A Robust Framework for Data Generative and Heart Disease Prediction Based on Efficient Deep Learning Models
Abstract
1. Introduction
- Propose two deep learning (DL) models based on one-dimensional convolutional neural network (1D-CNN) and bidirectional long short-term memory (Bi-LSTM) for HD diagnosis.
- Propose a generative adversarial network (GAN) model to augment the imbalanced and limited dataset to have a balanced distribution and larger dataset for training the predictive models (1D-CNN, and Bi-LSTM) and improving their performance.
- Reduce model complexity, computation time and dataset dimensionality for more quick diagnosis using a fine-tuning and dimension reduction technique.
- Evaluate the effectiveness of the proposed DL models using various performance measures and compare with conventional ML and DL models such as support vector machines (SVM) and artificial neural networks (ANNs).
2. Related Work
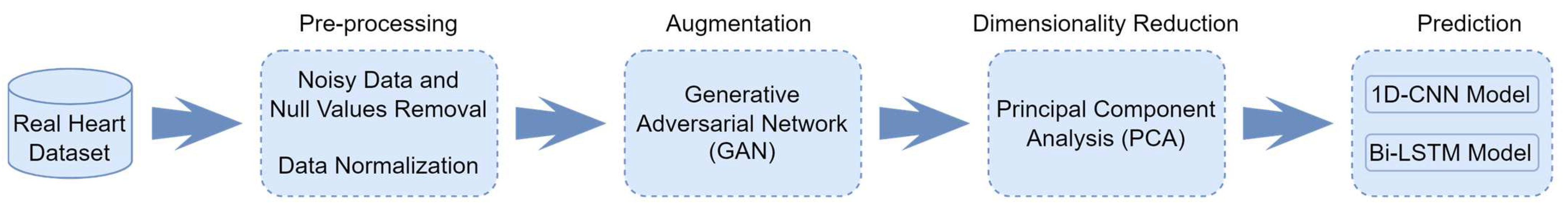
3. Materials and Methods
3.1. Data Sources and Pre-Processing
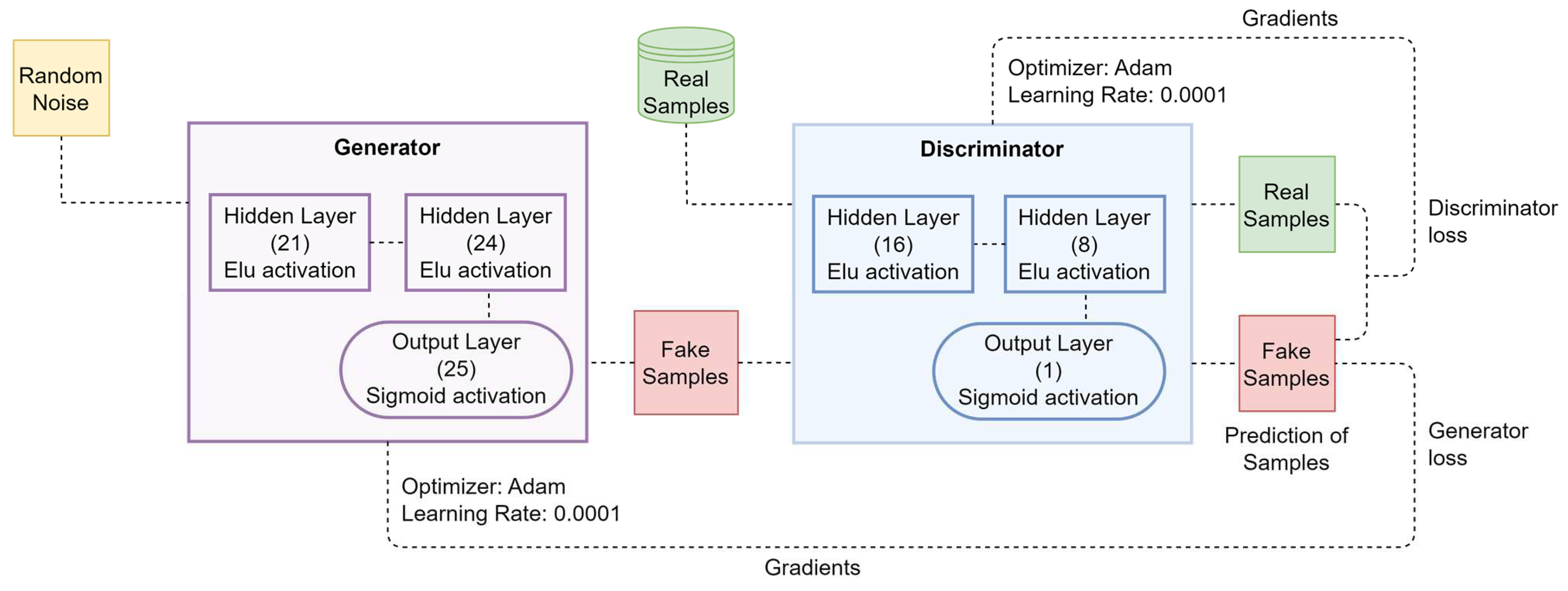
3.2. Data Augmentation Model
3.3. Dimensionality Reduction Method
3.4. HD Diagnosis DL Models
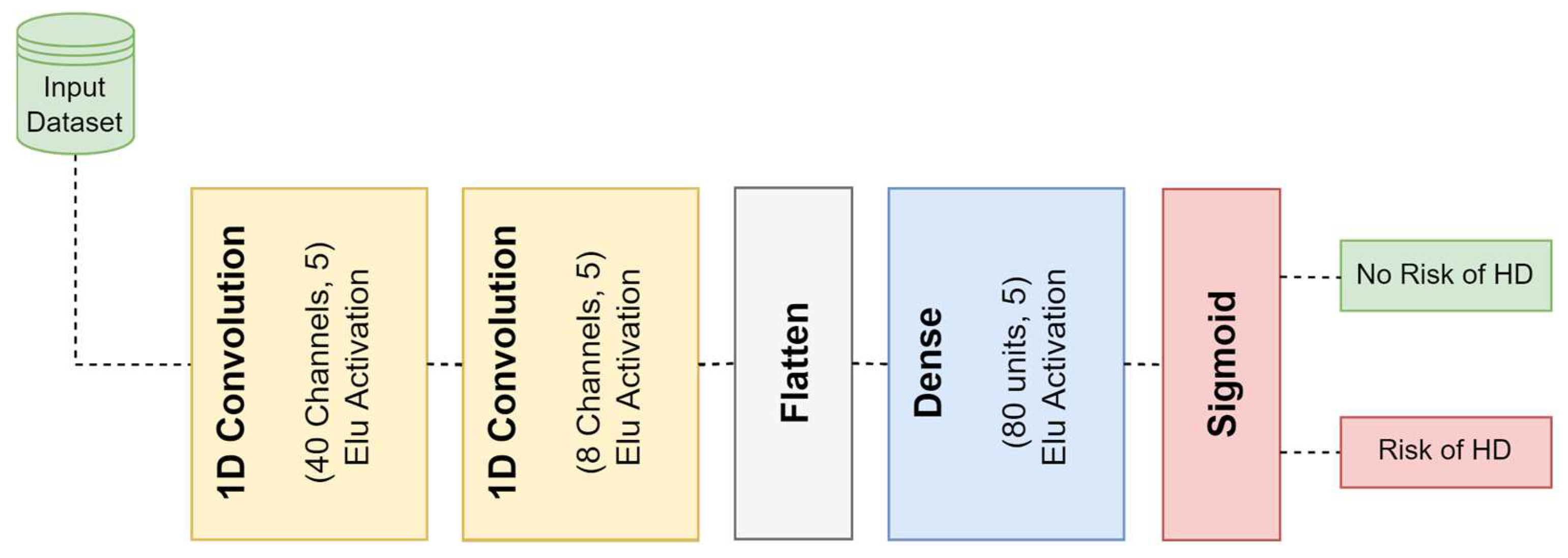
3.4.1. One-Dimensional Convolutional Neural Network (1D-CNN)
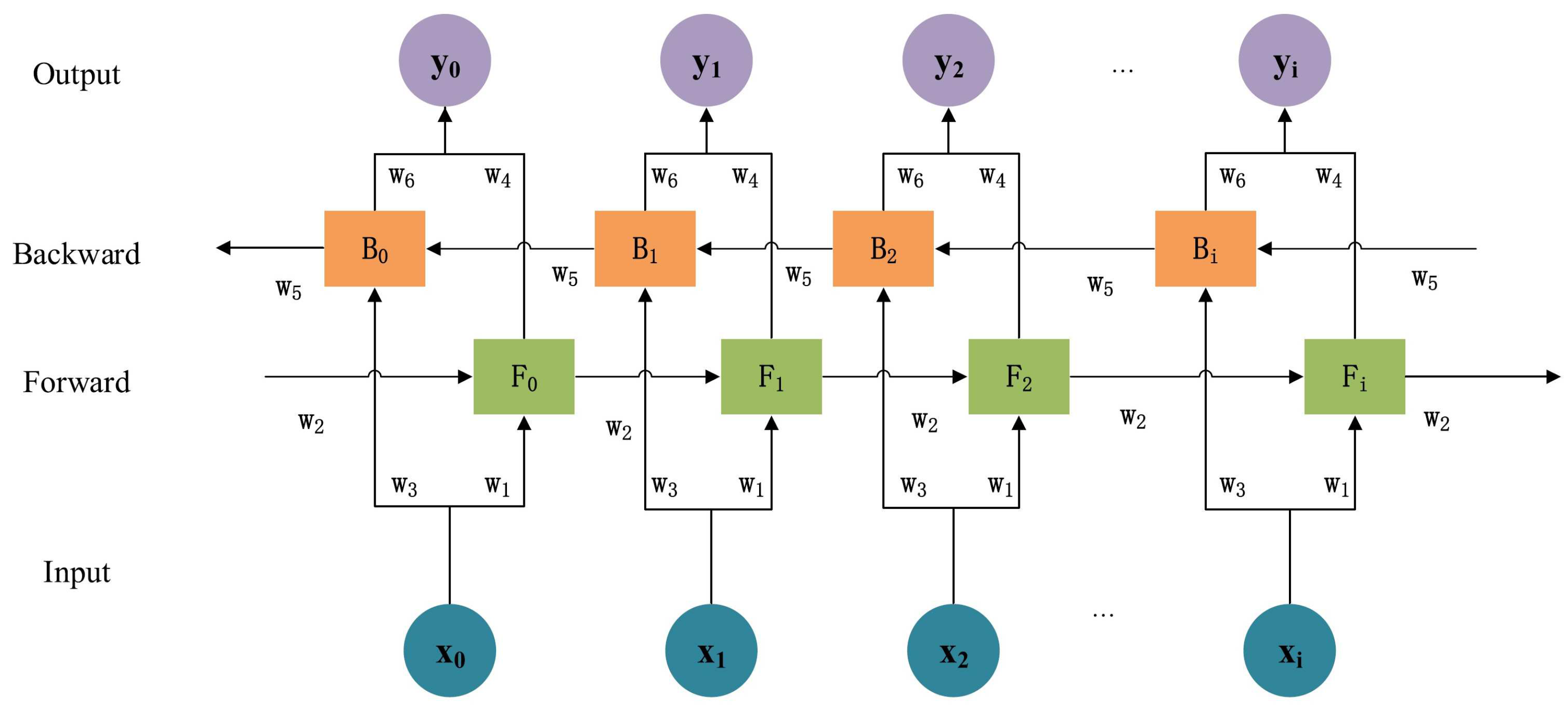
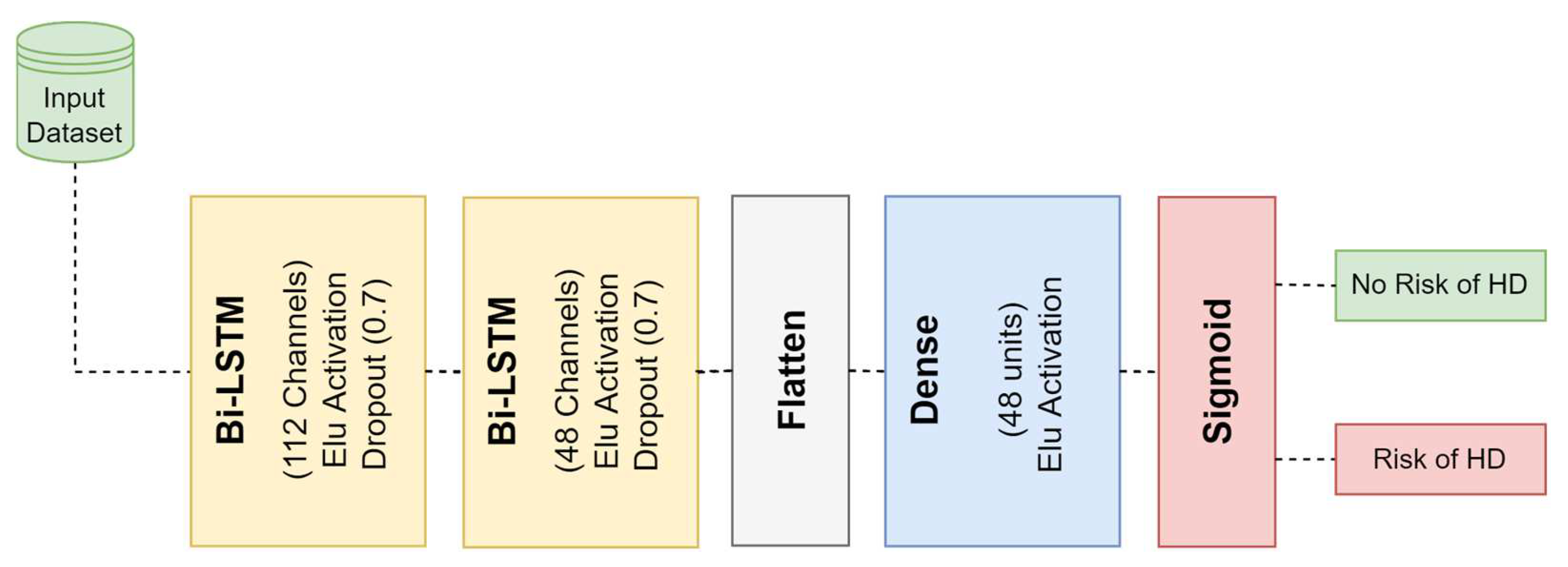
3.4.2. Bidirectional Long Short-Term Memory (Bi-LSTM)
3.5. Brief Outline of Traditional ML and DL Models
3.5.1. Support Vector Machine (SVM)
3.5.2. Artificial Neural Network (ANN)
4. Results and Discussion
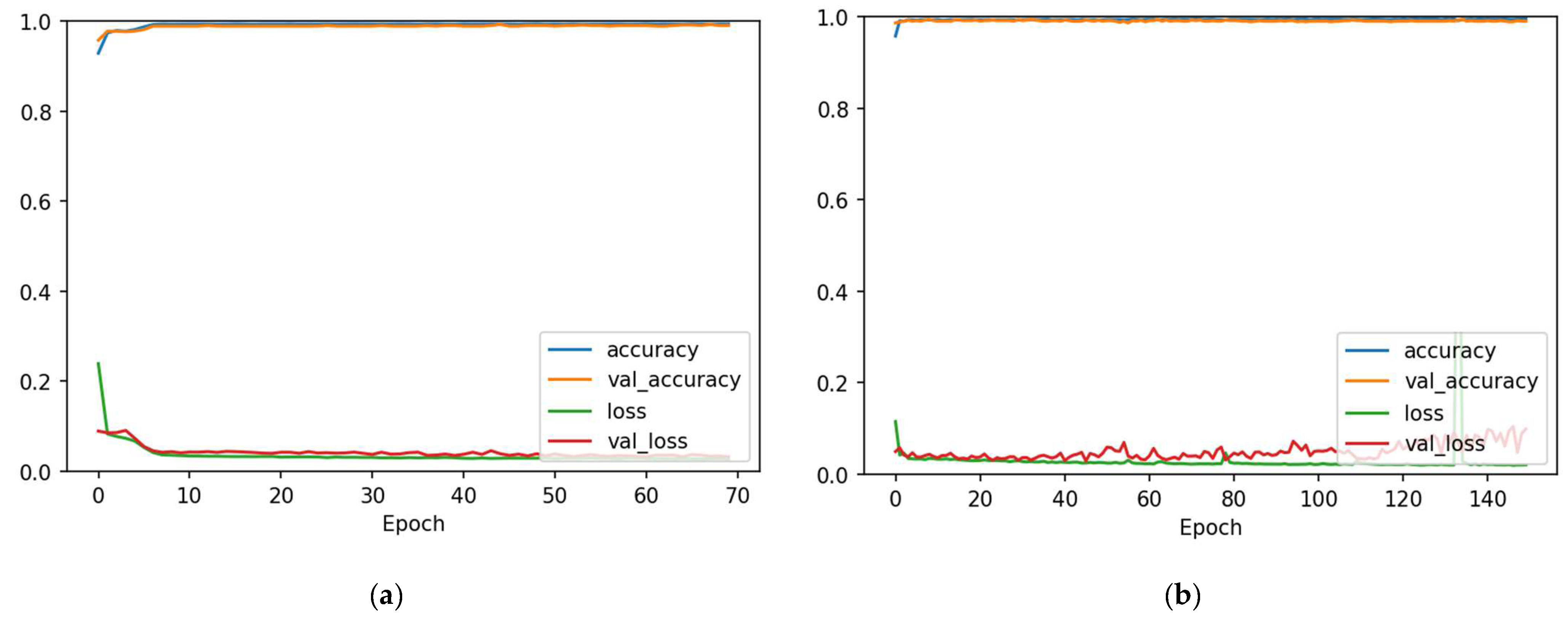
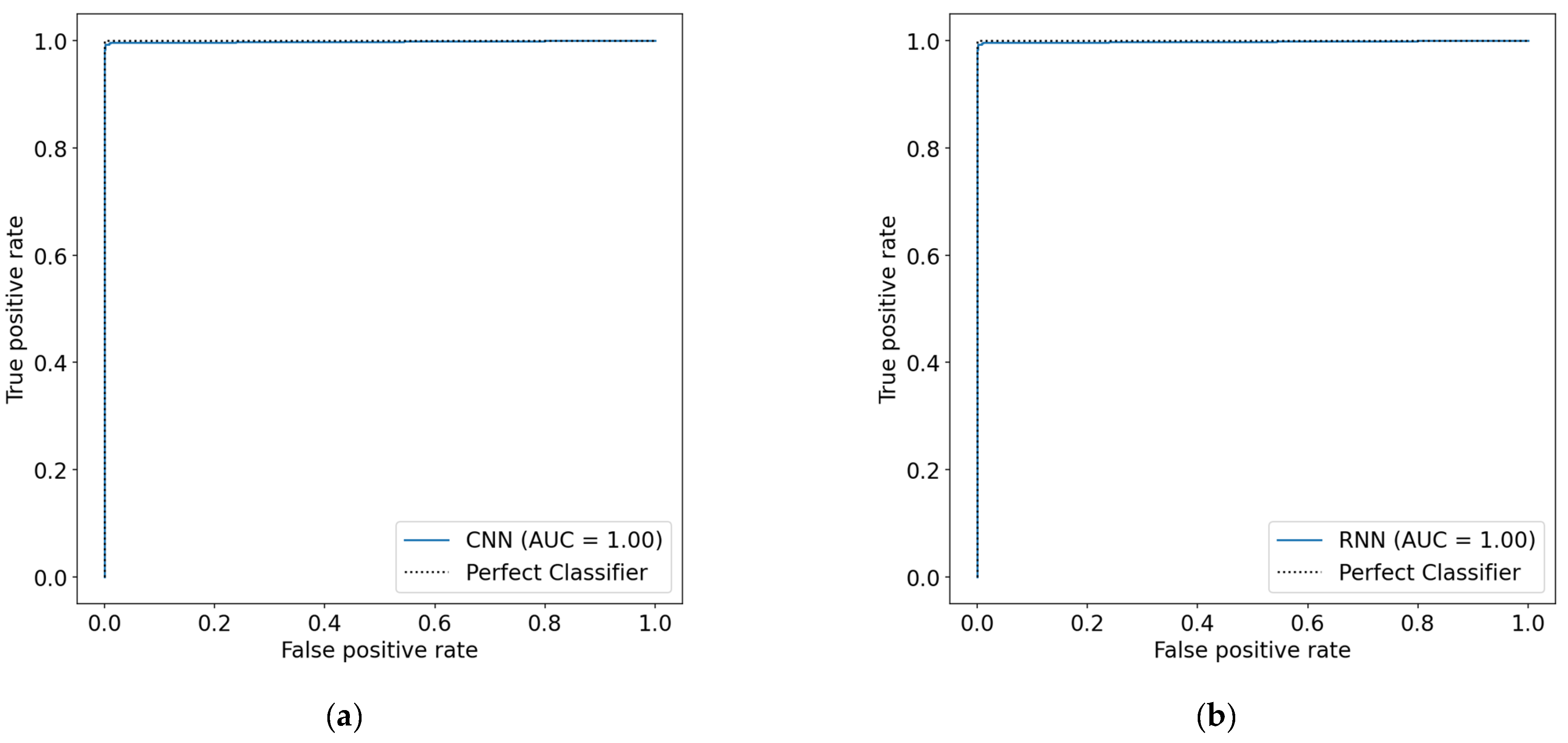
4.1. 1D-CNN and Bi-LSTM Models Performance before Data Augmentation
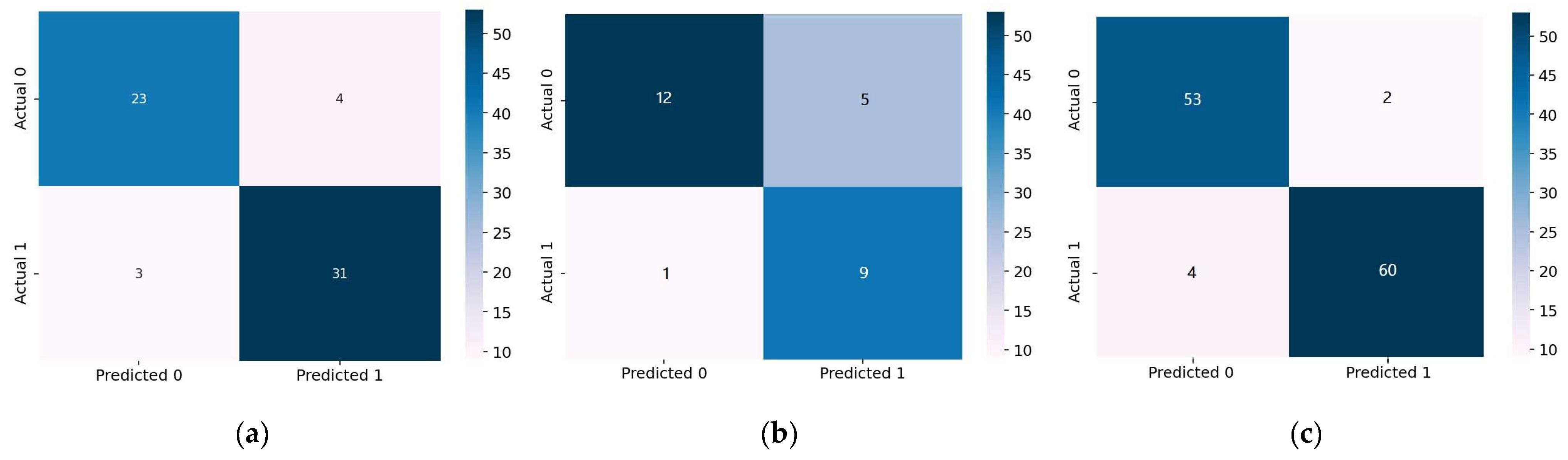
4.1.1. 1D-CNN Performance Evaluation
4.1.2. Bi-LSTM Performance Evaluation
4.2. 1D-CNN and Bi-LSTM Models Performance after Data Augmentation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alnemari, W.; Bakry, S.; Albagami, S.; AL-Zahrani, S.; Almousa, A.; Alsufyani, A.; Siddiqui, M. Awareness and knowledge of Rheumatic heart disease among medical students comparing to other health specialties students in Umm Al-Qura University, Makkah city, KSA-Analytic cross-sectional study. Med. Sci. 2022, 26, 1–7. [Google Scholar] [CrossRef]
- Elhoseny, M.; Mohammed, M.A.; Mostafa, S.A.; Abdulkareem, K.H.; Maashi, M.S.; Garcia-Zapirain, B.; Mutlag, A.A.; Maashi, M.S. A new multi-agent feature wrapper machine learning approach for heart disease diagnosis. Comput. Mater. Contin. 2021, 67, 51–71. [Google Scholar] [CrossRef]
- Sarra, R.R.; Dinar, A.M.; Mohammed, M.A. Enhanced accuracy for heart disease prediction using artificial neural network. Indones. J. Electr. Eng. Comput. Sci. 2022, 29, 375–383. [Google Scholar] [CrossRef]
- Odah, M.M.; Alfakieh, H.O.; Almathami, A.A.; Almuashi, I.M.; Awad, M.; Ewis, A.A. Public Awareness of Coronary Artery Disease and its Risk Factors Among Al-Qunfudah Governorate Population. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 34–38. [Google Scholar] [CrossRef]
- Sharma, M. ECG and Medical Diagnosis Based Recognition & Prediction of Cardiac Disease Using Deep Learning. J. Sci. Technol. Res. 2019, 8, 233–240. Available online: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85073992637&partnerID=40&md5=51e5526c27bba7420d6702548a9547f2 (accessed on 1 October 2022).
- Mhamdi, L.; Dammak, O.; Cottin, F.; Dhaou, I.B. Artificial Intelligence for Cardiac Diseases Diagnosis and Prediction Using ECG Images on Embedded Systems. Biomedicines 2022, 10, 2013. [Google Scholar] [CrossRef]
- Sarra, R.R.; Dinar, A.M.; Mohammed, M.A.; Abdulkareem, K.H. Enhanced Heart Disease Prediction Based on Machine Learning and χ2 Statistical Optimal Feature Selection Model. Designs 2022, 6, 87. [Google Scholar] [CrossRef]
- Rahman, A.U.; Saeed, M.; Mohammed, M.A.; Jaber, M.M.; Garcia-Zapirain, B. A novel fuzzy parameterized fuzzy hypersoft set and riesz summability approach based decision support system for diagnosis of heart diseases. Diagnostics 2022, 12, 1546. [Google Scholar] [CrossRef]
- Bizopoulos, P.; Koutsouris, D. Deep Learning in Cardiology. IEEE Rev. Biomed. Eng. 2019, 12, 168–193. [Google Scholar] [CrossRef]
- Rahman, A.U.; Saeed, M.; Mohammed, M.A.; Krishnamoorthy, S.; Kadry, S.; Eid, F. An Integrated Algorithmic MADM Approach for Heart Diseases’ Diagnosis Based on Neutrosophic Hypersoft Set with Possibility Degree-Based Setting. Life 2022, 12, 729. [Google Scholar] [CrossRef]
- Nasser, A.R.; Hasan, A.M.; Humaidi, A.J.; Alkhayyat, A.; Alzubaidi, L.; Fadhel, M.A.; Santamaría, J.; Duan, Y. IoT and Cloud Computing in Health-Care: A New Wearable Device and Cloud-Based Deep Learning Algorithm for Monitoring of Diabetes. Electronics 2021, 10, 2719. [Google Scholar] [CrossRef]
- Mathur, P.; Srivastava, S.; Xu, X.; Mehta, J.L. Artificial Intelligence, Machine Learning, and Cardiovascular Disease. Clin. Med. Insights. Cardiol. 2020, 14, 1179546820927404. [Google Scholar] [CrossRef] [PubMed]
- Ontor, M.Z.H.; Ali, M.M.; Ahmed, K.; Bui, F.M.; Al-Zahrani, F.A.; Mahmud, S.M.H.; Azam, S. Early-stage cervical cancerous cell detection from cervix images using yolov5. Comput. Mater. Contin. 2022, 74, 3727–3741. [Google Scholar]
- Subahi, A.F.; Khalaf, O.I.; Alotaibi, Y.; Natarajan, R.; Mahadev, N.; Ramesh, T. Modified Self-Adaptive Bayesian Algorithm for Smart Heart Disease Prediction in IoT System. Sustainability 2022, 14, 14208. [Google Scholar] [CrossRef]
- Sandhiya, S.; Palani, U. An effective disease prediction system using incremental feature selection and temporal convolutional neural network. J. Ambient. Intell. Humaniz. Comput. 2020, 11, 5547–5560. [Google Scholar] [CrossRef]
- Zamzmi, G.; Hsu, L.Y.; Li, W.; Sachdev, V.; Antani, S. Harnessing Machine Intelligence in Automatic Echocardiogram Analysis: Current Status, Limitations, and Future Directions. IEEE Rev. Biomed. Eng. 2021, 14, 181–203. [Google Scholar] [CrossRef]
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of Heart Disease Using a Combination of Machine Learning and Deep Learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef]
- Ali, L.; Rahman, A.; Khan, A.; Zhou, M.; Javeed, A.; Khan, J.A. An Automated Diagnostic System for Heart Disease Prediction Based on X2 Statistical Model and Optimally Configured Deep Neural Network. IEEE Access 2019, 7, 34938–34945. [Google Scholar] [CrossRef]
- Das, S.; Sharma, R.; Gourisaria, M.K.; Rautaray, S.S.; Pandey, M. Heart Disease Detection Using Core Machine Learning and Deep Learning Techniques: A Comparative Study. Int. J. Emerg. Technol. 2020, 11, 531–538. Available online: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85086388478&partnerID=40&md5=80e37a22acd2ffaa9a92d13f43111c5c (accessed on 1 October 2022).
- Mienye, I.D.; Sun, Y.; Wang, Z. Improved sparse autoencoder based artificial neural network approach for prediction of heart disease. Inform. Med. Unlocked 2020, 18, 100307. [Google Scholar] [CrossRef]
- Sajja, T.K.; Kalluri, H.K. A deep learning method for prediction of cardiovascular disease using convolutional neural network. Rev. D' Intell. Artif. 2020, 34, 601–606. [Google Scholar] [CrossRef]
- Krishnan, S.; Magalingam, P.; Ibrahim, R.B. Advanced Recurrent Neural Network with Tensorflow for Heart Disease Prediction. Int. J. Adv. Sci. 2020, 29, 966–977. Available online: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85083335150&partnerID=40&md5=f5697e2dfdb572a90e9cb7626e394456 (accessed on 1 October 2022).
- Ali, S.A.; Raza, B.; Malik, A.K.; Shahid, A.R.; Faheem, M.; Alquhayz, H.; Kumar, Y.J. An optimally configured and improved deep belief network (OCI-DBN) approach for heart disease prediction based on Ruzzo–Tompa and stacked genetic algorithm. IEEE Access 2020, 8, 65947–65958. [Google Scholar] [CrossRef]
- Janosi, A.; Steinbrunn, W.; Pfisterer, M.; Detrano, R. UCI Machine Learning Repository: Heart Disease Dataset. Available online: https://archive-beta.ics.uci.edu/ml/datasets/heart+disease (accessed on 1 March 2022).
- UCI Machine Learning Repository: Statlog (Heart). Available online: https://archive-beta.ics.uci.edu/ml/datasets/statlog+heart (accessed on 1 March 2022).
- Vijayashree, J.; Parveen Sultana, H. Heart disease classification using hybridized Ruzzo-Tompa memetic based deep trained Neocognitron neural network. Health Technol. 2020, 10, 207–216. [Google Scholar] [CrossRef]
- Heart Disease Dataset (Comprehensive). Available online: https://www.kaggle.com/datasets/sid321axn/heart-statlog-cleveland-hungary-final (accessed on 1 March 2022).
- Wang, J.; Liu, X.; Wang, F.; Zheng, L.; Gao, F.; Zhang, H.; Zhang, X.; Xie, W.; Wang, B. Automated interpretation of congenital heart disease from multi-view echocardiograms. Med. Image Anal. 2021, 69, 101942. [Google Scholar] [CrossRef] [PubMed]
- Baghel, N.; Dutta, M.K.; Burget, R. Automatic diagnosis of multiple cardiac diseases from PCG signals using convolutional neural network. Comput. Methods Programs Biomed. 2020, 197, 105750. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yang, L.; Cai, H.; Tang, R.; Chen, Y.; Li, W. Multi-classification of arrhythmias using a HCRNet on imbalanced ECG datasets. Comput. Methods Programs Biomed. 2021, 208, 106258. [Google Scholar] [CrossRef]
- Chandra, V.; Singh, V.; Sarkar, P.G. A Survey on the Role of Deep Learning in 2D Transthoracic Echocardiography. Int. J. Sci. Technol. Res. 2020, 9, 7060–7065. Available online: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85082954320&partnerID=40&md5=2c7158e2e70abc235bee836b8871e136 (accessed on 1 October 2022).
- Zhang, H. Heartbeat monitoring with an mm-wave radar based on deep learning: A novel approach for training and classifying heterogeneous signals. Remote Sens. Lett. 2020, 11, 993–1001. [Google Scholar] [CrossRef]
- Olaniyi, E.O.; Oyedotun, O.K.; Adnan, K. Heart Diseases Diagnosis Using Neural Networks Arbitration. Int. J. Intell. Syst. Appl. 2015, 7, 75–82. [Google Scholar]
- Amarbayasgalan, T.; Park, K.H.; Lee, J.Y.; Ryu, K.H. Reconstruction error based deep neural networks for coronary heart disease risk prediction. PLoS ONE 2019, 14, e0225991. [Google Scholar] [CrossRef] [PubMed]
- Samir, A.A.; Rashwan, A.R.; Sallam, K.M.; Chakrabortty, R.K.; Ryan, M.J.; Abohany, A.A. Evolutionary algorithm-based convolutional neural network for predicting heart diseases. Comput. Ind. Eng. 2021, 161, 107651. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn Res. 2012, 13, 281–305. [Google Scholar]
- Li, Z.; Zhou, D.; Wan, L.; Li, J.; Mou, W. Heartbeat classification using deep residual convolutional neural network from 2-lead electrocardiogram. J. Electrocardiol. 2020, 58, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Lih, O.S.; Jahmunah, V.; San, T.R.; Ciaccio, E.J.; Yamakawa, T.; Tanabe, M.; Kobayashi, M.; Faust, O.; Acharya, U.R. Comprehensive electrocardiographic diagnosis based on deep learning. Artif. Intell. Med. 2020, 103, 101789. [Google Scholar] [CrossRef]
- Alkhodari, M.; Fraiwan, L. Convolutional and recurrent neural networks for the detection of valvular heart diseases in phonocardiogram recordings. Comput. Methods Programs Biomed. 2021, 200, 105940. [Google Scholar] [CrossRef]
- Dang, H.; Sun, M.; Zhang, G.; Qi, X.; Zhou, X.; Chang, Q. A Novel Deep Arrhythmia-Diagnosis Network for Atrial Fibrillation Classification Using Electrocardiogram Signals. IEEE Access 2019, 7, 75577–75590. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, J.; Liu, Y.; Xie, Y.; Han, Z.; Yu, J. Recursion Enhanced Random Forest with an Improved Linear Model (RERF-ILM) for Heart Disease Detection on the Internet of Medical Things Platform. IEEE Access 2020, 8, 59247–59256. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Mastoi, Q.U.; Wah, T.Y.; Mohammed, M.A.; Iqbal, U.; Kadry, S.; Majumdar, A.; Thinnukool, O. Novel DERMA Fusion Technique for ECG Heartbeat Classification. Life 2022, 12, 842. [Google Scholar] [CrossRef]






| Attribute | Type | Summary |
|---|---|---|
| Age | Number | The number of years |
| Sex. | Category | 0: Female or 1: male |
| Cp. | Category | Chest pain of a specific type (1: typical angina, 2: atypical angina, 3: non anginal pain, 4: asymptomatic) |
| Trestbps | Number | Blood pressure at rest measured in mmHg |
| Chol | Number | Cholesterol level measured in mg/dL |
| Fbs | Category | Fasting glucose level over 120 mg/dL (0: false, 1: true) |
| Restecg. | Category | Electrocardiogram reading in resting state (0: normal, 1: ST-T wave abnormalities, and 2: left ventricular hypertrophy) |
| Thalach | Number | Results of a thallium stress test showing the highest possible heart rate |
| Exang. | Category | Angina during exercise (1 indicates yes, 0 indicates no) |
| Oldpeak | Number | Exercise ST depression versus rest |
| Slope | Category | ST segment inclination during exercise (1: up, 2: flat, 3: down) |
| Ca. | Category | Significant fluoroscopically colored vessel number |
| Thal | Category | Test results for thulium stress: 3: normal, 6: a fixed defect, and 7: a reversible defect |
| Num | Category | HD status (0 indicates less than 50% diameter narrowing, 1indicates more than 50% diameter narrowing) |
| Layer | Type | No. of Filters | Kernel Size | Activation Function |
|---|---|---|---|---|
| 1 | 1D-Convolution | 40 | 5 | Elu |
| 2 | 1D-Convolution | 8 | 5 | Elu |
| 3 | Flatten | - | - | - |
| 4 | Fully Connected | 80 | - | Elu |
| 5 | Fully Connected | 1 | - | Sigmoid |
| Layer | Type | No. of Neurons | Dropout Ratio | Function |
|---|---|---|---|---|
| 1 | Bi-LSTM | 112 | 0.7 | Elu |
| 2 | Bi-LSTM | 48 | 0.7 | Elu |
| 3 | Flatten | - | - | - |
| 4 | Fully Connected | 48 | - | Elu |
| 5 | Fully Connected | 1 | - | Sigmoid |
| Dataset | Records | Features | Accuracy | Specificity | Sensitivity | F1-Score |
|---|---|---|---|---|---|---|
| Cleveland | 303 | 14 | 87.10% | 86.97% | 86.97% | 86.97% |
| Statlog | 270 | 14 | 81.48% | 81.32% | 81.67% | 81.38% |
| Comprehensive | 1190 | 12 | 94.96% | 94.93% | 95.18% | 94.95% |
| Dataset | Records | Features | Accuracy | Specificity | Sensitivity | F1-Score |
|---|---|---|---|---|---|---|
| Cleveland | 303 | 14 | 88.52% | 89.08% | 87.80% | 88.21% |
| Statlog | 270 | 14 | 80.00% | 79.45% | 79.17% | 79.28% |
| Comprehensive | 1190 | 12 | 94.96% | 94.88% | 95.06% | 94.94% |
| Hyperparameter | Value | Description |
|---|---|---|
| Epoch | 50,000 | Number of training iterations |
| Batch size | 32 | Number of batch samples per iteration |
| Learning rate | 0.0001 | Learning rate |
| Optimizer | Adam | Optimization algorithm |
| Before Augmentation and PCA | After Augmentation and PCA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | Acc (%) | Spe (%) | Sen (%) | F1 (%) | Prediction Time (ms) | Acc (%) | Spe (%) | Sen (%) | F1 (%) | Prediction Time (ms) |
| 1D-CNN | 87.10 | 86.97 | 86.97 | 86.97 | 72.6 | 99.1 | 99.1 | 99.1 | 99.1 | 68.8 |
| Bi-LSTM | 88.52 | 89.08 | 87.80 | 88.21 | 80.4 | 99.3 | 99.2 | 99.3 | 99.2 | 74.8 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| SVM | 86.88% | 87.76% | 85.95% | 86.44% |
| ANN | 93.44% | 93.83% | 92.97% | 93.30% |
| Proposed GAN-CNN | 99.1% | 99.1% | 99.1% | 99.1% |
| Proposed GAN-Bi-LSTM | 99.3% | 99.2% | 99.3% | 99.2% |
| Study | Method | Accuracy | Dataset | Model Complexity |
|---|---|---|---|---|
| [18] | DNN | 94.2% | Cleveland | 6 layers (3 Dense, 2 Dropout, 1 Output) |
| [21] | SAE-ANN 1 | 90% | Framingham | SAE network (5 layers) |
| [20] | ANN | 92% | Cleveland | 2 layers (1 Dense, 1 Output) |
| [22] | CNN | 94.78% | Cleveland | 5 layers (2 Conv1D, 2 Dropout, 1 Output) |
| [23] | RNN-GRU 2 | 98.4% | Cleveland | 7 layers of GRUs |
| [24] | OCI-DBN 3 | 94.61% | Cleveland | 3 layers (2 Dense, 1 Output) |
| [19] | DNN | 93.33% | Cleveland | 3 layers (2 Dense, 1 Output) |
| Proposed | GAN-1D-CNN GAN-Bi-LSTM | 99.10% 99.30% | Cleveland | 4 layers (2 Conv1D, 1 Dense, 1 Output) 4 layers (2 Bi-LSTM, 1 Dense, 1 Output) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarra, R.R.; Dinar, A.M.; Mohammed, M.A.; Ghani, M.K.A.; Albahar, M.A. A Robust Framework for Data Generative and Heart Disease Prediction Based on Efficient Deep Learning Models. Diagnostics 2022, 12, 2899. https://doi.org/10.3390/diagnostics12122899
Sarra RR, Dinar AM, Mohammed MA, Ghani MKA, Albahar MA. A Robust Framework for Data Generative and Heart Disease Prediction Based on Efficient Deep Learning Models. Diagnostics. 2022; 12(12):2899. https://doi.org/10.3390/diagnostics12122899
Chicago/Turabian StyleSarra, Raniya R., Ahmed M. Dinar, Mazin Abed Mohammed, Mohd Khanapi Abd Ghani, and Marwan Ali Albahar. 2022. "A Robust Framework for Data Generative and Heart Disease Prediction Based on Efficient Deep Learning Models" Diagnostics 12, no. 12: 2899. https://doi.org/10.3390/diagnostics12122899
APA StyleSarra, R. R., Dinar, A. M., Mohammed, M. A., Ghani, M. K. A., & Albahar, M. A. (2022). A Robust Framework for Data Generative and Heart Disease Prediction Based on Efficient Deep Learning Models. Diagnostics, 12(12), 2899. https://doi.org/10.3390/diagnostics12122899








