Eplet-Predicted Antigens: An Attempt to Introduce Eplets into Unacceptable Antigen Determination and Calculated Panel-Reactive Antibody Calculation Facilitating Kidney Allocation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. HLA Typing
2.3. Anti-HLA Antibodies
2.4. Antibody-Verified Eplets
2.5. CPRA Calculation and Comparison
3. Results
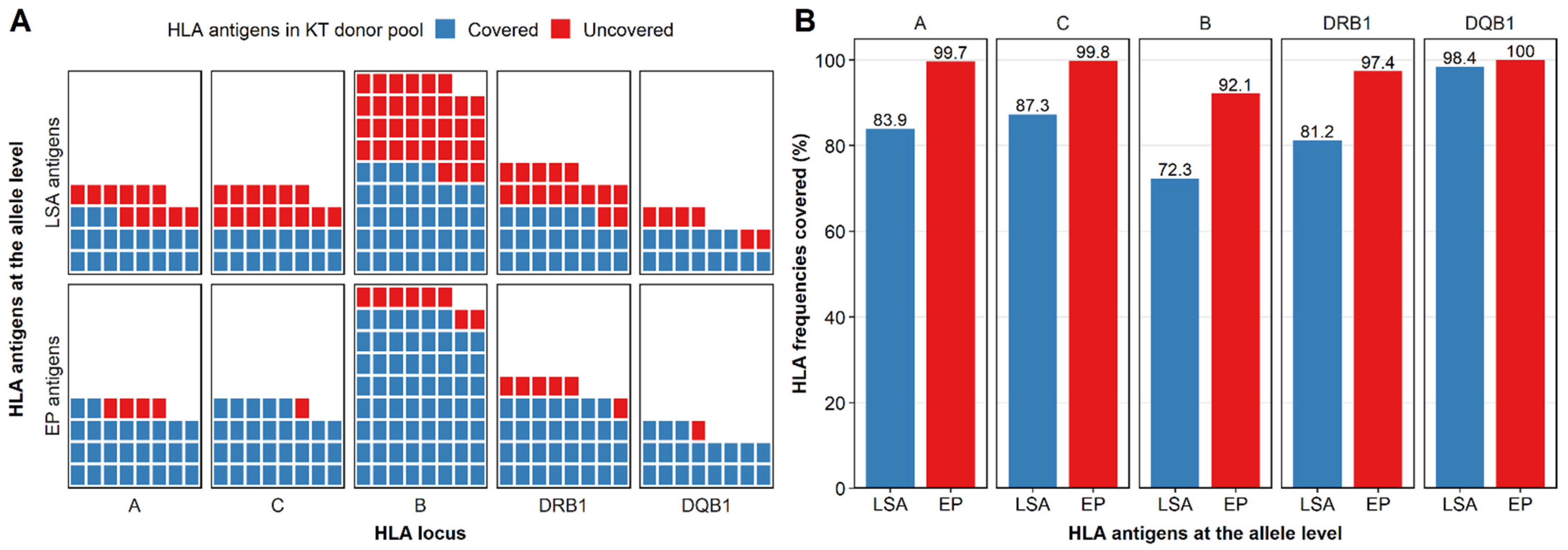
3.1. LSA Kits Covered Few HLA Antigens in the Donor Population
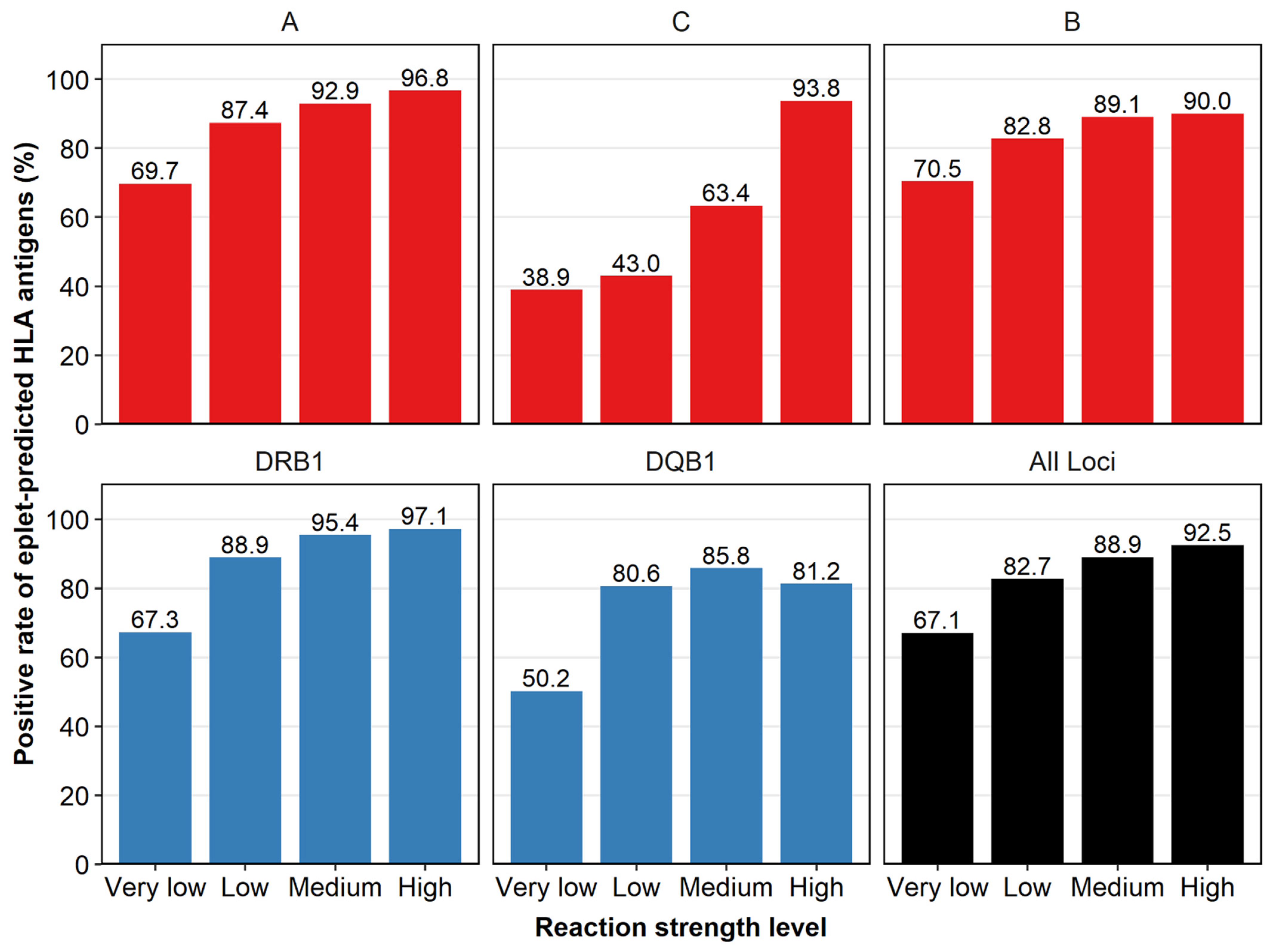
3.2. Reactivity of Non-LSA Antigens Could Be Predicted by LSA Antigens Based on Eplets
3.3. Reactivity Concordance Was Found between LSA Antigens and Their Eplet-Predicted Antigens
3.4. Eplet-Predicted HLA Antigens Covered More HLA Antigens than LSA Kits
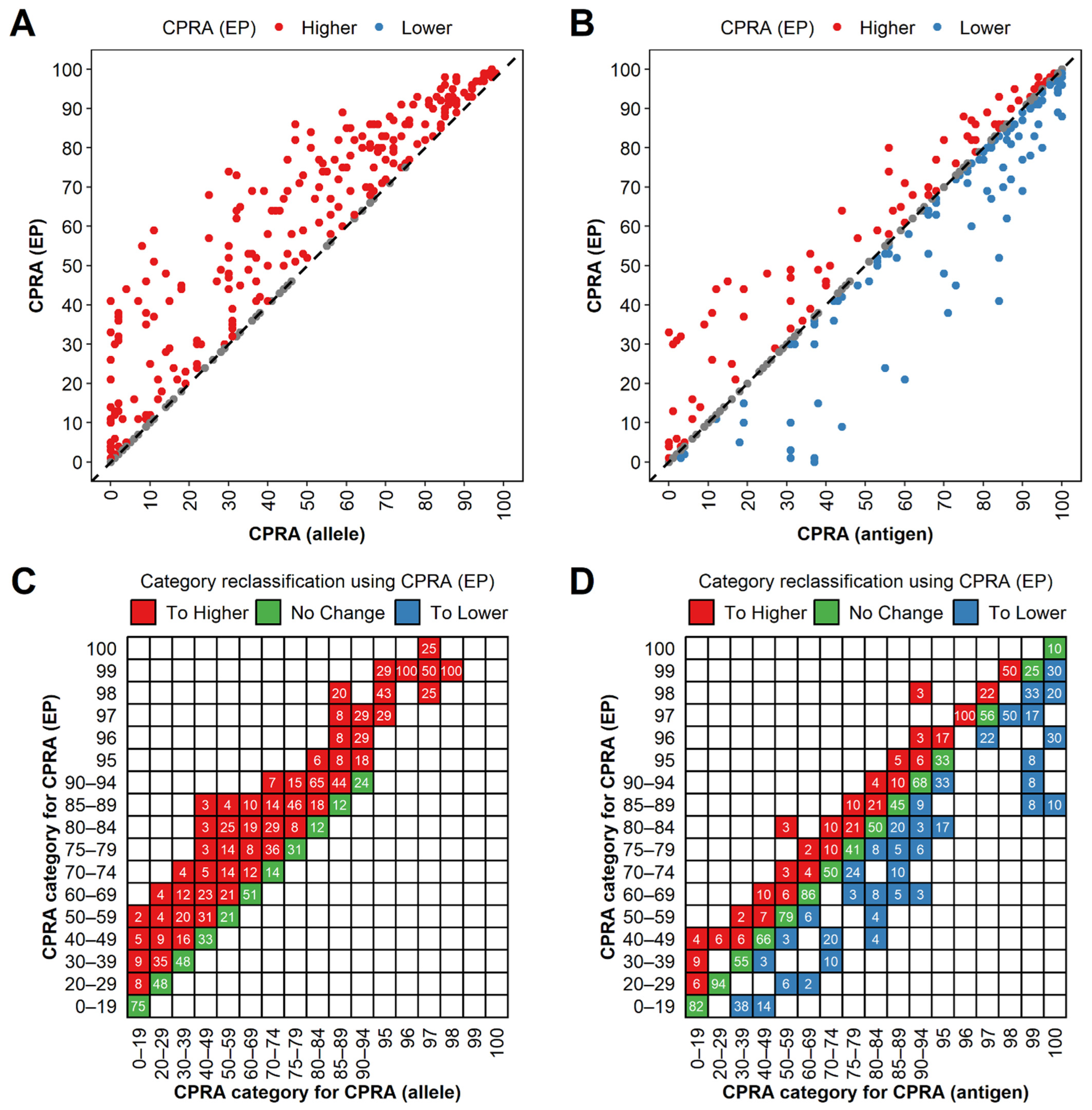
3.5. CPRA (Eplet-Predicted) Were Higher than CPRA (Allele) and Lower than CPRA (Antigen) Generally
3.6. Eplet-Predicted Antigens Facilitated UA Determination for Non-LSA Antigens and Avoided Acute Rejection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tonelli, M.; Wiebe, N.; Knoll, G.; Bello, A.; Browne, S.; Jadhav, D.; Klarenbach, S.; Gill, J. Systematic Review: Kidney Transplantation Compared With Dialysis in Clinically Relevant Outcomes. Am. J. Transplant. 2011, 11, 2093–2109. [Google Scholar] [CrossRef] [PubMed]
- Cecka, J.M. Calculated PRA (CPRA): The New Measure of Sensitization for Transplant Candidates: Sensitized Patients, PRA and CPRA. Am. J. Transplant. 2010, 10, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Kransdorf, E.P.; Pando, M.J.; Stewart, D.; Lindblad, K.; Bray, R.; Murphey, C.; Kaur, N.; Patel, J.K.; Kim, I.; Zhang, X.; et al. Stem cell donor HLA typing improves CPRA in kidney allocation. Am. J. Transplant. 2021, 21, 138–147. [Google Scholar] [CrossRef]
- Stewart, D.E.; Wilk, A.R.; Toll, A.E.; Harper, A.M.; Lehman, R.R.; Robinson, A.M.; Noreen, S.A.; Edwards, E.B.; Klassen, D.K. Measuring and monitoring equity in access to deceased donor kidney transplantation. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2018, 18, 1924–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duquesnoy, R.J.; Kamoun, M.; Baxter-Lowe, L.A.; Woodle, E.S.; Bray, R.A.; Claas, F.H.J.; Eckels, D.D.; Friedewald, J.J.; Fuggle, S.V.; Gebel, H.M.; et al. Should HLA Mismatch Acceptability for Sensitized Transplant Candidates Be Determined at the High-Resolution Rather Than the Antigen Level? Am. J. Transplant. 2015, 15, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Dinh, A.; Heron, S.; Gasiewski, A.; Kneib, C.; Mehler, H.; Mignogno, M.T.; Morlen, R.; Slavich, L.; Kentzel, E.; et al. Assessing the utilization of high-resolution 2-field HLA typing in solid organ transplantation. Am. J. Transplant. 2019, 19, 1955–1963. [Google Scholar] [CrossRef] [PubMed]
- Senev, A.; Emonds, M.; Van Sandt, V.; Lerut, E.; Coemans, M.; Sprangers, B.; Kuypers, D.; Naesens, M. Clinical importance of extended second field high-resolution HLA genotyping for kidney transplantation. Am. J. Transplant. 2020, 20, 3367–3378. [Google Scholar] [CrossRef]
- Picascia, A.; Grimaldi, V.; Napoli, C. From HLA typing to anti-HLA antibody detection and beyond: The road ahead. Transplant. Rev. 2016, 30, 187–194. [Google Scholar] [CrossRef]
- Cecka, J.M.; Reed, E.F.; Zachary, A.A. HLA high-resolution typing for sensitized patients: A solution in search of a problem? Am. J. Transplant. 2015, 15, 855–856. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Doss, S.A.; Stephen, S.; Pratheeba, M.; Jeyaseelan, L.; Daniel, D. The challenge of using the virtual crossmatch as a singular tool for the detection of Anti-HLA antibodies- A study from a tertiary care institute from South India. Transpl. Immunol. 2021, 65, 101349. [Google Scholar] [CrossRef] [PubMed]
- Pinelli, D.F.; Tambur, A.R. Virtual crossmatching for deceased donor transplantation: One size does not fit all. Kidney Int. 2020, 97, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Tambur, A.R.; Claas, F.H.J. HLA Epitopes as Viewed by Antibodies: What Is it All About? Am. J. Transplant. 2015, 15, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- Duquesnoy, R.J.; Askar, M. HLAMatchmaker: A Molecularly Based Algorithm for Histocompatibility Determination. V. Eplet Matching for HLA-DR, HLA-DQ, and HLA-DP. Hum. Immunol. 2007, 68, 12–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duquesnoy, R.J. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum. Immunol. 2006, 67, 847–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duquesnoy, R.J.; Marrari, M.; Sousa, L.C.D.D.M.; Barroso, J.R.P.D.M.; Aita, K.M.D.S.U.; da Silva, A.S.; Monte, S.J.H.D. Workshop report: A website for the antibody-defined HLA epitope registry. Int. J. Immunogenetics 2012, 40, 54–59. [Google Scholar] [CrossRef] [PubMed]
- US Department of Health and Human Services. OPTN: Organ Procurement and Transplantation Network. 2021. Available online: https://optn.transplant.hrsa.gov/policies-bylaws/policies (accessed on 1 October 2021).
- Chan, Y.P.; Wong, M.W.K.; Tang, L.W.M.; Guo, M.; Yang, W.; Ip, P.; Li, P.K.T.; Leung, C.B.; Chau, K.F.; Lam, J.C.K.; et al. A simplified method of calculating cPRA for kidney allocation application in Hong Kong: A retrospective study. Transpl. Int. 2017, 30, 1234–1242. [Google Scholar] [CrossRef] [Green Version]
- Huber, L.; Lachmann, N.; Niemann, M.; Naik, M.; Liefeldt, L.; Glander, P.; Schmidt, D.; Halleck, F.; Waiser, J.; Brakemeier, S.; et al. Pretransplant virtual PRA and long-term outcomes of kidney transplant recipients. Transpl. Int. 2015, 28, 710–719. [Google Scholar] [CrossRef]
- Kransdorf, E.P.; Kittleson, M.M.; Patel, J.K.; Pando, M.J.; Steidley, D.E.; Kobashigawa, J.A. Calculated panel-reactive antibody predicts outcomes on the heart transplant waiting list. J. Heart Lung Transplant. 2017, 36, 787–796. [Google Scholar] [CrossRef]
- Tague, L.K.; Witt, C.A.; Byers, D.E.; Yusen, R.D.; Aguilar, P.R.; Kulkarni, H.S.; Bain, K.B.; Fester, K.A.; Puri, V.; Kreisel, D.; et al. Association between Allosensitization and Waiting List Outcomes among Adult Lung Transplant Candidates in the United States. Ann. Am. Thorac. Soc. 2019, 16, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, M.; Heßler, N.; König, I.R.; Lachmann, N.; Dick, A.; Ditt, V.; Budde, K.; Reinke, P.; Eisenberger, U.; Suwelack, B.; et al. Unacceptable human leucocyte antigens for organ offers in the era of organ shortage: Influence on waiting time before kidney transplantation. Nephrol. Dial. Transplant. 2017, 32, 880–889. [Google Scholar] [CrossRef]
- Cecka, J.M.; Kucheryavaya, A.Y.; Reinsmoen, N.L.; Leffell, M.S. Calculated PRA: Initial Results Show Benefits for Sensitized Patients and a Reduction in Positive Crossmatches: Calculated PRA Benefits Sensitized Patients. Am. J. Transplant. 2011, 11, 719–724. [Google Scholar] [CrossRef]
- Debout, A.; Foucher, Y.; Trébern-Launay, K.; Legendre, C.; Kreis, H.; Mourad, G.; Garrigue, V.; Morelon, E.; Buron, F.; Rostaing, L.; et al. Each additional hour of cold ischemia time significantly increases the risk of graft failure and mortality following renal transplantation. Kidney Int. 2015, 87, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Ponticelli, C.E. The impact of cold ischemia time on renal transplant outcome. Kidney Int. 2015, 87, 272–275. [Google Scholar] [CrossRef] [Green Version]
- Keith, D.S. Parsing the 100% calculated panel reactive antibody kidney transplant candidates: Who gets transplanted? HLA 2020, 95, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Choi, J.; Toyoda, M.; Vo, A.; Peng, A.; Jordan, S.C. Desensitization: Overcoming the Immunologic Barriers to Transplantation. J. Immunol. Res. 2017, 2017, e6804678. [Google Scholar] [CrossRef] [PubMed]
- Caro-Oleas, J.L.; Gonzalez-Escribano, M.F.; Gonzalez-Roncero, F.M.; Acevedo-Calado, M.J.; Cabello-Chaves, V.; Gentil-Govantes, M.A.; Nunez-Roldan, A. Clinical relevance of HLA donor-specific antibodies detected by single antigen assay in kidney transplantation. Nephrol. Dial. Transplant. 2012, 27, 1231–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, B.H.; Choi, B.S.; Oh, E.J.; Park, C.W.; Kim, J.-I.; Moon, I.S.; Kim, Y.-S.; Yang, C.W. Clinical impact of the baseline donor-specific anti-human leukocyte antigen antibody measured by Luminex single antigen assay in living donor kidney transplant recipients after desensitization therapy. Transpl. Int. 2014, 27, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Bosanquet, J.P.; Witt, C.A.; Bemiss, B.C.; Byers, D.E.; Yusen, R.D.; Patterson, A.G.; Kreisel, D.; Mohanakumar, T.; Trulock, E.P.; Hachem, R.R. The impact of pre-transplant allosensitization on outcomes after lung transplantation. J. Heart Lung Transplant. 2015, 34, 1415–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tambur, A.R.; Audry, B.; Antoine, C.; Suberbielle, C.; Glotz, D.; Jacquelinet, C. Harnessing Scientific and Technological Advances to Improve Equity in Kidney Allocation Policies. Am. J. Transplant. 2017, 17, 3149–3158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duquesnoy, R.J.; Gebel, H.M.; Woodle, E.S.; Nickerson, P.; Baxter-Lowe, L.A.; Bray, R.A.; Claas, F.H.J.; Eckels, D.D.; Friedewald, J.J.; Fuggle, S.V.; et al. High-Resolution HLA Typing for Sensitized Patients: Advances in Medicine and Science Require Us to Challenge Existing Paradigms. Am. J. Transplant. 2015, 15, 2780–2781. [Google Scholar] [CrossRef]
- Zavyalova, D.; Abraha, J.; Rao, P.; Morris, G.P. Incidence and impact of allele-specific anti-HLA antibodies and high-resolution HLA genotyping on assessing immunologic compatibility. Hum. Immunol. 2021, 82, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Sypek, M.; Kausman, J.; Holt, S.; Hughes, P. HLA Epitope Matching in Kidney Transplantation: An Overview for the General Nephrologist. Am. J. Kidney Dis. 2018, 71, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Duquesnoy, R.J.; Marrari, M.; Mulder, A.; Sousa, L.C.D.D.M.; Da Silva, A.S.; Monte, S.J.H.D. First report on the antibody verification of HLA-ABC epitopes recorded in the website-based HLA Epitope Registry. Tissue Antigens 2014, 83, 391–400. [Google Scholar] [CrossRef]
- Duquesnoy, R.J.; Marrari, M.; Tambur, A.R.; Mulder, A.; Sousa, L.C.D.D.M.; da Silva, A.S.; Monte, S.J.D. First report on the antibody verification of HLA-DR, HLA-DQ and HLA-DP epitopes recorded in the HLA Epitope Registry. Hum. Immunol. 2014, 75, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Gebel, H.M.; Bray, R.A. HLA antibody detection with solid phase assays: Great expectations or expectations too great? Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2014, 14, 1964–1975. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sanchez, C.; Usenko, C.Y.; Herrera, N.D.; Tambur, A.R. The shared epitope phenomenon—A potential impediment to virtual crossmatch accuracy. Clin. Transplant. 2020, 34, e13906. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, R.; Reverberi, L. Factors affecting the antigen-antibody reaction. Blood Transfus. 2007, 5, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Opelz, G. Non-HLA transplantation immunity revealed by lymphocytotoxic antibodies. The Lancet 2005, 365, 1570–1576. [Google Scholar] [CrossRef]
- Robinson, J.; Guethlein, L.A.; Cereb, N.; Yang, S.Y.; Norman, P.J.; Marsh, S.G.E.; Parham, P. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet. 2017, 13, e1006862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schinstock, C.A.; Gandhi, M.J.; Stegall, M.D. Interpreting Anti-HLA Antibody Testing Data: A Practical Guide for Physicians. Transplantation 2016, 100, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Sapir-Pichhadze, R.; Zhang, X.; Ferradji, A.; Madbouly, A.; Tinckam, K.J.; Gebel, H.M.; Blum, D.; Marrari, M.; Kim, S.J.; Fingerson, S.; et al. Epitopes as characterized by antibody-verified eplet mismatches determine risk of kidney transplant loss. Kidney Int. 2020, 97, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadhassanzadeh, H.; Oualkacha, K.; Zhang, W.; Klement, W.; Bourdiec, A.; Lamsatfi, J.; Yi, Y.; Foster, B.; Keown, P.; Gebel, H.M.; et al. On Path to Informing Hierarchy of Eplet Mismatches as Determinants of Kidney Transplant Loss. Kidney Int. Rep. 2021, 6, 1567–1579. [Google Scholar] [CrossRef] [PubMed]
- Senev, A.; Coemans, M.; Lerut, E.; Van Sandt, V.; Kerkhofs, J.; Daniëls, L.; Driessche, M.V.; Compernolle, V.; Sprangers, B.; Van Loon, E.; et al. Eplet Mismatch Load and De Novo Occurrence of Donor-Specific Anti-HLA Antibodies, Rejection, and Graft Failure after Kidney Transplantation: An Observational Cohort Study. J. Am. Soc. Nephrol. JASN 2020, 31, 2193–2204. [Google Scholar] [CrossRef] [PubMed]
- Reed, E.F.; Rao, P.; Zhang, Z.; Gebel, H.; Bray, R.A.; Guleria, I.; Lunz, J.; Mohanakumar, T.; Nickerson, P.; Tambur, A.R.; et al. Comprehensive Assessment and Standardization of Solid Phase Multiplex-Bead Arrays for the Detection of Antibodies to HLA. Am. J. Transplant. 2013, 13, 1859–1870. [Google Scholar] [CrossRef] [Green Version]
- Visentin, J.; Marroc, M.; Guidicelli, G.; Bachelet, T.; Nong, T.; Moreau, J.-F.; Lee, J.-H.; Merville, P.; Couzi, L.; Taupin, J.-L. Clinical impact of preformed donor-specific denatured class I HLA antibodies after kidney transplantation. Clin. Transplant. 2015, 29, 393–402. [Google Scholar] [CrossRef]
- Malheiro, J.; Tafulo, S.; Dias, L.; Martins, L.S.; Fonseca, I.; Beirão, I.; Castro-Henriques, A.; Cabrita, A. Analysis of preformed donor-specific anti-HLA antibodies characteristics for prediction of antibody-mediated rejection in kidney transplantation. Transpl. Immunol. 2015, 32, 66–71. [Google Scholar] [CrossRef]
- Amrouche, L.; Aubert, O.; Suberbielle, C.; Rabant, M.; Van Huyen, J.-P.D.; Martinez, F.; Sberro-Soussan, R.; Scemla, A.; Tinel, C.; Snanoudj, R.; et al. Long-term Outcomes of Kidney Transplantation in Patients With High Levels of Preformed DSA: The Necker High-Risk Transplant Program. Transplantation 2017, 101, 2440–2448. [Google Scholar] [CrossRef]
- Tambur, A.R. HLA-Epitope Matching or Eplet Risk Stratification: The Devil Is in the Details. Front. Immunol. 2018, 9, 2010. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhang, H.; Tan, J.; Fu, Q.; Li, J.; Wu, C.; Huang, H.; Xu, B.; Ling, L.; Liu, L.; et al. Eplet-Predicted Antigens: An Attempt to Introduce Eplets into Unacceptable Antigen Determination and Calculated Panel-Reactive Antibody Calculation Facilitating Kidney Allocation. Diagnostics 2022, 12, 2983. https://doi.org/10.3390/diagnostics12122983
Wu W, Zhang H, Tan J, Fu Q, Li J, Wu C, Huang H, Xu B, Ling L, Liu L, et al. Eplet-Predicted Antigens: An Attempt to Introduce Eplets into Unacceptable Antigen Determination and Calculated Panel-Reactive Antibody Calculation Facilitating Kidney Allocation. Diagnostics. 2022; 12(12):2983. https://doi.org/10.3390/diagnostics12122983
Chicago/Turabian StyleWu, Wenrui, Huanxi Zhang, Jinghong Tan, Qian Fu, Jun Li, Chenglin Wu, Huiting Huang, Bowen Xu, Liuting Ling, Longshan Liu, and et al. 2022. "Eplet-Predicted Antigens: An Attempt to Introduce Eplets into Unacceptable Antigen Determination and Calculated Panel-Reactive Antibody Calculation Facilitating Kidney Allocation" Diagnostics 12, no. 12: 2983. https://doi.org/10.3390/diagnostics12122983




