Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks
Abstract
1. Introduction
2. Materials and Methods
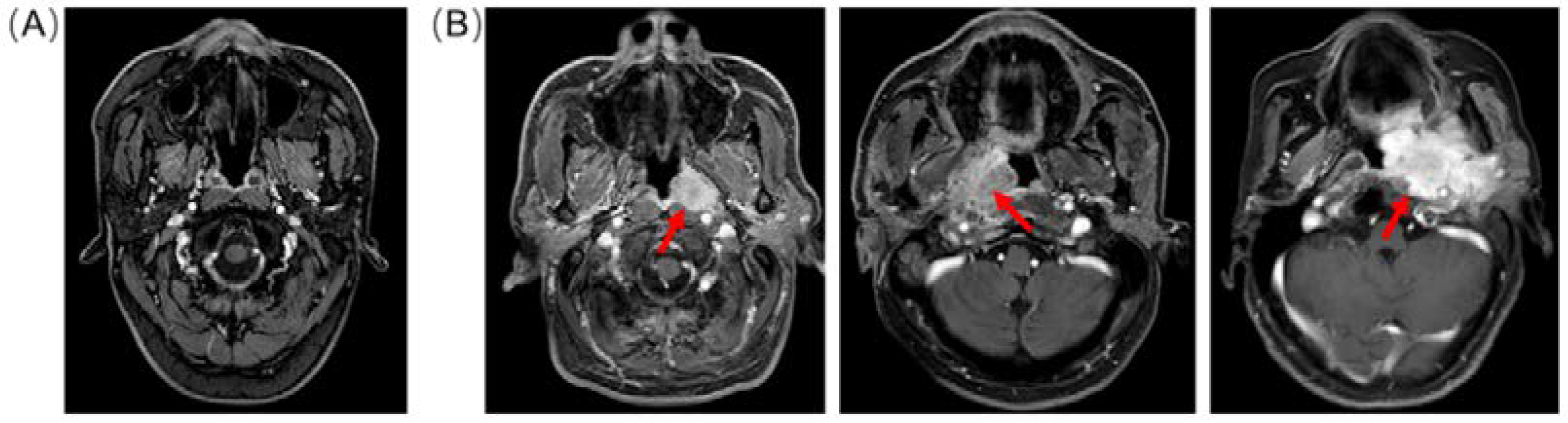
2.1. Dataset Preparation
2.2. Choice of Splitter
2.3. Evaluated Shallow and Hierarchical Convolutional Neural Network
2.4. Hyper-parameter Optimization
2.5. Data Analysis
3. Results
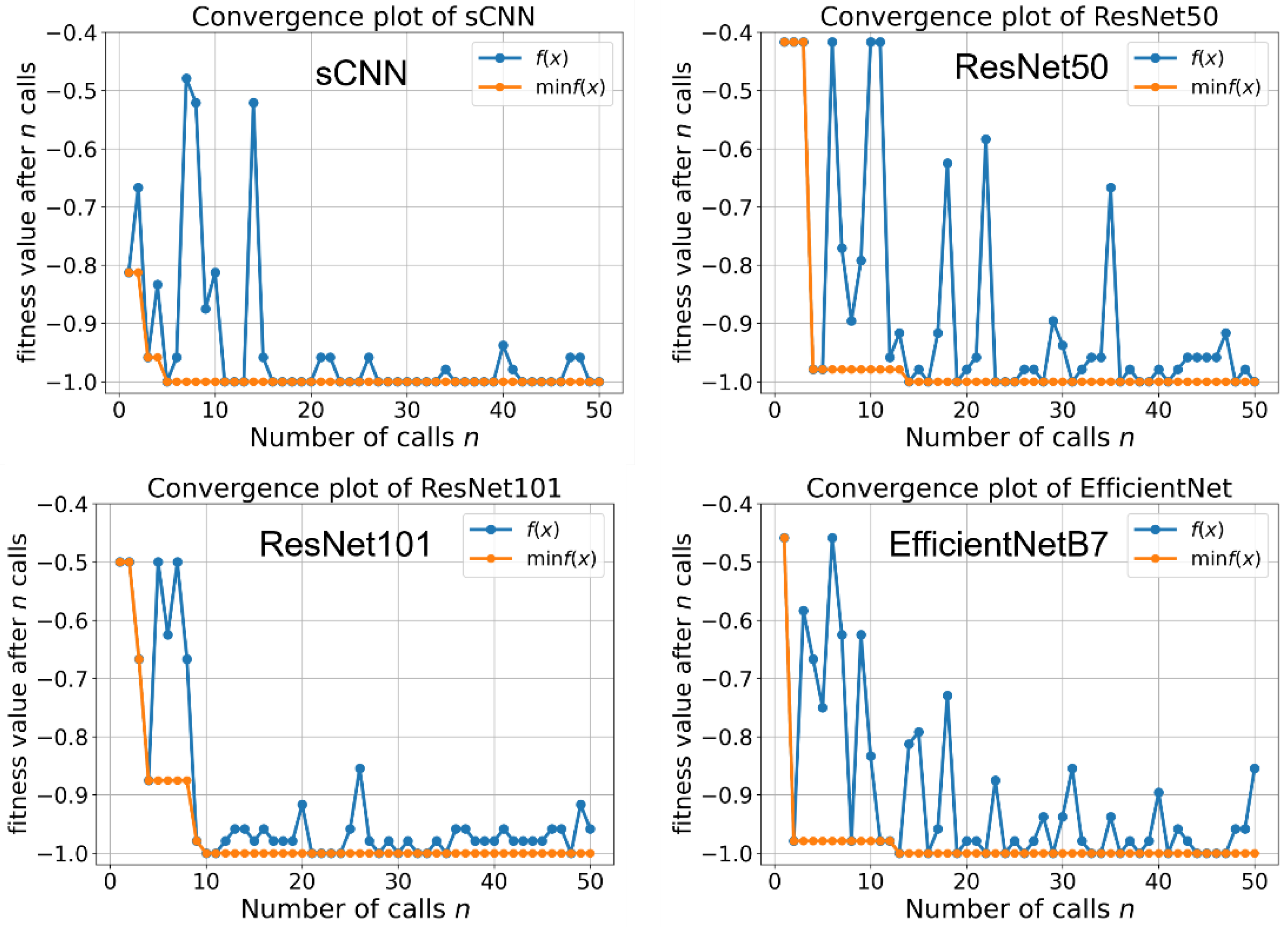
3.1. Hyper-Parameter Optimization
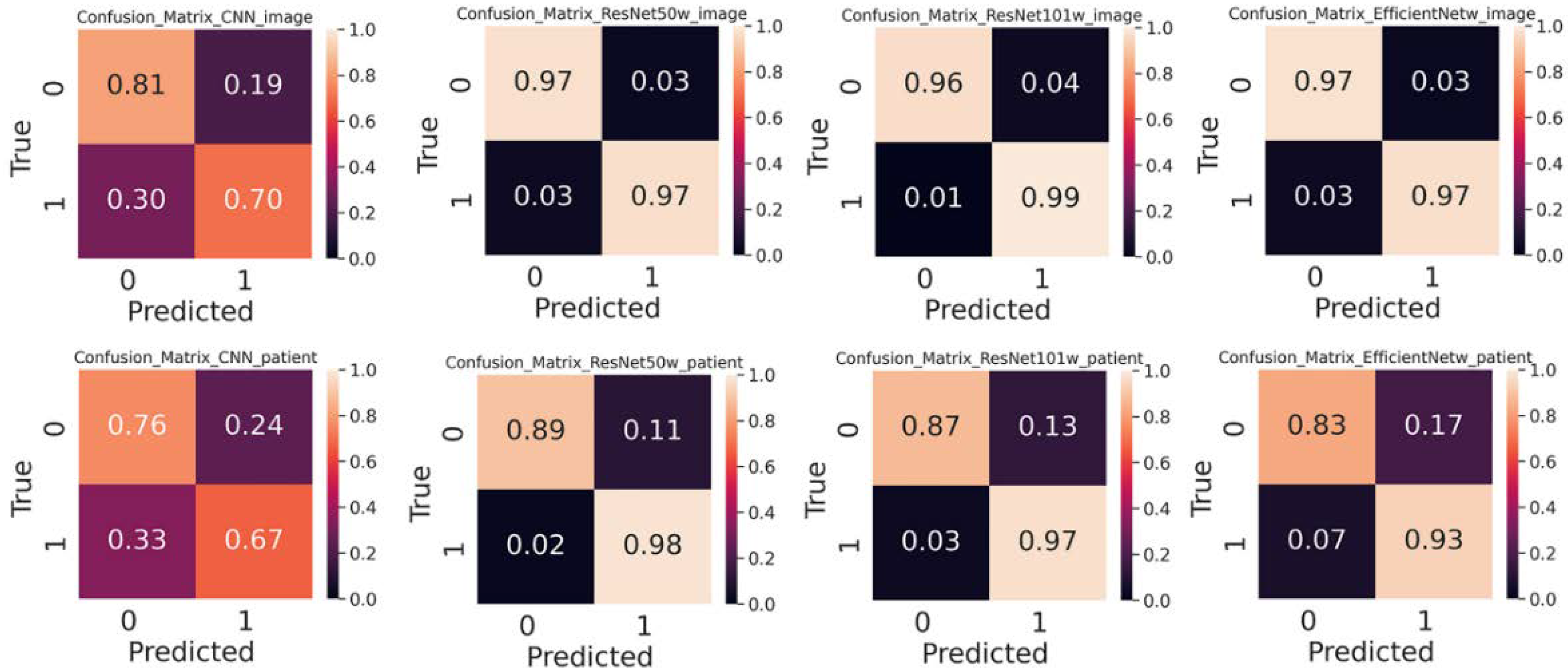
3.2. Performance of Shallow and Hierarchical Learning
3.3. Choice of Splitter Affects Performance
3.4. Pre-trained Weight Improves Performance
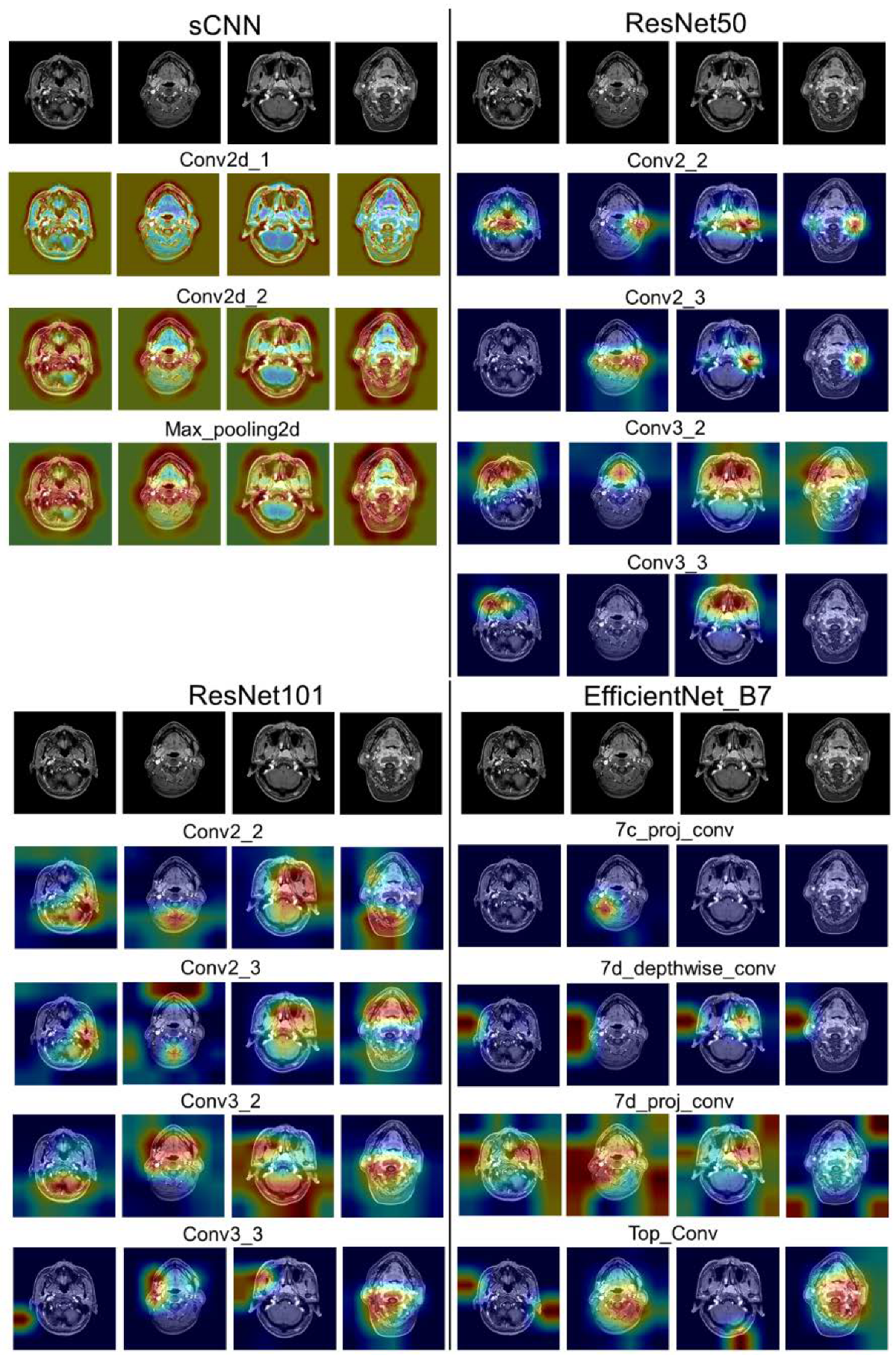
3.5. Interpretability of Shallow and Hierarchical Learning
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.W.; Hui, E.P.; Lo, K.-W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.-F.; King, A.D.; et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, C.; Pan, L. Nasopharyngeal carcinoma: A review of current updates. Exp. Ther. Med. 2018, 15, 3687–3692. [Google Scholar] [CrossRef] [PubMed]
- Bakkalci, D.; Jia, Y.; Winter, J.R.; Lewis, J.E.; Taylor, G.S.; Stagg, H.R. Risk factors for Epstein Barr virus-associated cancers: A systematic review, critical appraisal, and mapping of the epidemiological evidence. J. Glob. Health 2020, 10, 010405. [Google Scholar] [CrossRef] [PubMed]
- Razek, A.A.K.A.; King, A. MRI and CT of Nasopharyngeal Carcinoma. Am. J. Roentgenol. 2012, 198, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y. Learning Deep Architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Wang, M.; Liang, Y.; Hu, Z.; Chen, S.; Shi, B.; Heidari, A.A.; Zhang, Q.; Chen, H.; Chen, X. Lupus nephritis diagnosis using enhanced moth flame algorithm with support vector machines. Comput. Biol. Med. 2022, 145, 105435. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, D.; Yu, F.; Heidari, A.A.; Bano, Y.; Ibrohimov, A.; Liu, Y.; Cai, Z.; Chen, H.; Chen, X. An optimized machine learning framework for predicting intradialytic hypotension using indexes of chronic kidney disease-mineral and bone disorders. Comput. Biol. Med. 2022, 145, 105510. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Yun, J.; Cho, Y.; Shin, K.; Jang, R.; Bae, H.-J.; Kim, N. Deep Learning in Medical Imaging. Neurospine 2019, 16, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Ginneken, B.V.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Hwang, D.K.; Hsu, C.C.; Chang, K.J.; Chao, D.; Sun, C.H.; Jheng, Y.C.; Yarmishyn, A.A.; Wu, J.C.; Tsai, C.Y.; Wang, M.L.; et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics 2019, 9, 232–245. [Google Scholar] [CrossRef]
- Li, L.; Wei, M.; Liu, B.; Atchaneeyasakul, K.; Zhou, F.; Pan, Z.; Kumar, S.A.; Zhang, J.Y.; Pu, Y.; Liebeskind, D.S.; et al. Deep Learning for Hemorrhagic Lesion Detection and Segmentation on Brain CT Images. IEEE J. Biomed. Health Inform. 2021, 25, 1646–1659. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Bozorgtabar, B.; Mahapatra, D.; von Tengg-Kobligk, H.; Poellinger, A.; Ebner, L.; Thiran, J.-P.; Reyes, M. Informative sample generation using class aware generative adversarial networks for classification of chest Xrays. Comput. Vis. Image Underst. 2019, 184, 57–65. [Google Scholar] [CrossRef]
- Yun, J.; Park, J.E.; Lee, H.; Ham, S.; Kim, N.; Kim, H.S. Radiomic features and multilayer perceptron network classifier: A robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci. Rep. 2019, 9, 5746. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Zhang, L.; Xie, L.; Kong, R.; Zhang, H.; Chang, S. COVID-19 Imaging-based AI Research—A Literature Review. Curr. Med. Imaging 2022, 18, 496–508. [Google Scholar]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. 2021, 4, 65. [Google Scholar] [CrossRef] [PubMed]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Feng, H.; Lv, W.; Ashrafinia, S.; Yuan, Q.; Wang, Q.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; et al. Machine Learning Methods for Optimal Radiomics-Based Differentiation Between Recurrence and Inflammation: Application to Nasopharyngeal Carcinoma Post-therapy PET/CT Images. Mol. Imaging Biol. 2020, 22, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; He, X.; Ouyang, F.; Gu, D.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Tian, J.; Zhang, S. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017, 403, 21–27. [Google Scholar] [CrossRef]
- Xie, C.-Y.; Hu, Y.-H.; Ho, J.W.-K.; Han, L.-J.; Yang, H.; Wen, J.; Lam, K.-O.; Wong, I.Y.-H.; Law, S.Y.-K.; Chiu, K.W.-H.; et al. Using Genomics Feature Selection Method in Radiomics Pipeline Improves Prognostication Performance in Locally Advanced Esophageal Squamous Cell Carcinoma-A Pilot Study. Cancers 2021, 13, 2145. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abd Ghani, M.K.; Hamed, R.I.; Ibrahim, D.A. Review on Nasopharyngeal Carcinoma: Concepts, methods of analysis, segmentation, classification, prediction and impact: A review of the research literature. J. Comput. Sci. 2017, 21, 283–298. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Ghani, M.K.A.; Hamed, R.I.; Ibrahim, D.A. Analysis of an electronic methods for nasopharyngeal carcinoma: Prevalence, diagnosis, challenges and technologies. J. Comput. Sci. 2017, 21, 241–254. [Google Scholar] [CrossRef]
- Ng, W.T.; But, B.; Choi, H.C.W.; de Bree, R.; Lee, A.W.M.; Lee, V.H.F.; López, F.; Mäkitie, A.A.; Rodrigo, J.P.; Saba, N.F.; et al. Application of Artificial Intelligence for Nasopharyngeal Carcinoma Management—A Systematic Review. Cancer Manag. Res. 2022, 14, 339–366. [Google Scholar] [CrossRef]
- Abd Ghani, M.K.; Mohammed, M.A.; Arunkumar, N.; Mostafa, S.A.; Ibrahim, D.A.; Abdullah, M.K.; Jaber, M.M.; Abdulhay, E.; Ramirez-Gonzalez, G.; Burhanuddin, M.A. Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques. Neural Comput. Appl. 2020, 32, 625–638. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abd Ghani, M.K.; Hamed, R.I.; Ibrahim, D.A.; Abdullah, M.K. Artificial neural networks for automatic segmentation and identification of nasopharyngeal carcinoma. J. Comput. Sci. 2017, 21, 263–274. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Xie, L.; Xu, L.; Kong, R.; Chang, S.; Xu, X. Improvement of Prediction Performance With Conjoint Molecular Fingerprint in Deep Learning. Front. Pharmacol. 2020, 11, 606668. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, Nevada, 3–6 December 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Chuang, W.Y.; Chang, S.H.; Yu, W.H.; Yang, C.K.; Yeh, C.J.; Ueng, S.H.; Liu, Y.J.; Chen, T.D.; Chen, K.H.; Hsieh, Y.Y.; et al. Successful Identification of Nasopharyngeal Carcinoma in Nasopharyngeal Biopsies Using Deep Learning. Cancers 2020, 12, 507. [Google Scholar] [CrossRef]
- Xu, J.; Wang, J.; Bian, X.; Zhu, J.Q.; Tie, C.W.; Liu, X.; Zhou, Z.; Ni, X.G.; Qian, D. Deep Learning for nasopharyngeal Carcinoma Identification Using Both White Light and Narrow-Band Imaging Endoscopy. Laryngoscope 2022, 132, 999–1007. [Google Scholar] [CrossRef]
- Bai, X.; Hu, Y.; Gong, G.; Yin, Y.; Xia, Y. A deep learning approach to segmentation of nasopharyngeal carcinoma using computed tomography. Biomed. Signal Process. Control 2021, 64, 102246. [Google Scholar] [CrossRef]
- Lin, L.; Dou, Q.; Jin, Y.M.; Zhou, G.Q.; Tang, Y.Q.; Chen, W.L.; Su, B.A.; Liu, F.; Tao, C.J.; Jiang, N.; et al. Deep Learning for Automated Contouring of Primary Tumor Volumes by MRI for Nasopharyngeal Carcinoma. Radiology 2019, 291, 677–686. [Google Scholar] [CrossRef]
- Soniya; Paul, S.; Singh, L. A review on advances in deep learning. In Proceedings of the 2015 IEEE Workshop on Computational Intelligence: Theories, Applications and Future Directions (WCI), Kanpur, India, 14–17 December 2015; pp. 1–6. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Cheplygina, V. Cats or CAT scans: Transfer learning from natural or medical image source data sets? Curr. Opin. Biomed. Eng. 2019, 9, 21–27. [Google Scholar] [CrossRef]
- Studer, L.; Alberti, M.; Pondenkandath, V.; Goktepe, P.; Kolonko, T.; Fischer, A.; Liwicki, M.; Ingold, R. A Comprehensive Study of ImageNet Pre-Training for Historical Document Image Analysis. In Proceedings of the 2019 International Conference on Document Analysis and Recognition (ICDAR), Sydney, Australia, 20–25 September 2019; pp. 720–725. [Google Scholar]
- Li, C.; Jing, B.; Ke, L.; Li, B.; Xia, W.; He, C.; Qian, C.; Zhao, C.; Mai, H.; Chen, M.; et al. Development and validation of an endoscopic images-based deep learning model for detection with nasopharyngeal malignancies. Cancer Commun. 2018, 38, 59. [Google Scholar] [CrossRef]
- Wong, L.M.; Ai, Q.Y.H.; Mo, F.K.F.; Poon, D.M.C.; King, A.D. Convolutional neural network in nasopharyngeal carcinoma: How good is automatic delineation for primary tumor on a non-contrast-enhanced fat-suppressed T2-weighted MRI? Jpn. J. Radiol. 2021, 39, 571–579. [Google Scholar] [CrossRef]
- Wong, L.M.; King, A.D.; Ai, Q.Y.H.; Lam, W.K.J.; Poon, D.M.C.; Ma, B.B.Y.; Chan, K.C.A.; Mo, F.K.F. Convolutional neural network for discriminating nasopharyngeal carcinoma and benign hyperplasia on MRI. Eur. Radiol. 2021, 31, 3856–3863. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]




| Accuracy | Precision | F1-Score | AUC | |
|---|---|---|---|---|
| sCNN | 0.72 | 0.75 | 0.72 | 0.73 |
| ResNet50 | 0.64 | 0.67 | 0.59 | 0.64 |
| ResNet50-Weight | 0.97 | 0.97 | 0.97 | 0.97 |
| ResNet101 | 0.62 | 0.68 | 0.57 | 0.63 |
| ResNet101-Weight | 0.98 | 0.98 | 0.98 | 0.98 |
| EfficientNet | 0.56 | 0.60 | 0.52 | 0.56 |
| EfficientNet-Weight | 0.97 | 0.97 | 0.97 | 0.97 |
| Accuracy | Precision | F1-Score | AUC | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
| sCNN | 0.67 | 0.72 | 0.64 | 0.66 | 0.78 | 0.54 |
| ResNet50 | 0.74 | 0.82 | 0.71 | 0.76 | 0.66 | 0.86 |
| ResNet50-Weight | 0.93 | 0.94 | 0.93 | 0.94 | 0.90 | 0.98 |
| ResNet101 | 0.61 | 0.66 | 0.55 | 0.63 | 0.76 | 0.97 |
| ResNet101-Weight | 0.91 | 0.93 | 0.91 | 0.98 | 0.87 | 0.97 |
| EfficientNet | 0.75 | 0.79 | 0.73 | 0.76 | 0.72 | 0.80 |
| EfficientNet-Weight | 0.87 | 0.88 | 0.87 | 0.87 | 0.83 | 0.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, L.; Mao, R.; Wu, J.; Ge, C.; Xiao, F.; Xu, X.; Xie, L.; Gu, X. Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks. Diagnostics 2022, 12, 2478. https://doi.org/10.3390/diagnostics12102478
Ji L, Mao R, Wu J, Ge C, Xiao F, Xu X, Xie L, Gu X. Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks. Diagnostics. 2022; 12(10):2478. https://doi.org/10.3390/diagnostics12102478
Chicago/Turabian StyleJi, Li, Rongzhi Mao, Jian Wu, Cheng Ge, Feng Xiao, Xiaojun Xu, Liangxu Xie, and Xiaofeng Gu. 2022. "Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks" Diagnostics 12, no. 10: 2478. https://doi.org/10.3390/diagnostics12102478
APA StyleJi, L., Mao, R., Wu, J., Ge, C., Xiao, F., Xu, X., Xie, L., & Gu, X. (2022). Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks. Diagnostics, 12(10), 2478. https://doi.org/10.3390/diagnostics12102478






