 0.001 | Categorical smooth loss | 598,403 | 14,714,688 | 15,313,091 | Adam | Early stop |
| InceptResNetV2 | 0.001 | Categorical smooth loss | 393,475 | 54,336,736 | 54,730,211 | Adam | Early stop |
| Xception | 0.001 | Categorical smooth loss | 1,179,907 | 20,861,480 | 22,041,387 | Adam | Early stop |
| Ensemble | 0.001 | Categorical smooth loss | 43,872,899 | 33,036,672 | 76,909,571 | Adam | Early stop |
| DEEP_Pachi | 0.001 | Categorical smooth loss | 766,291 | 33,036,848 | 33,803,139 | Adam | Early stop |
| Nos. of Pre-Trained Network | Nos. of Self-Attention Heads | Learning Rate | Nos. of Epoch | Accuracy (%) | Precision (%) | F1_Score (%) |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 × 10−3 | 50 | 0.96 | 0.96 | 0.96 |
| 2 | 2 | 3 × 10−3 | 50 | 0.96 | 0.97 | 0.96 |
| 3 | 2 | 3 × 10−3 | 50 | 0.97 | 0.97 | 0.97 |
| 1 | 4 | 3 × 10−3 | 50 | 0.96 | 0.97 | 0.96 |
| 2 | 4 | 3 × 10−3 | 50 | 0.97 | 0.98 | 0.97 |
| 3 | 4 | 3 × 10−3 | 50 | 0.98 | 0.97 | 0.97 |
| 1 | 8 | 3 × 10−3 | 50 | 0.96 | 0.97 | 0.97 |
| 2 | 8 | 3 × 10−3 | 50 | 0.97 | 0.99 | 0.98 |
| 3 | 8 | 3 × 10−3 | 50 | 0.98 | 0.98 | 0.98 |
| 1 | 16 | 3 × 10−3 | 50 | 0.98 | 0.98 | 0.98 |
| 2 | 16 | 3 × 10−3 | 50 | 0.99 | 1.0 | 0.98 |
| 3 | 16 | 3 × 10−3 | 50 | 1.0 | 0.98 | 0.99 |
| Models | ACC (%) | SEN (%) | SPE (%) | PRE (%) | F1_Score (%) | AUC (%) |
|---|---|---|---|---|---|---|
| 40× Magnification-Benign | ||||||
| DenseNet201 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| InceptionResNet | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.99 |
| VGG16 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Xception | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 100× Magnification-Benign | ||||||
| DenseNet201 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| InceptionResNet | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| VGG16 | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.99 |
| Xception | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.99 |
| 200× Magnification-Benign | ||||||
| DenseNet201 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| InceptionResNet | 0.99 | 0.98 | 0.99 | 0.99 | 0.98 | 0.98 |
| VGG16 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Xception | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 400× Magnification Benign | ||||||
| DenseNet201 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| InceptionResNet | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| VGG16 | 0.99 | 0.98 | 0.99 | 0.99 | 0.98 | 0.98 |
| Xception | 0.99 | 0.98 | 0.99 | 0.99 | 0.98 | 0.98 |
| 40× Magnification Malignant | ||||||
| DenseNet201 | 0.98 | 0.99 | 0.99 | 0.95 | 0.97 | 0.99 |
| InceptionResNet | 0.94 | 0.95 | 0.97 | 0.83 | 0.88 | 0.96 |
| VGG16 | 0.94 | 0.93 | 0.96 | 0.82 | 0.86 | 0.94 |
| Xception | 0.94 | 0.93 | 0.96 | 0.82 | 0.86 | 0.94 |
| 100× Magnification Malignant | ||||||
| DenseNet201 | 0.97 | 0.98 | 0.98 | 0.91 | 0.94 | 0.98 |
| InceptionResNet | 0.94 | 0.95 | 0.97 | 0.83 | 0.88 | 0.96 |
| VGG16 | 0.94 | 0.94 | 0.96 | 0.83 | 0.87 | 0.95 |
| Xception | 0.96 | 0.96 | 0.97 | 0.86 | 0.90 | 0.97 |
| 200× Magnification Malignant | ||||||
| DenseNet201 | 0.98 | 0.97 | 0.98 | 0.94 | 0.95 | 0.98 |
| InceptionResNet | 0.93 | 0.94 | 0.96 | 0.80 | 0.85 | 0.95 |
| VGG16 | 0.92 | 0.93 | 0.95 | 0.79 | 0.84 | 0.94 |
| Xception | 0.95 | 0.95 | 0.97 | 0.85 | 0.89 | 0.96 |
| 400× Magnification Malignant | ||||||
| DenseNet201 | 0.98 | 0.98 | 0.98 | 0.92 | 0.95 | 0.98 |
| InceptionResNet | 0.96 | 0.97 | 0.98 | 0.88 | 0.92 | 0.97 |
| VGG16 | 0.97 | 0.96 | 0.98 | 0.90 | 0.93 | 0.97 |
| Xception | - | - | - | - | - | - |
| Models | ACC (%) | SEN (%) | SPE (%) | PRE (%) | F1_Score | AUC |
|---|---|---|---|---|---|---|
| 100× Magnification | ||||||
| Backbone Network | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 400× Magnification | ||||||
| Network Backbone | 0.95 | 0.93 | 0.93 | 0.95 | 0.94 | 0.93 |
| DEEP_Pachi | 0.96 | 0.96 | 0.96 | 0.97 | 0.95 | 0.96 |
| Models | ACC (%) | SEN (%) | SPE (%) | PRE (%) | F1_Score (%) | AUC (%) |
|---|---|---|---|---|---|---|
| 40× Magnification-Benign | ||||||
| Network Backbone | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 100× Magnification-Benign | ||||||
| Network Backbone | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 200× Magnification-Benign | ||||||
| Network Backbone | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 400× Magnification Benign | ||||||
| Network Backbone | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 40× Magnification Malignant | ||||||
| Network Backbone | 0.97 | 0.98 | 0.98 | 0.92 | 0.94 | 0.98 |
| DEEP_Pachi | 0.99 | 1.0 | 1.0 | 0.96 | 0.98 | 0.98 |
| 100× Magnification Malignant | ||||||
| Network Backbone | 0.97 | 0.98 | 0.98 | 0.91 | 0.94 | 0.98 |
| DEEP_Pachi | 0.99 | 1.0 | 1.0 | 0.94 | 0.98 | 0.98 |
| 200× Magnification Malignant | ||||||
| Network Backbone | 0.96 | 0.96 | 0.98 | 0.90 | 0.92 | 0.97 |
| DEEP_Pachi | 0.99 | 0.99 | 0.99 | 0.95 | 0.98 | 0.98 |
| 400× Magnification Malignant | ||||||
| Network Backbone | 0.98 | 0.98 | 0.98 | 0.92 | 0.95 | 0.98 |
| DEEP_Pachi | 1.0 | 1.0 | 1.0 | 0.97 | 0.99 | 0.99 |
| Ref/Year | Approach | Data Type | Classification Type | Accuracy (%) | ||||
|---|---|---|---|---|---|---|---|---|
| 40× | 100× | 200× | 400× | Binary | ||||
| [112] 2018 | Ensemble (CNN + LSTM) | BreaKHis | 88.7 | 85.3 | 88.6 | 88.4 | ||
| [113] 2018 | DenseNet CNN | BreaKHis | 93.6 | 97.4 | 95.9 | 94.7 | ||
| [77] 2018 | Xception | BreaKHis | 95.3 | 93.4 | 93.1 | 91.7 | ||
| [114] 2018 | KAZE features + Bag of Features | BreaKHis | 85.9 | 80.4 | 78.1 | 71.1 | ||
| [102] 2019 | CNN | BreaKHis | 77.2 | |||||
| CNN + DA | 76.7 | |||||||
| CGANs based DA | 77.3 | |||||||
| DA + CGANs based DA | 75.2 | |||||||
| CNN | 75.4 | |||||||
| CNN + DA | 75.9 | |||||||
| CGANs based DA | 78.5 | |||||||
| DA + CGANs based DA | 78.7 | |||||||
| [115] 2019 | Deep ResNet + CBAM | BreaKHis | 91.2 | 91.7 | 92.6 | 88.9 | ||
| [103] 2019 | Transfer Learning (VGG16 + VGG19 + CNN) | 98.2 | 98.3 | 98.2 | 97.5 | |||
| 98.1 | ||||||||
| [116] 2019 | IRRCNN | BreaKHis | 98.0 | 97.6 | 97.3 | 97.4 | ||
| [85] 2019 | Inception_V3 | BreaKHis | Multiclass | 90.3 | 85.4 | 84.0 | 82.1 | |
| Binary | 97.7 | 94.2 | 87.2 | 96.7 | ||||
| Inception_ResNet_V2 | Multiclass | 98.4 | 98.7 | 97.9 | 97.4 | |||
| Binary | 99.9 | 99.9 | 1.0 | 99.9 | ||||
| [80] 2019 | BHCNet-6 + ERF | BreaKHis | Multiclass | 94.4 | 94.5 | 92.3 | 91.1 | |
| CNN +SE-ResNet | Binary | 98.9 | 99.0 | 99.3 | 99.0 | |||
| [117] 2020 | Deep CNN | BreaKHis | 73.4 | 76.8 | 83.2 | 75.8 | ||
| [94] 2020 | VGG16 + SVM (Balanced + DA) | BreaKHis | 94.0 | 92.9 | 91.2 | 91.8 | ||
| Ensemble (VGG16 + VGG19 + ResNet 50) + RF Classifier | 90.3 | 90.1 | 87.4 | 86.6 | ||||
| Ensemble (VGG16 + VGG19 + ResNet 50) + SVM Classifier | 82.2 | 87.6 | 86.5 | 83.0 | ||||
| [78] 2020 | ResHist (RL Based 152-layer CNN) | BreaKHis | 86.4 | 87.3 | 91.4 | 86.3 | ||
| [64] 2020 | VGGNET16-RF | BreaKHis | 92.2 | 93.4 | 95.2 | 92.8 | ||
| VGGNET16-SVM | 94.1 | 95.1 | 97.0 | 93.4 | ||||
| [118] 2020 | CNN + spectral–spatial features | BreaKHis | Malignant | 97.6 | 97.4 | 97.3 | 97.0 | |
| [100] 2020 | NucTraL+BCF | BreaKHis | 96.9 | |||||
| [119] 2020 | ResNet50 + KWE LM | BreaKHis | Malignant | 88.4 | 87.1 | 90.0 | 84.1 | |
| [93] 2020 | AlexNet + SVM | BreaKHis | 84.1 | 87.5 | 89.4 | 85.2 | ||
| VGG16 + SVM | 86.4 | 87.8 | 86.8 | 84.4 | ||||
| VGG19+SVM | 86.6 | 88.1 | 85.8 | 81.7 | ||||
| GoogleNet + SVM | 81.0 | 84.5 | 82.5 | 79.8 | ||||
| ResNet18 + SVM | 84.0 | 84.3 | 82.5 | 79.8 | ||||
| ResNet50 + SVM | 87.7 | 87.8 | 90.1 | 83.7 | ||||
| ResNet101 + SVM | 86.4 | 88.9 | 90.1 | 83.2 | ||||
| ResNetInceptionV2 + SVM | 86.3 | 86.3 | 87.1 | 81.4 | ||||
| InceptionV3 + SVM | 85.8 | 84.7 | 86.8 | 82.9 | ||||
| SqueezeNet + SVM | 81.2 | 83.7 | 84.2 | 77.5 | ||||
| [120] 2020 | Optimized CNN | BreaKHis | 80.8 | 76.6 | 79.9 | 74.2 | ||
| [110] 2020 | InceptionV3 + BCNNs | BreaKHis | 95.7 | 94.7 | 94.8 | 94.5 | ||
| 96.1 | ||||||||
| [105] 2020 | VGG16 + SVM | BreaKHis | 78.6 | 85.2 | 82.0 | 79.6 | ||
| VGG19 + SVM | 77.3 | 79.1 | 83.0 | 79.1 | ||||
| Xception + SVM | 81.6 | 82.9 | 78.4 | 76.1 | ||||
| ResNet50 + SVM | 86.4 | 86.0 | 84.3 | 82.9 | ||||
| VGG16 + LR | 78.8 | 85.2 | 81.2 | 79.1 | ||||
| VGG19 + LR | 77.6 | 82.4 | 82.2 | 77.8 | ||||
| Xception + LR | 82.4 | 79.6 | 79.4 | 83.1 | ||||
| ResNet50 + LR | 83.1 | 86.7 | 84.0 | 80.1 | ||||
| [107] 2020 | Shearlet-based features | BreaKHis | 89.4 | 88.0 | 86.0 | 83.0 | ||
| Histogram-based features. | 92.6 | 93.9 | 95.0 | 94.7 | ||||
| Concatenating all features | 98.2 | 97.2 | 97.8 | 97.3 | ||||
| [104] 2021 | MA-MIDN | BreaKHis | 96.3 | 95.7 | 97.0 | 95.4 | ||
| [108] 2021 | AhoNet (Resnet18 + ECA + MPN-COV) | BreaKHis | 97.5 | 97.3 | 99.2 | 97.1 | ||
| [109] 2021 | 3PCNNB-Net | BreaKHis | 92.3 | 93.1 | 97.0 | 92.1 | ||
| [121] 2021 | APVEC | BreaKHis | 92.1 | 90.2 | 95.0 | 92.8 | ||
| [111] 2021 | Stochastic Dilated Residual Ghost Model | BreaKHis | 98.4 | 98.4 | 96.3 | 97.4 | ||
| [105] 2021 | Transfer Learning via Fine-tuning Strategy | BreaKHis | 99.3 | 99.0 | 98.1 | 98.8 | ||
| 98.4 | ||||||||
| [122] 2021 | BCHisto-Net | BreaKHis | 100× Magnification | 89 | ||||
| Ours | DEEP_Pachi | BreaKHis | 99.8 | 99.8 | 99.8 | 1.0 | 99.8 | |
| Ref/Year | Approach | Data Type | Accuracy (%) |
|---|---|---|---|
| [18] 2018 | DCNN + SVM | BACH | 77.8 |
| [123] 2018 | Pre-trained VGG-16 | BACH | 83.0 |
| Ensemble of three DCNNs | 87.0 | ||
| [124] 2018 | Ensemble (DenseNet 169 + Denseness 201 + ResNet 34) | BACH | 90.0 |
| [20] 2019 | All Patches in One Decision | BACH | 90% 92.5 |
| [125] 2019 | Ensemble (DenseNet 161+ ResNet 152 + ResNet 101) | BACH | 91.8 |
| [126] 2020 | Hybrid Features + SVM | BACH | 92.2 |
| Hybrid Features + MLP | 85.2 | ||
| Hybrid Features + RF | 80.2 | ||
| Hybrid Features + XGBoost | 82.7 | ||
| [87] 2020 | Attention Guided CNN | BACH | 93.0 |
| [99] 2020 | Random Forest | BACH | 91.2 |
| SVM | 95.0 | ||
| XGBoost | 42.5 | ||
| MLP | 91.0 | ||
| [104] 2021 | MA-MIDN | BACH | 93.57 |
| [108] 2021 | AhoNet (Resnet18 + ECA + MPN-COV) | BACH | 85.0 |
| [101] 2021 | Inception V3 + XGBoost | BACH | 87.0 |
| [127] 2022 | DSAGu-CNN | BACH | 96.47 |
| Ours | DEEP_Pachi | BACH | 99.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ukwuoma, C.C.; Hossain, M.A.; Jackson, J.K.; Nneji, G.U.; Monday, H.N.; Qin, Z. Multi-Classification of Breast Cancer Lesions in Histopathological Images Using DEEP_Pachi: Multiple Self-Attention Head. Diagnostics 2022, 12, 1152. https://doi.org/10.3390/diagnostics12051152
Ukwuoma CC, Hossain MA, Jackson JK, Nneji GU, Monday HN, Qin Z. Multi-Classification of Breast Cancer Lesions in Histopathological Images Using DEEP_Pachi: Multiple Self-Attention Head. Diagnostics. 2022; 12(5):1152. https://doi.org/10.3390/diagnostics12051152
Chicago/Turabian StyleUkwuoma, Chiagoziem C., Md Altab Hossain, Jehoiada K. Jackson, Grace U. Nneji, Happy N. Monday, and Zhiguang Qin. 2022. "Multi-Classification of Breast Cancer Lesions in Histopathological Images Using DEEP_Pachi: Multiple Self-Attention Head" Diagnostics 12, no. 5: 1152. https://doi.org/10.3390/diagnostics12051152
APA StyleUkwuoma, C. C., Hossain, M. A., Jackson, J. K., Nneji, G. U., Monday, H. N., & Qin, Z. (2022). Multi-Classification of Breast Cancer Lesions in Histopathological Images Using DEEP_Pachi: Multiple Self-Attention Head. Diagnostics, 12(5), 1152. https://doi.org/10.3390/diagnostics12051152









