Cervical Cell/Clumps Detection in Cytology Images Using Transfer Learning
Abstract
1. Introduction
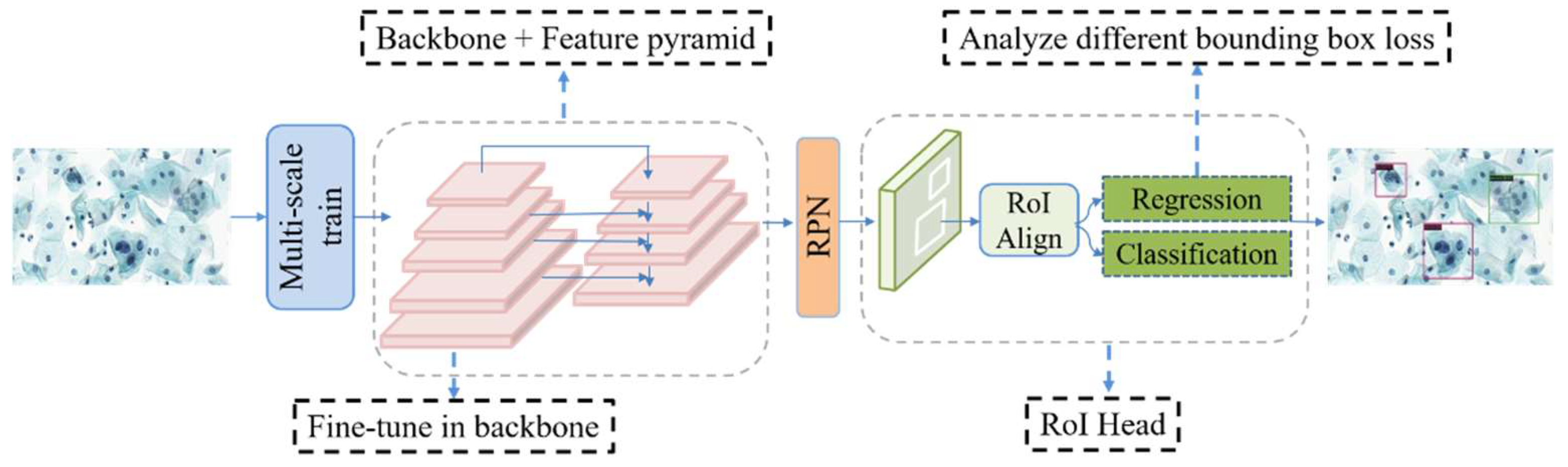
2. Materials and Methods
2.1. CNN-Based Object Detection
2.2. Transfer Learning
2.3. Multi-Scale Training
2.4. Bounding Box Loss
- (1)
- L1 loss:
- (2)
- L2 loss:
- (3)
- SmoothL1 loss:
- (4)
- IoU loss:
- (5)
- GIoU loss:
- (6)
- DIoU loss:
- (7)
- CIoU loss:
| Symbol | Explanation |
|---|---|
| The difference between the predicted value and the true value | |
| Prediction box | |
| Ground truth | |
| For and , find the smallest enclosing convex object | |
| Central point of | |
| Center point of | |
| Euclidean distance | |
| the diagonal length of the smallest enclosing box covering and | |
| positive trade-off parameter | |
| Measure the consistency of aspect ratio | |
| The width of | |
| The height of | |
| The width of | |
| The height of |
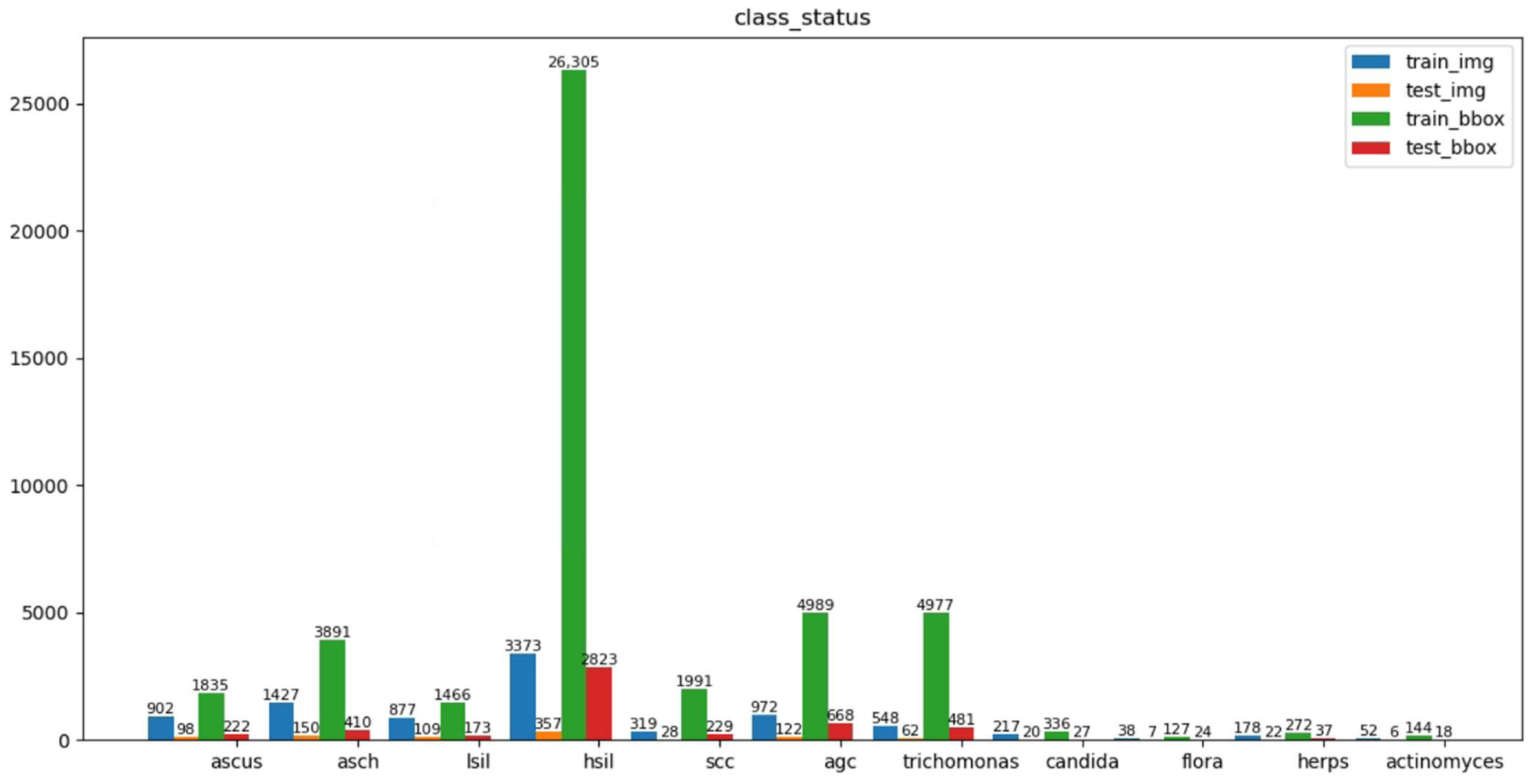
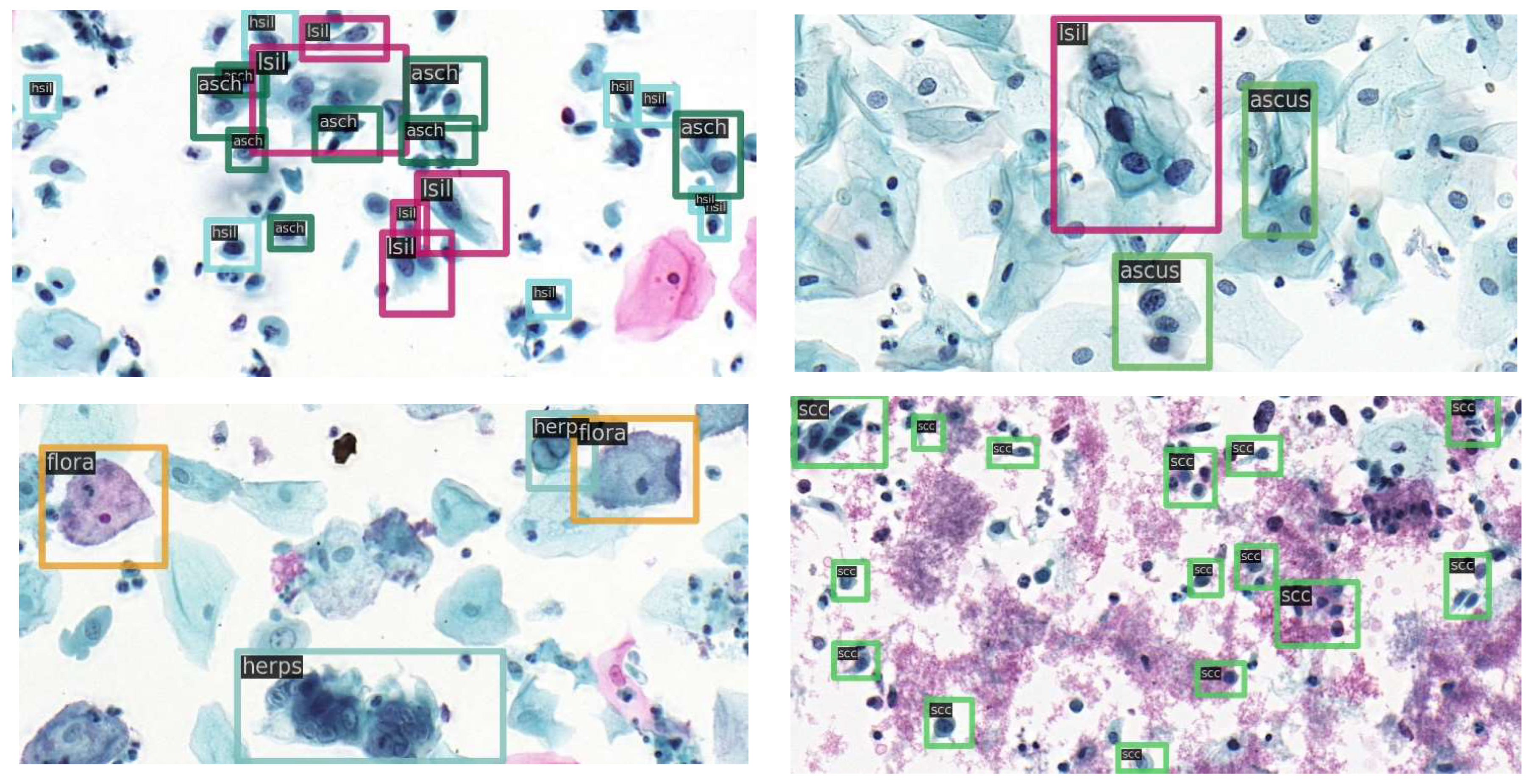
2.5. The Dataset
2.6. Experimental Setups
3. Results
3.1. Comparison of Transfer Learning for Different Source Data Domains
3.2. Multi-Scale Training
3.3. Fine-Tuning
3.4. Only Initialize the Backbone Network Parameters
3.5. Different Bounding Box Loss Experiments
3.6. Using Different Means and Stds for Multi-Scale Training
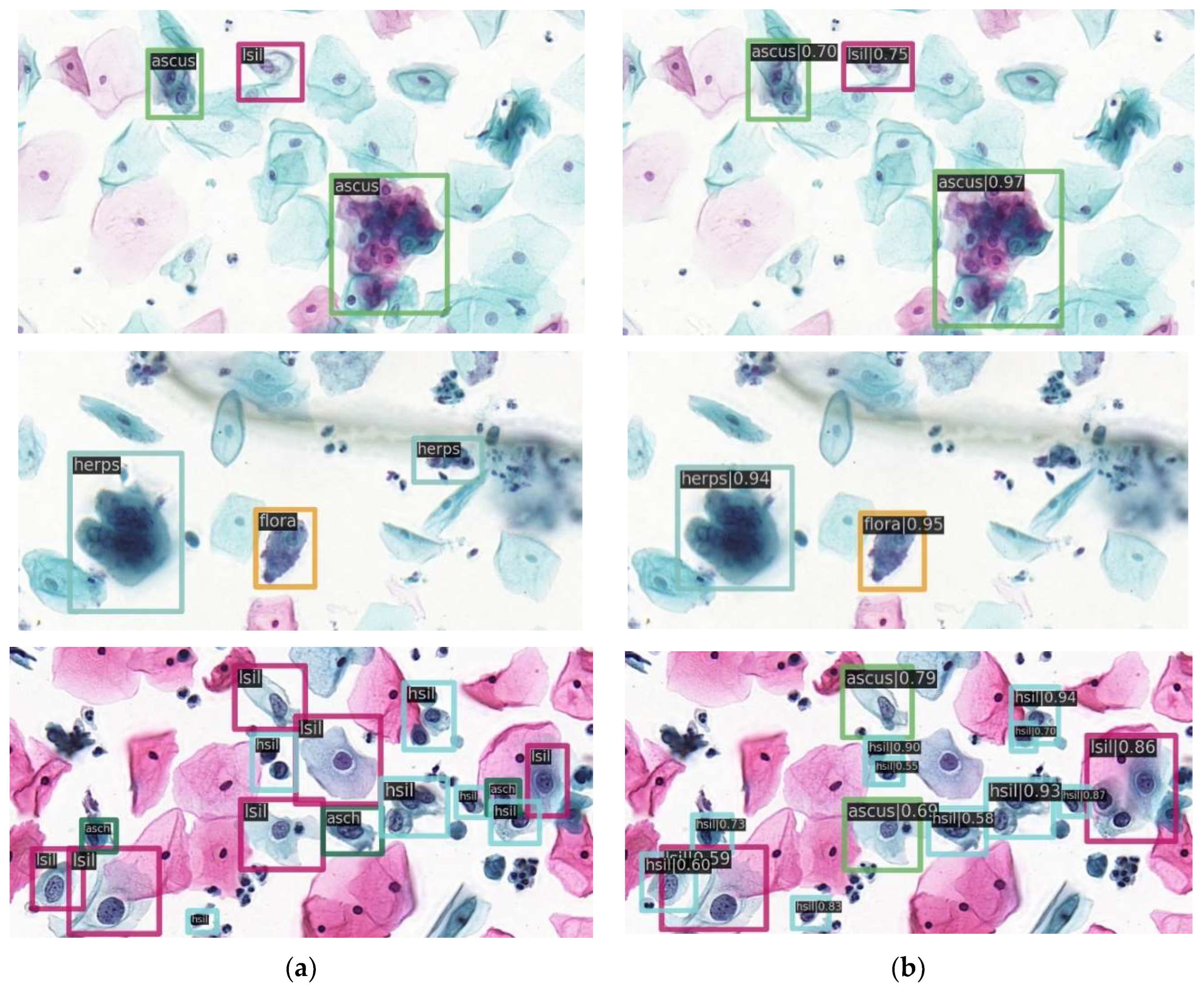
3.7. The Results with State-of-the-Art Methods
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahaman, M.M.; Li, C.; Wu, X.; Yao, Y.; Hu, Z.; Jiang, T.; Li, X.; Qi, S. A survey for cervical cytopathology image analysis using deep learning. IEEE Access 2020, 8, 61687–61710. [Google Scholar] [CrossRef]
- World Health Organization. WHO Guidelines for the Use of Thermal Ablation for Cervical Pre-Cancer Lesions; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018. [Google Scholar]
- Chaturvedi, A.K. Epidemiology and clinical aspects of HPV in head and neck cancers. Head Neck Pathol. 2012, 6, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Shiraz, A.; Crawford, R.; Egawa, N.; Griffin, H.; Doorbar, J. The early detection of cervical cancer. The current and changing landscape of cervical disease detection. Cytopathology 2020, 31, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Saslow, D.; Solomon, D.; Lawson, H.W.; Killackey, M.; Kulasingam, S.L.; Cain, J.; Garcia, F.A.R.; Moriarty, A.T.; Waxman, A.G.; Wilbur, D.C.; et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am. J. Clin. Pathol. 2012, 137, 516–542. [Google Scholar] [CrossRef]
- Parkin, D.M. Global cancer statistics in the year 2000. Lancet Oncol. 2001, 2, 533–543. [Google Scholar] [CrossRef]
- Richardson, L.; Tota, J.; Franco, E.L. Optimizing technology for cervical cancer screening in high-resource settings. Expert Rev. Obstet. Gynecol. 2011, 6, 343–353. [Google Scholar] [CrossRef]
- Huang, S. Research on Cervical Cancer Cell Detection Technology Based on Deep Learning. Master’s Thesis, Shanghai Normal University, Shanghai, China, 2021. [Google Scholar]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis-A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Pal, D.; Reddy, P.B.; Roy, S. Attention UW-Net: A fully connected model for automatic segmentation and annotation of chest X-ray. Comput. Biol. Med. 2022, 150, 106083. [Google Scholar] [CrossRef]
- Jafarbigloo, S.K.; Danyali, H. Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI Trans. Intell. Technol. 2021, 6, 426–439. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sadhukhan, B.; Sarkar, N.; Roy, D.; De, S. Stock market prediction using deep learning algorithms. CAAI Trans. Intell. Technol. 2021, 1–13. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, L.; Nogues, I.; Summers, R.M.; Liu, S.; Yao, J. DeepPap: Deep convolutional networks for cervical cell classification. IEEE J. Biomed. Health Inform. 2017, 21, 1633–1643. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Ma, Q.; Tan, L.; Wu, Y.; Xiao, J. Artificial classification of cervical squamous lesions in ThinPrep cytologic tests using a deep convolutional neural network. Oncol. Lett. 2020, 20, 113. [Google Scholar] [CrossRef]
- Lin, H.; Hu, Y.; Chen, S.; Yao, J.; Zhang, L. Fine-grained classification of cervical cells using morphological and appearance based convolutional neural networks. IEEE Access 2019, 7, 71541–71549. [Google Scholar] [CrossRef]
- Hussain, E.; Mahanta, L.B.; Das, C.R.; Talukdar, R.K. A comprehensive study on the multi-class cervical cancer diagnostic prediction on pap smear images using a fusion-based decision from ensemble deep convolutional neural network. Tissue Cell 2020, 65, 101347. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abdurahman, F.; Ayalew, Y.A. Single-cell conventional pap smear image classification using pre-trained deep neural network architectures. BMC Biomed. Eng. 2021, 3, 11. [Google Scholar] [CrossRef]
- Albuquerque, T.; Cruz, R.; Cardoso, J.S. Ordinal losses for classification of cervical cancer risk. PeerJ Comput. Sci. 2021, 7, e457. [Google Scholar] [CrossRef]
- Manna, A.; Kundu, R.; Kaplun, D.; Sinitca, A.; Sarkar, R. A fuzzy rank-based ensemble of CNN models for classification of cervical cytology. Sci. Rep. 2021, 11, 14538. [Google Scholar] [CrossRef]
- Rahaman, M.; Li, C.; Yao, Y.; Kulwa, F.; Wu, X.; Li, X.; Wang, Q. DeepCervix: A deep learning-based framework for the classification of cervical cells using hybrid deep feature fusion techniques. Comput. Biol. Med. 2021, 136, 104649. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, S.; Wang, B.; Dun, H.; Xu, L.; Huang, X.; Shi, E.; Feng, X. Generative adversarial network based data augmentation to improve cervical cell classification model. Math. Biosci. Eng. 2021, 18, 1740–1752. [Google Scholar] [CrossRef]
- Shi, J.; Wang, R.; Zheng, Y.; Jiang, Z.; Zhang, H.; Yu, L. Cervical cell classification with graph convolutional network. Comput. Methods Programs Biomed. 2021, 198, 105807. [Google Scholar] [CrossRef]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q.; Yin, Y. Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci. Rep. 2018, 38, BSR20181769. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dou, Q.; Yang, H.; Liu, J.; Fu, L.; Zhang, Y.; Zheng, L.; Zhang, D. Cervical cell multi-classification algorithm using global context information and attention mechanism. Tissue Cell 2022, 74, 101677. [Google Scholar] [CrossRef] [PubMed]
- Jantzen, J.; Norup, J.; Dounias, G.; Bjerregaard, B. Pap-smear benchmark data for pattern classification. Nat. Inspired Smart Inf. Syst. NiSIS 2005, 1–9. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. NIPS 2014, 27, 3320–3328. [Google Scholar]
- Xiang, Y.; Sun, W.; Pan, C.; Yan, M.; Yin, Z.; Liang, Y. A novel automation-assisted cervical cancer reading method based on convolutional neural network. Biocybern. Biomed. Eng. 2020, 40, 611–623. [Google Scholar] [CrossRef]
- Liang, Y.; Pan, C.; Sun, W.; Liu, Q.; Du, Y. Global context-aware cervical cell detection with soft scale anchor matching. Comput. Methods Programs Biomed. 2021, 204, 106061. [Google Scholar] [CrossRef]
- Xueyu, L.; Qinghua, L. Detection and classification of cervical exfoliated cells based on faster R-CNN. In Proceedings of the 11th International Conference on Advanced Infocomm Technology (ICAIT), Jinan, China, 18–20 October 2019. [Google Scholar]
- Pirovano, A.; Almeida, L.G.; Ladjal, S.; Bloch, I.; Berlemont, S. Computer-aided diagnosis tool for cervical cancer screening with weakly supervised localization and detection of abnormalities using adaptable and explainable classifier. Med. Image Anal. 2021, 73, 102167. [Google Scholar] [CrossRef]
- Chai, Z.; Luo, L.; Lin, H.; Chen, H.; Han, A.; Heng, P.-A. Deep Semi-supervised Metric Learning with Dual Alignment for Cervical Cancer Cell Detection. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022. [Google Scholar]
- Xie, Y.; Xing, F.; Shi, X.; Kong, X.; Su, H.; Yang, L. Efficient and robust cell detection: A structured regression approach. Med. Image Anal. 2018, 44, 245–254. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, D.; Wang, L.; Li, Y.; Chen, X.; Luo, R.; Che, S.; Liang, H.; Li, Y.; Liu, S.; et al. DCCL: A benchmark for cervical cytology analysis. In International Workshop on Machine Learning in Medical Imaging; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Liang, Y.; Tang, Z.; Yan, M.; Chen, J.; Liu, Q.; Xiang, Y. Comparison detector for cervical cell/clumps detection in the limited data scenario. Neurocomputing 2021, 437, 195–205. [Google Scholar] [CrossRef]
- Cheng, S.; Liu, S.; Yu, J.; Rao, G.; Xiao, Y.; Han, W.; Zhu, W.; Lv, X.; Li, N.; Cai, J.; et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun. 2021, 12, 5639. [Google Scholar] [CrossRef]
- Cao, L.; Yang, J.; Rong, Z.; Li, L.; Xia, B.; You, C.; Lou, G.; Jiang, L.; Du, C.; Meng, H.; et al. A novel attention-guided convolutional network for the detection of abnormal cervical cells in cervical cancer screening. Med. Image Anal. 2021, 73, 102197. [Google Scholar] [CrossRef]
- Li, X.; Xu, Z.; Shen, X.; Zhou, Y.; Xiao, B.; Li, T.-Q. Detection of cervical cancer cells in whole slide images using deformable and global context aware faster RCNN-FPN. Curr. Oncol. 2021, 28, 3585–3601. [Google Scholar] [CrossRef]
- Wei, Z.; Cheng, S.; Liu, X.; Zeng, S. An efficient cervical whole slide image analysis framework based on multi-scale semantic and spatial deep features. arXiv 2021, arXiv:2106.15113. [Google Scholar]
- Chen, T.; Zheng, W.; Ying, H.; Tan, X.; Li, K.; Li, X.; Chen, D.Z.; Wu, J. A Task Decomposing and Cell Comparing Method for Cervical Lesion Cell Detection. IEEE Trans. Med. Imaging 2022, 41, 2432–2442. [Google Scholar]
- Zhang, M.; Shen, L. Cervical Cell Detection Benchmark with Effective Feature Representation. In Proceedings of the International Conference on Cognitive Systems and Signal Processing (ICCSIP 2020), Zhuhai, China, 25–27 December 2020, Communications in Computer and Information Science; Springer: Singapore, 2020; Volume 1397. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Zitnick, C.L.; Dollár, P. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014. [Google Scholar]
- Yu, J.; Jiang, Y.; Wang, Z.; Cao, Z.; Huang, T. Unitbox: An advanced object detection network. In Proceedings of the 24th ACM international conference on Multimedia, Amsterdam, The Netherlands, 15–19 October 2016. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU loss: Faster and better learning for bounding box regression. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Processing Syst. 2015, 28. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Redmon, J.; Ali, F. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Redmon, J.; Ali, F. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Tian, Z.; Shen, C.; Chen, H.; He, T. Fcos: Fully convolutional one-stage object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019. [Google Scholar]
- Jiang, H.; Zhou, Y.; Lin, Y.; Chan, R.C.; Liu, J.; Chen, H. Deep Learning for Computational Cytology: A Survey. arXiv 2022, arXiv:2202.05126. [Google Scholar]
- Hongyi, D. Deep Learning of PyTorch Object Detection in Practice, 1st ed.; Mechanical Industry Press: Beijing, China, 2020; ISBN 978-7-111-64174-2. [Google Scholar]
- Liu, Z.; Gao, G.; Sun, L.; Fang, L. IPG-net: Image pyramid guidance network for small object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Plissiti, M.E.; Dimitrakopoulos, P.; Sfikas, G.; Nikou, C.; Krikoni, O.; Charchanti, A. SIPAKMED: A new dataset for feature and image based classification of normal and pathological cervical cells in Pap smear images. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018. [Google Scholar]
- Lu, Z.; Carneiro, G.; Bradley, A.P.; Ushizima, D.; Nosrati, M.S.; Bianchi, A.G.C.; Carneiro, C.M.; Hamarneh, G. Evaluation of three algorithms for the segmentation of overlapping cervical cells. IEEE J. Biomed. Health Inform. 2016, 21, 441–450. [Google Scholar] [CrossRef]
- Amorim, J.G.A.; Macarini, L.A.B.; Matias, A.V. A novel approach on segmentation of AgNOR-stained cytology images using deep learning. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020. [Google Scholar]
- Chen, K.; Wang, J.; Pang, J.; Cao, Y.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Xu, J.; et al. MMDetection: Open mmlab detection toolbox and benchmark. arXiv 2019, arXiv:1906.07155. [Google Scholar]

| Resize Input Image |
|---|
|
|
|
|
| Backbone | Initialization | mAP (%) | AR (%) |
|---|---|---|---|
| ResNet50 | None | 8.2 | 41.9 |
| ResNet50 | ImageNet | 51.9 | 83.3 |
| ResNet50 | Faster_rcnn_r50_fpn_1x_coco.pth | 58.4 | 84.1 |
| ResNet50 | Faster_rcnn_r50_fpn_2x_coco.pth | 57.6 | 85.9 |
| ResNet101 | None | 7.2 | 37.5 |
| ResNet101 | ImageNet | 58.3 | 86.0 |
| ResNet101 | Faster_rcnn_r101_fpn_1x_coco.pth | 58.5 | 84.7 |
| ResNet101 | Faster_rcnn_r101_fpn_2x_coco.pth | 59.8 | 84.0 |
| Backbone | mstrain | Initialization | mAP (%) | AR (%) |
|---|---|---|---|---|
| ResNet50 | n | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | 58.7 | 85.7 |
| ResNet50 | y | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | 60.9 | 87.2 |
| ResNet50 | y | Faster_rcnn_r50_fpn_1x_coco.pth | 60.0 | 86.3 |
| ResNet50 | y | Faster_rcnn_r50_fpn_2x_coco.pth | 60.5 | 86.4 |
| ResNet101 | n | Faster_rcnn_r101_fpn_mstrain_3x_coco.pth | 59.4 | 85.2 |
| ResNet101 | y | Faster_rcnn_r101_fpn_mstrain_3x_coco.pth | 62.2 | 86.9 |
| ResNet101 | y | Faster_rcnn_r101_fpn_1x_coco.pth | 61.3 | 88.2 |
| ResNet101 | y | Faster_rcnn_r101_fpn_2x_coco.pth | 61.7 | 87.3 |
| Backbone | Frozen_Stages | mAP (%) | AR (%) | Params (M) |
|---|---|---|---|---|
| ResNet50 | no | 59.4 | 87.1 | 41.4 |
| ResNet50 | Stem | 59.5 | 87.4 | 41.39 |
| ResNet50 | Stem + 1st | 60.9 | 87.2 | 41.17 |
| ResNet50 | Stem + first 2 | 59.9 | 86.6 | 39.95 |
| ResNet50 | Stem + first 3 | 54.9 | 84.9 | 32.86 |
| ResNet50 | Stem + first 4 | 49.4 | 84.5 | 17.89 |
| ResNet50 | Stem + 2nd | 59.4 | 87.4 | 40.17 |
| ResNet50 | Stem + 3rd | 58.5 | 86.7 | 34.29 |
| ResNet50 | Stem + 4th | 58.8 | 86.9 | 26.43 |
| Backbone | Frozen_Stages | mAP (%) | AR (%) | Params (M) |
|---|---|---|---|---|
| ResNet101 | no | 60.2 | 87.2 | 60.39 |
| ResNet101 | Stem | 61.5 | 86.8 | 60.38 |
| ResNet101 | Stem + 1st | 62.2 | 86.9 | 60.17 |
| ResNet101 | Stem + first 2 | 61.9 | 86.4 | 58.95 |
| ResNet101 | Stem + first 3 | 55.7 | 85.3 | 32.86 |
| ResNet101 | Stem + first 4 | 46.5 | 81.6 | 17.89 |
| Backbone | Initialization | mAP (%) | AR (%) |
|---|---|---|---|
| ResNet50 | None | 8.9 | 43.2 |
| ResNet50 | ImageNet | 57.4 | 86.0 |
| ResNet50 | faster_rcnn_r50_fpn_mstrain_3x_coco_only_backbone.pth | 57.5 | 86.5 |
| ResNet101 | None | 8.4 | 42.7 |
| ResNet101 | ImageNet | 57.9 | 87.0 |
| ResNet101 | faster_rcnn_r101_fpn_mstrain_3x_coco_only_backbone.pth | 59.8 | 86.8 |
| Model | Initialization | Bbox Loss | mAP (%) | AR (%) |
|---|---|---|---|---|
| ResNet50 | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | L1 | 60.9 | 87.2 |
| ResNet50 | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | SmoothL1 | 61.1 | 86.9 |
| ResNet50 | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | IoU | 60.4 | 87.3 |
| ResNet50 | Faster_rcnn_r50_fpn_mstrain_3x_coco.pth | GIou | 60.1 | 87.9 |
| Model | Initialization | Bbox Loss | mAP(%) | AR(%) |
|---|---|---|---|---|
| ResNet50 | Faster_rcnn_r50_fpn_1x _coco.pth | L1 | 58.4 | 84.1 |
| ResNet50 | Faster_rcnn_r50_fpn_1x _coco.pth | SmoothL1 | 59.2 | 84.3 |
| ResNet50 | Faster_rcnn_r50_fpn_iou_1x_coco.pth | IoU | 59.0 | 86.1 |
| ResNet50 | Faster_rcnn_r50_fpn_giou_1x_coco.pth | GIou | 59.7 | 84.8 |
| Backbone | Box Loss | Mean, std from Data | mAP (%) | AR (%) |
|---|---|---|---|---|
| ResNet50 | SmoothL1 | ImageNet | 61.1 | 86.9 |
| ResNet50 | SmoothL1 | coco | 61.2 | 87.7 |
| ResNet50 | SmoothL1 | self | 61.6 | 87.7 |
| Model | Comparison Detector [35] | Faster R-CNN [46] [45] | * Faster R-CNN | RetinaNet [56] | * RetinaNet |
|---|---|---|---|---|---|
| Initialization | ImageNet | ImageNet | COCO | ImageNet | COCO |
| mAP(%) | 48.8 | 51.9 | 61.6 | 53.8 | 57.2 |
| AR(%) | 64 | 83.3 | 87.7 | 81.4 | 88.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Li, M.; Li, G.; Zhang, Y.; Sun, C.; Bai, N. Cervical Cell/Clumps Detection in Cytology Images Using Transfer Learning. Diagnostics 2022, 12, 2477. https://doi.org/10.3390/diagnostics12102477
Xu C, Li M, Li G, Zhang Y, Sun C, Bai N. Cervical Cell/Clumps Detection in Cytology Images Using Transfer Learning. Diagnostics. 2022; 12(10):2477. https://doi.org/10.3390/diagnostics12102477
Chicago/Turabian StyleXu, Chuanyun, Mengwei Li, Gang Li, Yang Zhang, Chengjie Sun, and Nanlan Bai. 2022. "Cervical Cell/Clumps Detection in Cytology Images Using Transfer Learning" Diagnostics 12, no. 10: 2477. https://doi.org/10.3390/diagnostics12102477
APA StyleXu, C., Li, M., Li, G., Zhang, Y., Sun, C., & Bai, N. (2022). Cervical Cell/Clumps Detection in Cytology Images Using Transfer Learning. Diagnostics, 12(10), 2477. https://doi.org/10.3390/diagnostics12102477





