1. Introduction
In human DNA sequences, change due to certain reasons is called a mutation [
1,
2]. Mutations can be good or bad and are used by scientists to study human health, body cell development, etc. Health scientists already have conducted a lot of work to identify mutations in human beings [
3], because this identification could work as a foundation for personalized medicine [
4]. Furthermore, genetic engineering can play an important role to prevent disease, as well as making an early diagnosis to control the death rate [
5,
6].
Gene mutations play a vital role in cancerous cell generation [
7], while genetic engineering is capable to predict deadly diseases such as cancer even before the symptoms of the disease are produced in the human body [
8,
9]. For this purpose, many techniques are available, but the traditional method used is lab evaluation, which is time-consuming, as well as expensive [
10,
11]. Cancer is a disease that can affect all the different body systems and tissues in human beings. Malignancy of cancer has more than 100 types and, back in 2010 alone, cancer was responsible for 1 out of 8 deaths worldwide [
12]. Kidney malignancy is one of the different types of malignant cancers, but it is not a single disease, actually, it is a combination of many types, for example, clear cell, type 1 papillary, type 2 papillary, chromophobe, TFE3, TFEB, and oncocytoma [
13].
Among all these types, the prevalence of renal cell carcinoma (RCC) or kidney cancer is all over the world, but most of the cases are reported in North America. Gender and age play an important role in the occurrence of the disease, as males above 65 years old are considered to be more prone to this disease, and obesity is also one of the main causes of the disease [
14,
15]. It initially starts from the outer walls of the kidney [
16]. Based on some specific characteristics, RCC is divided into three main types of RCC. Clear cell RCCs—these are the most usual type of renal cell carcinoma. Researcher call them clear cells because most of the time cells within the tumor are clear. Papillary cell RCCs—these are the second most common type of kidney cancer. Papillary cells are distinguished by small, rounded protuberances on their surface. These tumors present as either Type 1 or the more aggressive Type 2 forms. Chromophobe cell RCCs—these are the third most common form of kidney malignancy. Scientists call them chromophobe because these cells do not acquire colored stains easily [
17].
Various studies have been reported previously which targeted the prediction of cancer-driver gene mutations. Luo, in 2019, proposed deepdriver, which used a deep convolution neural network approach for cancer driver mutation prediction. The proposed method can predict breast and colorectal cancer. The AUC scores of deepdriver on cancer and colorectal cancer are 0.98 and 0.97, respectively [
18]. In 2018, Wand et al. proposed a Bayesian hierarchical modeling algorithm for cancer driver mutation prediction named rDriver. They examined 3080 samples of 8 different kinds of cancer, in which the rDriver predicted 1389 affected samples. The evaluation process of the rDriver predictions method is conducted by using engineered cell line models and gives good results. The value results are a positive predictive value of 0.94 in PIK3CA genes [
19].
In 2017, Pi-Jung et al. proposed a CNV method for cancer driver mutation prediction. For simulation and results, they used four TCGA datasets (BRCA, HNSC, KIRC, and THCA). They covered breast, head, neck, thyroid, and kidney cancer genes. They also discovered rare driver genes in their work. They did all the work with the help of gene sequence length [
20]. A driver mutation provided growth advantages to the affected tumor cell. In 2013, Mao et. Al. introduced the candrA tool for the prediction of cancer driver mutations. The proposed method was based on a set of 95 structural and evolutionary features by using 10 functional prediction algorithms such as CHASM, SIFT, and Mutation Assessor. They used two mutation datasets, GBM and OVC [
21].
The majority of the work carried out for the identification of RCCC driver genes uses experimental lab procedures, and those who used computational approaches lacked in performance due to fewer data, such as Kocak et al. [
22]. The focus of their study was only on the
PBRM1 gene, and the sample size for training was also not good. Thus, to address this problem, the present study aims to propose a prediction model for renal clear cell carcinoma mutations in gene sequences using machine learning algorithms. We curated a comparatively large dataset from the IntOgen and NCBI databases, and after meticulous and thorough feature extraction, we trained different machine learning classifiers. After an exhaustive evaluation of machine learning classifiers, the best-performing classifier was selected as the final model and was compared with existing methods. The proposed method is easy to use, efficient and accurate for obtaining results, scientists and researchers only need sequence information and can avoid hectic experimentations. To do so, the intended clinical use is that scientists, researchers, and others in the clinical community can opt for a system developed based on the proposed method and obtain results by inputting the gene sequence. The sequence could be from human biological samples. The system will help them classify it as a potential RCCC gene or not.
3. Results
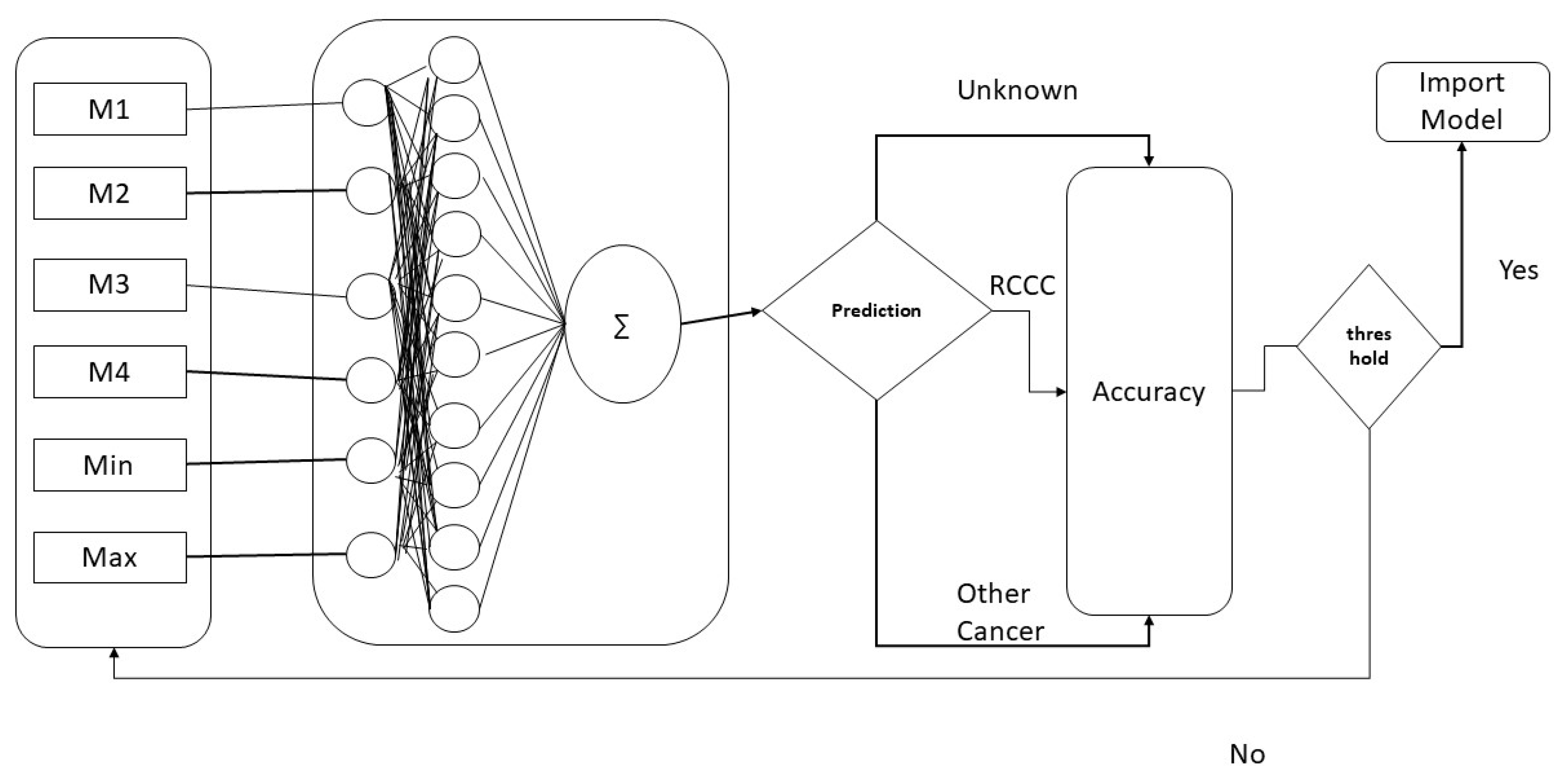
In the proposed method, after data collection, the first step in the pre-processing layer was to perform the mutation and save the results. Later on, for all sequential data, features were computed and fed to machine learning classifiers for evaluation. For evaluating the training accuracy, self-consistency testing was performed in which the same training and testing data were used. To evaluate and validate the outcome of the prediction model, the proposed system was tested by using three different techniques, i.e., independent testing, K-cross-validation testing, and Jackknife testing.
3.1. Training Accuracy
The training accuracy was evaluated using self-consistency testing [
44]. For this purpose, the same training and testing data were used. The evaluation scores are shown in
Table 2.
Table 2 illustrate that the model was trained accurately, identifying all positive/negative samples correctly.
3.2. Validation of the Model through 10-Fold Cross-Validation and Jackknife Testing
The validation of results was performed using various evaluation metrics such as accuracy, specificity, sensitivity, and Matthews Correlation Coefficient. The test methods adapted were independent testing, K-cross-validation testing, and Jackknife testing [
3].
In case of the unavailability of a separate test dataset, the best approach to test any predictive model is k-fold cross-validation [
45]. Using k-fold cross-validation, the dataset is split into k-disjoint folds, and the model is validated k-times. In each iteration, k-1 folds are chosen for the training model, while the remaining 1-fold is used for testing. This testing fold is chosen separately in each iteration [
46]. For the evaluation of the proposed model, the value of k was chosen as 10. The overall mean accuracy score is 0.98, and scores of the remaining measures, as well as for all folds, are shown in
Table 3. The box plot graph is shown in
Figure 3, while the ROC curves for all classes using 10-fold cross-validation are shown in
Figure 4.
To further elaborate on the 10-fold cross-validation, training and validation loss for each fold is shown in
Figure 5.
In
Figure 5, the curves are plotted between the number of iterations and losses. The neural network was trained till convergence by using an early stopping criterion, while max iterations were set as 3000. However, it was observed that for all 10 iterations of k-fold, the model converged on an average of 20–30 epochs. This is the reason that, after such epochs, we can observe that the loss curves converged at a point and turned into straight lines.
For further exhaustive validation of the model, the Jackknife test was opted for. The Jackknife test is also referred to as Leave-One-Out cross-validation, and works on the same principle as k-fold cross-validation, with the value of k = the number of samples in the dataset [
2]. It is known to be the least arbitrary method which can yield unique output for a given benchmark. After testing, the accuracy metrics were computed to evaluate the quality of the proposed algorithm. The results for Jackknife validation are shown in
Table 4, while the ROC curve is shown in
Figure 6.
Using the scores of Jackknife validation, RCCC_Pred was compared with a few existing methods. The results are shown in
Table 5.
Herein, we compared the results of Jackknife testing of the present study with this method and observed a lack of performance. It could be observed that the proposed method outperformed the existing method by Kocak et al. [
22] in terms of all accuracy measures.
3.3. Independent Dataset Validation
For any new predictor, it is of great importance to test its ability to predict against unknown data. Here, unknown data are referred to as the data which the model has not seen or observed during the training process [
1,
2,
39]. Therefore, keeping in view the importance of this test, it was performed for the evaluation of the model proposed in the present study. As the whole dataset was created manually in this study, all possible data samples available at that time were already retrieved. Here, we trained the model from scratch using 70% of the data as described in the Methods while testing it for the remaining 30% of samples. The scores were computed along with the ROC curve and are represented in
Table 6 and
Figure 7, respectively.
3.4. Comparison with Other Classifiers
Besides comparing the proposed method with previously existing methods, we have also run experiments to compare the performance of our method with different other classifiers. The benchmark dataset of the proposed method is also trained with other machine learning algorithms, but Hist Gradient boosting shows the best results. The details of other proposed models are shown in
Table 7.
Table 7 shows the accuracy scores for three different kinds of tests, which are Jackknife, independent, and cross-validation, while these tests are performed using five classifiers. To further elaborate on performance, the ROC curve for all five classifiers for the independent dataset testing is added in
Figure 8.
The decision tree showed better performance in independent dataset testing, however, overall, the performance of Hist Gradient boosting was better as compared with the other classifiers. Based on these results, the Hist Gradient Boosting was considered as the final model for the proposed RCCC_Pred classifier.
4. Discussion
Herein, we proposed a prediction model named RCCC_Pred for renal clear cell carcinoma mutations in gene sequences using machine learning algorithms. We curated a dataset from IntOgen and NCBI databases and, after meticulous feature extraction, we trained different machine learning classifiers. After a thorough evaluation, the best-performing classifier was selected as the final model and was compared with existing methods. The decision tree showed better performance in independent dataset testing, however, overall, the performance of Hist Gradient boosting outperformed other classifiers. Based on these results, the Hist Gradient boosting was considered as the final model for the proposed RCCC_Pred classifier.
Previously, a few studies were proposed for renal clear cell carcinoma or other cancers [
47]. A few machine-learning-based and experimental approaches have been proposed to study multicellular complexity and tissue specificity [
48], as well as to study molecular interactions in cancer [
49]. The majority of the work conducted for the identification of RCCC driver genes uses experimental lab procedures. A few research studies used AI-based approaches to predict cancer driver mutations, but their methods used a very limited amount of data. In a previous study reported by Kocak et al. [
22], the researchers proposed a machine-learning-based algorithm for kidney cancer prediction at the level of the gene. The sample set for the machine learning consists of 161 label examples of augmented data, from which 74 mutations are recorded in the PBRM1 gene, and the other 87 occurred outside the PBRM1 gene. The focus of the study is only on one gene code, which is
PBRM1, and the sample size for training is also not good. Due to the tiny dataset, the possibility of overfitting occurrence is very high, which affects the accuracy of the system.
In 2020, Kocak et al. [
50] further extended their work by considering BAP1 mutation in clear cell renal cell carcinoma. However, again, the dataset was limited, comprising only 65 samples. Here, authors used a Random Forest classifier and correctly classified samples with 84.6% accuracy and an area under the curve of only 0.897. For similar BAP1 mutation status with 54 samples, Feng et al. [
51] reported an accuracy of 83% for Jackknife testing using Random Forest. Using image data for clear cell renal cell carcinoma, Acosta et al. proposed a deep-learning-based method for analyzing intertumoral heterogeneity and considered the three most frequently mutated genes, which were BAP1, PBRM1, and SETD2. Overall, the authors achieved an area under the receiver operating characteristic curve of around 0.89.
By considering the importance of clear cell renal cell carcinoma, Chen et al. [
52] proposed a deep learning algorithm for the prediction of prognosis and immunotherapeutic response. The authors used data from 3 different cohorts, and samples were around 730 after performing pre-processing. After training the deep learning model for 100 epochs, the authors achieved a sensitivity of 0.71 and a specificity of 0.68.
The proposed method of the present study is trained on 10706 genes and 27685 instances, from which 1513 are other tumor drivers, 1272 are RCCC tumor drivers, and the rest are passenger gene mutation instances. The proposed system covers all the kidney genes and a huge number of tumor driver mutations, so it gives more accurate and reliable results after deployment.
5. Conclusions
The kidney is one of the vital organs in the human body, as it is responsible for blood cleaning, removing waste and poisonous substances from the blood, and balancing the electrolytes in the body. Kidney cancer is the most popular cancer in developing countries because of a lot of reasons, one of which is the huge amount of alcohol consumption. In this research work, a machine-learning-based efficient automated method is introduced for the prediction of kidney cancer before the development of kidney cancer. For this purpose, the proposed approach maintains the record of cancer driver mutations in the human body, and for this reason, the statistical position sensation calculation is performed. It then validates the approach with different types of testing techniques, which are Jackknife, independent dataset, and cross-validation. The Jackknife, cross-validation, and independent test accuracies of the system are 100%, 98%, and 83%, respectively.