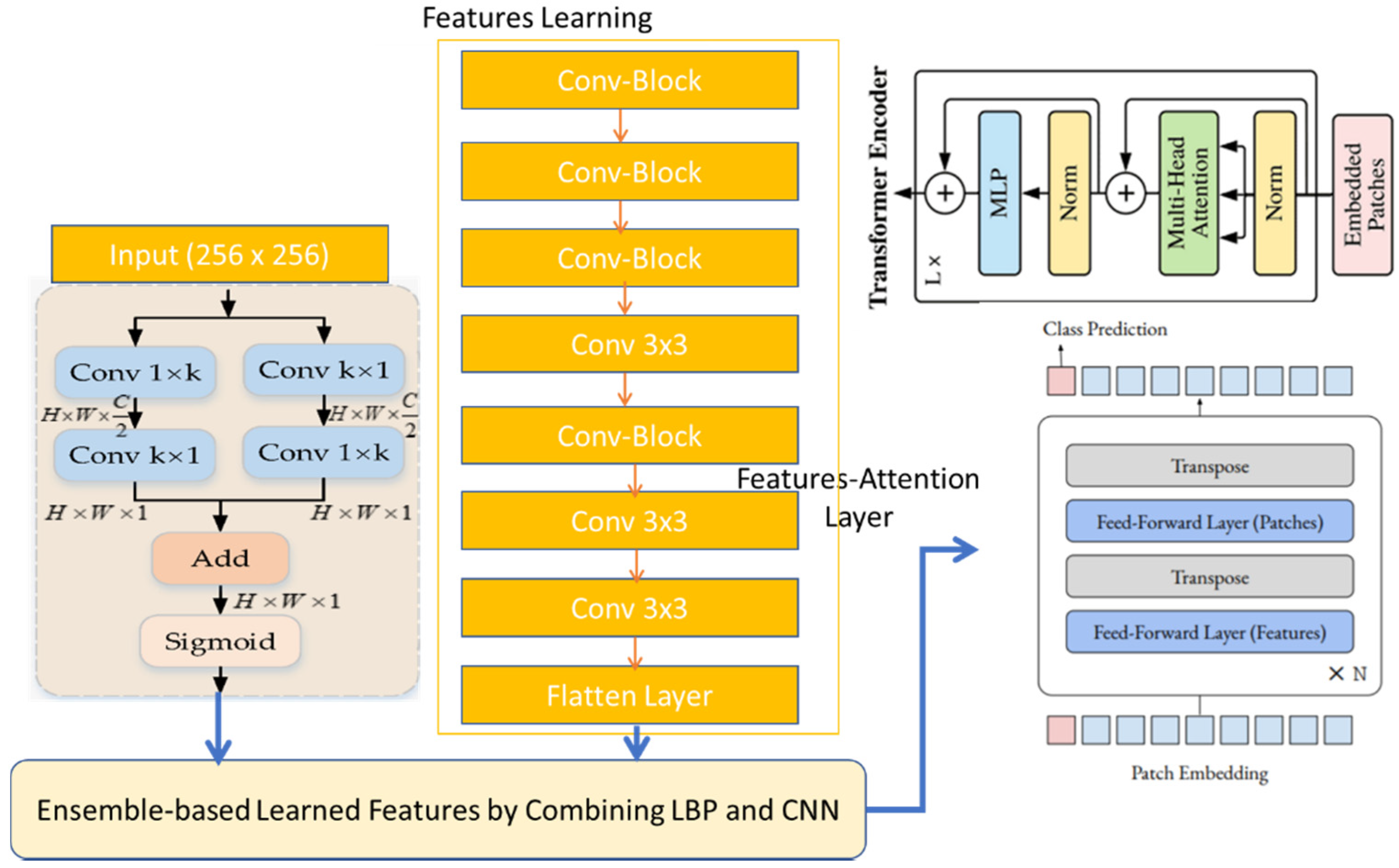
PCG signals frequently experience noise from sources, such as lung noises, power frequency interference, electromagnetic interference from the environment, and interference from electrical signals with human body signals. The diagnosis of PCG recordings is made difficult, if not impossible, by these diverse noise components. To remove such noises, we employed a multi-resolution and windowing technique, by using the continuous wavelet transform (CWT) along with a collection of high-pass and low-pass filters, which exhibited exceptional performance in signal denoising. To test and train the proposed system, we used 2400 PCG sound signals, which were doubled by using data augmentation techniques. Afterwards, the time–frequency content of PCG signals was represented most clearly among the three time–frequency representations (short-time Fourier transform (STFT)) to transfer 1D signals into 2D spectrograms. We applied the characteristics of convolutional neural networks (CNNs) to the Vit architecture [
44] to recognize five classes of CVD diseases, while retaining the benefits of transformers. A machine with 32 GB of RAM and an Intel Core i7 6700K processor was used for the experiments. The Keras platform and TensorFlow backend, powered by GPUs, were used by the suggested deep learning model.
4.1. Performance Evalaution
Metrics were used to measure the proposed solutions and the existing solutions. The first measurement was
Accuracy (ACC). This was the metric used to evaluate the classification models, a way of measuring how well the algorithm classified the data. Optimizing the model’s accuracy lowered the cost of errors, which can be huge. In addition, it could be calculated as follows:
True Positive (
TP) and True Negative (
TN) showed that the algorithm correctly predicted the data, either true or false. As for false, a False Positive (
FP) and False Negative (
FN) showed that the algorithm predicted the data incorrectly, whether true or false. The second measurement was Sensitivity (SE), which was defined as the percent of actual positive instances that were virtually predicted as positive, indicating that there was a percent of actual positive instances that were misclassified as negative. It is worth noting that this measurement implied that a low
FN rate almost always accompanied a high recall. Sensitivity is also known as Recall, True Positive Rate and Hit Rate. The following formula can be used to compute it:
The third measurement was
Specificity (SP), defined as the percent of true negative to total negative in the data. The formula for this measurement is as follows:
The fourth measurement was
Precision, which refers to the ability to correctly identify positive categories among entire expected positive classes, as measured by the proportion of all correctly predicted positive categories to all accurately expected positive categories. Mathematically, it can be represented as follows:
The fifth measurement was the
F1-
Score. This metric is important for determining the exactness and robustness of a classifier. The
F1-
Score is a critical metric for performance assessment that considers both recall and precision. It can be calculated as follows:
4.4. Computational Analysis
To measure the performance of the proposed CVT-Trans system, we computed the running time. To show that the suggested technique was a computationally efficient model, it was developed and trained on a GPU-based system, rather than a CPU-based system. For all datasets, the durations for STFT calculation, CVT with time domain input, and CVT-Trans with frequency input were computed. On average, this step took 0.4 Ms. Overall, a CWT spectrogram transformed steps from original 1-D PCG signals taking, on average, 1.2 MS. This point-of-view is visually explained in
Figure 5a. An attention-based CVT transformer architecture was created in this study to categorize the spectrogram into five groups. On average, this step took 1.2 MS. The key benefits of the proposed technique were quick classification and STFT computation, excellent accuracy acquired by utilizing all datasets, and a minimal number of layers. The original TL models contain more parameters and FLOPs (given in
Figure 5b), compared to the suggested CVT-Trans model.
The outcome was a compact model that required fewer computational support systems for implementation. Training time complexity = O (n × m × d), where, the parameter n is the input dimension, d is the batch size, and m is the output dimension. In general, the proposed CVT-Trans model took linear running time calculated as O (n × m × d). By utilizing tensor processing units (TPUs), which were offered by the Google cloud, this time complexity could be further decreased. In actual use, the TPUs significantly increased DL model speeds while using less power. This viewpoint will also be covered in further studies.
4.5. Results Analysis
The authors’ approaches only considered a single representation of heart sound signals, a spectrogram from the preprocessed heart signals that were captured through PCG signals. The time–frequency domain image of a sound signal is called a spectrogram. The inputted heart sound waves were transformed into the appropriate spectrogram before being further categorized, using transfer learning models, into five categories (
Table 2).
Table 3 lists the various approaches that do not include transfer learning. The various classifications of cardiac disorders demonstrate the accuracy of alternative methods in comparison to transfer learning models. On the same database, these various methodologies were compared against one another. However, the outcome of
Table 4 demonstrates that the accuracy was up-to-the-mark.
Figure 6 displays the accuracy with loss of the proposed training and testing in the enriched-PCG and the original-PCG databases, respectively, for one transfer learning model. The 10-fold cross-validation method was used to conduct the experiment.
Table 3 shows these measures for each fold, including training and testing samples: training accuracy (Acc), testing accuracy (Val Acc), training loss (Loss), and testing loss (Val Loss). The results of 10-fold cross-validation on the expanded-PCG database using data augmentation are shown in
Table 3. The results of 10-fold cross-validation on the original database are shown in
Table 4, using two categories. The categorization of five classes using the proposed approach, both with supplemented data training and with original data training, achieved an average accuracy of 98%, as shown in
Table 3 and
Table 4. The PCG database’s results demonstrated the effectiveness of this approach. We additionally assessed the effects of the PCG augmentation technique using additional background deformation.
A machine with 32 GB of RAM and an Intel Core i7 6700K processor was used for the experiments. The Keras platform and TensorFlow backend, powered by GPUs, were used by the suggested deep learning model. The efficacy of the suggested strategy was tested in a few trials. The 10-fold cross-validation technique, which reduced the variance of the estimate for the classifiers, was used to evaluate the classification outcomes. The training and testing portions of the fragmented data were separated. The data set was split into ten subsets for the 10-fold cross-validation, making sure that each subset contained the same number of observations with a certain categorical value. The other night subgroups were combined to create a training set each time, with one of the 10 subsets being used as the test set. As a result, each fold was utilized ten times for training purposes and once for testing purposes. The average of the 10 implementations was the outcome. It confirmed whether the suggested method outperformed others.
Table 2 demonstrates the results obtained by the proposed CVT-Trans system to classify PCG signals into five classes.
Different strategies were employed in the past to classify PCG signals into AS, MR, MS, MVP, and N categories by using machine learning, feature engineering, and deep learning techniques. However, a thorough investigation of the efficacy of classification across huge datasets was not considered. Therefore, it was more desirable to determine if the suggested technique could classify PCG signals that contained various combinations of datasets. Seven distinct classification examples were offered from the datasets to handle this issue.
Table 4 tests all centers on identifying normal and abnormal PCG signals by using a 10-fold cross-validation test. We achieved the best classification results for two classes of normal and abnormal PCG signals by using data augmentation techniques.
Figure 7 and
Figure 8 show the categorization outcomes for five cases using a 10-fold cross-validation methodology. The classification results obtained by the classifier for binary and multi-class classification problems were thoroughly distributed in a confusion matrix. The 10-fold cross-validation’s overall confusion matrices are illustrated in
Figure 7 and
Figure 8. As shown in
Table 4 and
Table 5, our study showed that accuracy increased when distinguishing between binary and multi-class PCG signals. Overall classification accuracy for the experiment’s five binary classification cases was 100%, 99.00%, 99.75%, 99.75%, and 99.60%, respectively. The robustness of our model was verified and had the benefit of discriminating features. In addition, the features were automatically retrieved through the CVT-Trans model, which provided a low misclassification rate.
Comparisons were also performed using state-of-the-art approaches, such as those of Wang RNN et al. [
17] and Cheng RF et al. [
19]. The authors [
17] used LSTM and RNN deep learning models with the CWT-spectrogram technique to recognize cardiovascular disease categories. Whereas in [
19] the authors used different heart sound features to recognize two classes of PCG signals, we selected [
17,
19] because they were easy to implement, compared to other systems.
Figure 8 demonstrates the results obtained by these two techniques compared to the proposed CVT-Trans system. The proposed CVT-Trans system outperformed both techniques.
Additionally, for both the four-class and five-class classifications, we were able to obtain a total classification accuracy of 98.87% and 98.10%, respectively. In comparison to cutting-edge methods, the proposed (PRS) system could effectively distinguish between various classes of PCG signals. In another experiment, we compared recent studies, such as those of Z.H. Wang et al. [
17], and A.M. Alqudah [
18], as shown in
Table 5. The obtained result demonstrated that our CVT-Trans outperformed the rest.
Several studies with comparable data sizes to ours have achieved results that were less accurate than ours. Our model had slightly better accuracy, sensitivity, and specificity values. Moreover, the past studies developed mostly two-classes of PCG signals by using hand-crafted features. We employed five-classes of heart sound classification, and the features were not manually extracted. In our work discriminating characteristics were automatically picked up by the model, rather than the more laborious process of retrieving and selecting features manually. Additionally, our model classified each sample in less than a millisecond. Our approach was, thus, quicker and more suited for quick diagnosis. Our large data size also required 10-fold cross-validation, whereas 5-fold cross-validation was employed in previous studies. The size difference between the training set and resampling subsets became smaller with a bigger k value of 10, which lowered the bias of our suggested method. This demonstrated that our proposed strategy was more dependable. The confusion matrix revealed that each class’s misclassification rates were less than 3%. This low misclassification rate attested to the model’s great accuracy.
The suggested CWT spectral loss may be used to train a high-quality model, according to experimental data. It is difficult to balance the time and frequency (TF) resolutions in STFT, since they depend on the frameshift and analysis window design. The CWT may be used to do time–frequency (TF) analysis at various temporal and frequency resolutions. It is also feasible to modify the CWT’s time–frequency analysis in order to take into account scales resembling those used in human vision. In this study, employing both STFT and CWT spectral losses, we examined the training of high-quality features from waveform models. CWT can consider temporal and frequency resolutions that are closer to human perception scales than in STFT. The nonlinear nature of PCG signals cannot be adequately analyzed by the methods now in use in any of these fields. This study, in which the continuous wavelet transform (CWT), and STFT spectrogram methods were employed together to analyze nonlinear and non-stationary aspects of the PCG signals, overcomes these restrictions. To find time–frequency maps, two prominent techniques were used: the short-time Fourier transform (STFT) and the continuous wavelet transform (CWT). This illustration demonstrated how the CWT analyzed signals concurrently in time and frequency, as seen in
Figure 9. The example demonstrated how the CWT worked better than the short-time Fourier transform (STFT) when localizing transients. The example also demonstrated how to approximate time–frequency localized signals using the inverse CWT. The phase information of the signal under analysis was not provided by the CWT. In this paper, we used the CWT and STFT algorithms with the intention of comparing them to earlier STFT findings. The scale factor and the mother wavelet selection served as adjustment parameters. When the settings were changed, a combination of CWT and STFT was used to automatically diagnose CVD disease. Finally, a comparison between STFT and CWT was suggested by considering the precision of detection and time processing.
Figure 9 illustrates a time–frequency map made possible by CWT and STFT spectrograms.
Figure 10 shows the comparisons of different CNN-based pretrained models with the proposed CVT-Trans technique for classification of five categories of cardiovascular disease.
Figure 10a shows the testing/validation/training loss, and
Figure 10b represents the performance measures with computation time. This figure shows the efficiency of the proposed CVT-Trans model. The relationship between the true positive rate (TPR), which is known as sensitivity, and the false positive rate (FPR), which is known as 100% specificity for various classifications, is explained by the receiver operating characteristic (ROC) curve. The total area under the ROC curve (AUC) can be used to assess how well the categorization performed. In this work, the AUC was determined for each iteration and provided as a complete quantitative evaluation of classification performance using 10-fold cross-validation. The ROC curves and associated AUC for all seven instances are shown in
Figure 11 to better clarify the 1D CNN classifier’s performance. Overall, our classification strategy performed well. The suggested pattern recognition system (PRS) could successfully distinguish between various PCG signal classes using CWT, spectrogram, and CVT techniques.
We also performed experiments to show the importance of the proposed CVT-Trans system compared to state-of-the-art approaches, such as Cheng-FRED [
19], Rath-RF-MFO-XGB [
20], Li-PCA-TSVM [
21], Khan-ANN-LSTM [
22], Saputra-NN-PSO [
24], and Arsalan-RF [
25], in terms of SE, SP, F1-score, RL, PR, and ACC measures. The standard hyper-parameters were defined, as presented in the corresponding studies. Firstly, the comparisons were performed between the proposed method and state-of-the-art techniques by using data augmentation and a 10-fold cross-validation test, in terms of two stages of CVD disease, as shown in
Table 6. As displayed in
Table 6, the Khan-ANN-LSTM [
22] system obtained good classification results (SE of 0.88, SP of 0.87, F1-score of 0.88, RL of 85%, PR of 88%, and ACC of 86%) for CVD heart disease, compared to the other approaches. However, the proposed CVT-Trans method achieved 100% ACC. Secondly, we also measured the performance of the CVT-Trans system in terms of the recognition of the five stages of CVD heart disease. The results are mentioned in
Table 7. As mentioned in
Table 7, the proposed CVT-Trans system outperformed (SE of 0.99, SP of 0.98, F1-score of 0.99, RL of 98%, PR of 98%, and ACC of 100%) the state-of-the-art approaches. The good results obtained were because we developed a convolutional vision transformer (CVT) architecture based on local and global attention mechanisms in a continuous wavelet transform-based spectrogram (CWTS) strategy. The developed strategy was effective and efficient compared to many state-of-the-art systems.