The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment
Abstract
1. Introduction
2. Diagnosis
3. Prognosis
3.1. Evolution of Prognostic Factors over the Last Decades
3.2. Prognostic Markers
3.2.1. Rai and Binet Staging Systems
3.2.2. Serum Markers
3.2.3. IGHV Status and Stereotypes
3.2.4. Immunophenotypic Markers
3.2.5. Chromosomal Alterations
3.2.6. TP53 and ATM Alterations
3.2.7. Novel Gene Mutations and Clonal Evolution
3.2.8. Noncoding Alterations and Epigenetics
3.3. Risk Scoring Systems
4. Treatment
4.1. Treatment Evolution on the Last Decades
4.1.1. Chemoimmunotherapy
4.1.2. Bruton Kinase Inhibitors
4.1.3. BCL-2 Inhibitors
4.1.4. PI3K Inhibitors
4.1.5. Immunotherapy
4.1.6. Combinations of Novel Agents
4.1.7. Cellular Therapy
4.2. Current Treatment Strategies
4.3. Drug Resistance
4.3.1. Ibrutinib Resistance
4.3.2. Venetoclax Resistance
4.4. COVID-19 and CLL Treatment
5. Current Challenges
- -
- How valid are the prognostic scores in the era of TA?
- -
- Will high-risk early-stage patients benefit from early treatment?
- -
- What are the practical implications of complex karyotype for treatment selection?
- -
- Which are the subclones responsible for disease evolution, and how do they acquire an expansion benefit?
- -
- What is the meaning of low-burden clonal and subclonal mutations?
- -
- Is there still a role for CIT alone or in combination with TA in frontline young, mutated CLL patients?
- -
- Will TA combinations be the best treatment option as the first-line therapy for all patients?
- -
- What is the optimal sequence of TA in the treatment of CLL patients?
- -
- What are the best options to overcome an acquired resistance with TA?
- -
- Will the acquired resistant mutations detected early evolve into an overt resistance in all patients?
- -
- What is the optimal timing for allo-TPH and/or CAR-T cells?
- -
- Should patients with targeted therapies discontinue treatment if they get a SARS-CoV-2 infection?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [PubMed]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Niemann, C.; Kater, A.; Gregor, M.; Cymbalista, F.; Buske, C.; Hillmen, P.; et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 23–33. [Google Scholar] [CrossRef]
- Burger, J.A.; O’Brien, S. Evolution of CLL treatment—From chemoimmunotherapy to targeted and individualized therapy. Nat. Rev. Clin. Oncol. 2018, 15, 510–527. [Google Scholar] [CrossRef]
- Kipps, T.J.; Stevenson, F.K.; Wu, C.J.; Croce, C.M.; Packham, G.; Wierda, W.G.; O’Brien, S.; Gribben, J.; Rai, K. Chronic lymphocytic leukaemia. Nat. Rev. Dis. Prim. 2017, 3, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Chiorazzi, N.; Rai, K.R.; Ferrarini, M. Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 352, 804–815. [Google Scholar] [CrossRef]
- Rodríguez-Vicente, A.E.; Díaz, M.G.; Hernández-Rivas, J.M. Chronic lymphocytic leukemia: A clinical and molecular heterogenous disease. Cancer Genet. 2013, 206, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Mattsson, M.; Stamatopoulos, K.; Rosenquist, R. Prognostic indices in chronic lymphocytic leukaemia: Where do we stand how do we proceed? J. Intern. Med. 2015, 279, 347–357. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Rossi, D.; Gaidano, G. Richter syndrome: Pathogenesis and management. Semin. Oncol. 2016, 43, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Juliusson, G.; Gahrton, G. Chromosome Aberrations in B-Cell Chronic Lymphocytic Leukemia. Pathogenetic and Clinical Implications. Cancer Genet. Cytogenet. 1990, 45, 143–160. [Google Scholar] [CrossRef]
- Döhner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Kröber, A.; Bullinger, L.; Döhner, K.; Bentz, M.; Lichter, P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.S.; Böttcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nat. Cell Biol. 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nat. Cell Biol. 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Guièze, R.; Wu, C.J. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood 2015, 126, 445–453. [Google Scholar] [CrossRef]
- Baliakas, P.; Jeromin, S.; Iskas, M.; Puiggros, A.; Plevova, K.; Nguyen-Khac, F.; Davis, Z.; Rigolin, G.M.; Visentin, A.; Xochelli, A.; et al. Cytogenetic complexity in chronic lymphocytic leukemia: Definitions, associations, and clinical impact. Blood 2019, 133, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Bosch, F.; Dalla-Favera, R. Chronic lymphocytic leukaemia: From genetics to treatment. Nat. Rev. Clin. Oncol. 2019, 16, 684–701. [Google Scholar] [CrossRef]
- Cheson, B.; Bennett, J.; Grever, M.; Kay, N.; Keating, M.; O’Brien, S.; Rai, K. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: Revised guidelines for diagnosis and treatment. Blood 1996, 87, 4990–4997. [Google Scholar] [CrossRef] [PubMed]
- Cheson, B.D.; Bennett, J.M.; Rai, K.R.; Grever, M.R.; Kay, N.E.; Schiffer, C.A.; Oken, M.M.; Keating, M.J.; Boldt, D.H.; Kempin, S.J.; et al. Guidelines for clinical protocols for chronic lymphocytic leukemia: Recommendations of the national cancer institute-sponsored working group. Am. J. Hematol. 1988, 29, 152–163. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Rozman, C.; Montserrat, E.; Rodríguez-Fernández, J.M.; Ayats, R.; Vallespí, T.; Parody, R.; Ríos, A.; Prados, D.; Morey, M.; Gomis, F. Bone Marrow Histologic Pattern—The Best Single Prognostic Parameter in Chronic Lymphocytic Leukemia: A Multivariate Survival Analysis of 329 Cases. Blood 1984, 64, 642–648. [Google Scholar] [CrossRef]
- Melo, J.V.; Catovsky, D.; Galton, D.A. The Relationship between Chronic Lymphocytic Leukaemia and Prolymphocytic Leukaemia. II. Patterns of Evolution of “prolymphocytoid” Transformation. Br. J. Haematol. 1986, 64, 77–86. [Google Scholar] [CrossRef]
- Matutes, E.; Owusu-Ankomah, K.; Morilla, R.; Marco, J.G.; Houlihan, A.; Que, T.H.; Catovsky, D. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994, 8, 1640–1645. [Google Scholar] [PubMed]
- Moreau, E.J.; Matutes, E.; A’Hern, M.R.P.; Morilla, A.M.; Morilla, M.R.M.; Owusu-Ankomah, K.A.; Seon, B.K.; Catovsky, D. Improvement of the Chronic Lymphocytic Leukemia Scoring System with the Monoclonal Antibody SN8(CD79b). Am. J. Clin. Pathol. 1997, 108, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Dragovic-Ivancevic, T.; Kraguljac-Kurtovic, N.; Knezevic, V.; Bogdanovic, A.; Mihaljevic, B.; Bozic, B.; Gotic, M. The role of immunophenotyping in differential diagnosis of chronic lymphocytic leukemia. Srp. Arh. Celok. Lek. 2014, 142, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Matutes, E. New additions to antibody panels in the characterisation of chronic lymphoproliferative disorders. J. Clin. Pathol. 2002, 55, 180–183. [Google Scholar] [CrossRef]
- Delgado, J.; Matutes, E.; Morilla, A.M.; Morilla, M.R.M.; Rafiq-Mohammed, M.F.; Del Giudice, I.; Catovsky, D.D.; Owusu-Ankomah, K.A. Diagnostic Significance of CD20 and FMC7 Expression in B-Cell Disorders. Am. J. Clin. Pathol. 2003, 120, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.J.M.; Lhermitte, L.; Böttcher, S.; Almeida, J.R.S.; Van Der Velden, V.H.J.; Floresmontero, J.; Rawstron, A.C.; Asnafi, V.; Lecrevisse, Q.; Lucio, P.; et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012, 26, 1908–1975. [Google Scholar] [CrossRef]
- Köhnke, T.; Wittmann, V.K.; Bücklein, V.L.; Lichtenegger, F.; Pasalic, Z.; Hiddemann, W.; Spiekermann, K.; Subklewe, M. Diagnosis of CLL revisited: Increased specificity by a modified five-marker scoring system including CD. Br. J. Haematol. 2017, 179, 480–487. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Kreuzer, K.-A.; Soosapilla, A.; Spacek, M.; Stehlikova, O.; Gambell, P.; McIver-Brown, N.; Villamor, N.; Psarra, K.; Arroz, M.; et al. Reproducible diagnosis of chronic lymphocytic leukemia by flow cytometry: An European Research Initiative on CLL (ERIC) & European Society for Clinical Cell Analysis (ESCCA) Harmonisation project. Cytom. Part B Clin. Cytom. 2018, 94, 121–128. [Google Scholar] [CrossRef]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Hillmen, P.; Hallek, M.; Buske, C. ESMO Guidelines Committee Chronic Lymphocytic Leukaemia: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2015, 26 (Suppl. 5), v78–v84. [Google Scholar] [CrossRef]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef] [PubMed]
- Marti, G.E.; Rawstron, A.C.; Ghia, P.; Hillmen, P.; Houlston, R.S.; Kay, N.; Schleinitz, T.A.; Caporaso, N. Diagnostic Criteria for Monoclonal B-Cell Lymphocytosis. Br. J. Haematol. 2005, 130, 325–332. [Google Scholar] [CrossRef]
- Strati, P.; Shanafelt, T.D. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: Diagnosis, natural history, and risk stratification. Blood 2015, 126, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Maitre, E.; Troussard, X. Monoclonal B-cell lymphocytosis. Best Pract. Res. Clin. Haematol. 2019, 32, 229–238. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Bennett, F.L.; O’Connor, S.J.M.; Kwok, M.; Fenton, J.A.L.; Plummer, M.; De Tute, R.; Owen, R.G.; Richards, S.J.; Jack, A.S.; et al. Monoclonal B-Cell Lymphocytosis and Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2008, 359, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Gaidano, G. Richter syndrome: Molecular insights and clinical perspectives. Hematol. Oncol. 2009, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.-M.; Keating, M.J. Richter Syndrome: Biology, Incidence, and Therapeutic Strategies. Cancer 2005, 103, 216–228. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Deambrogi, C.; Rasi, S.; Laurenti, L.; Stamatopoulos, K.; Arcaini, L.; Lucioni, M.; Rocque, G.B.; Xu-Monette, Z.Y.; et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood 2011, 117, 3391–3401. [Google Scholar] [CrossRef] [PubMed]
- Rozman, C.; Montserrat, E. Chronic Lymphocytic Leukemia. N. Engl. J. Med. 1995, 333, 1052–1057. [Google Scholar] [CrossRef]
- Rigolin, G.M.; Saccenti, E.; Bassi, C.; Lupini, L.; Quaglia, F.M.; Cavallari, M.; Martinelli, S.; Formigaro, L.; Lista, E.; Bardi, M.A.; et al. Extensive next-generation sequencing analysis in chronic lymphocytic leukemia at diagnosis: Clinical and biological correlations. J. Hematol. Oncol. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Rai, K.R.; Jain, P. Chronic lymphocytic leukemia (CLL)-Then and now. Am. J. Hematol. 2016, 91, 330–340. [Google Scholar] [CrossRef]
- Rai, K.R.; Sawitsky, A.; Cronkite, E.P.; Chanana, A.D.; Levy, R.N.; Pasternack, B.S. Clinical Staging of Chronic Lymphocytic Leukemia. Blood 1975, 46, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Binet, J.L.; Auquier, A.; Dighiero, G.; Chastang, C.; Piguet, H.; Goasguen, J.; Vaugier, G.; Potron, G.; Colona, P.; Oberling, F.; et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 1981, 48, 198–206. [Google Scholar] [CrossRef]
- Viñolas, N.; Reverter, J.C.; Urbano-Ispizua, A.; Montserrat, E.; Rozman, C. Lymphocyte Doubling Time in Chronic Lymphocytic Leukemia: An Update of Its Prognostic Significance. Blood Cells 1987, 12, 457–470. [Google Scholar]
- Rozman, C.; Hernandez-Nieto, L.; Montserrat, E.; Brugués, R. Prognostic Significance of Bone-Marrow Patterns in Chronic Lymphocytic Leukaemia. Br. J. Haematol. 1981, 47, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Wanders, L.; Ostwald, M.; Busch, R.; Senekowitsch, R.; Stern, S.; Schick, H.-D.; Kuhn-Hallek, I.; Emmerich, B. Serum β2-Microglobulin and Serum Thymidine Kinase are Independent Predictors of Progression-Free Survival in Chronic Lymphocytic Leukemia and Immunocytoma. Leuk. Lymphoma 1996, 22, 439–447. [Google Scholar] [CrossRef]
- Wierda, W.G.; O’Brien, S.; Wang, X.; Faderl, S.; Ferrajoli, A.; Do, K.-A.; Cortes, J.; Thomas, D.; Garcia-Manero, G.; Koller, C.; et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood 2007, 109, 4679–4685. [Google Scholar] [CrossRef]
- Wierda, W.G.; O’Brien, S.; Wang, X.; Faderl, S.; Ferrajoli, A.; Do, K.-A.; Garcia-Manero, G.; Cortes, J.; Thomas, D.; Koller, C.; et al. Characteristics Associated with Important Clinical End Points in Patients with Chronic Lymphocytic Leukemia at Initial Treatment. J. Clin. Oncol. 2009, 27, 1637–1643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Magnac, C.; Porcher, R.; Davi, F.; Nataf, J.; Payelle-Brogard, B.; Tang, R.P.; Oppezzo, P.; Levy, V.; Dighiero, G.; Ajchenbaum-Cymbalista, F. Predictive value of serum thymidine kinase level for Ig-V mutational status in B-CLL. Leukemia 2003, 17, 133–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pratt, G.; Thomas, P.; Marden, N.; Alexander, D.; Davis, Z.; Hussey, D.; Parry, H.; Harding, S.; Catovsky, D.; Begley, J.; et al. Evaluation of serum markers in the LRF CLL4 trial: β2-microglobulin but not serum free light chains, is an independent marker of overall survival. Leuk. Lymphoma 2016, 57, 2342–2350. [Google Scholar] [CrossRef]
- Autore, F.; Strati, P.; Innocenti, I.; Corrente, F.; Trentin, L.; Cortelezzi, A.; Visco, C.; Coscia, M.; Cuneo, A.; Gozzetti, A.; et al. Elevated Lactate Dehydrogenase Has Prognostic Relevance in Treatment-Naïve Patients Affected by Chronic Lymphocytic Leukemia with Trisomy. Cancers 2019, 11, 896. [Google Scholar] [CrossRef]
- Pflug, N.; Bahlo, J.; Shanafelt, T.D.; Eichhorst, B.F.; Bergmann, M.A.; Elter, T.; Bauer, K.; Malchau, G.; Rabe, K.G.; Stilgenbauer, S.; et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood 2014, 124, 49–62. [Google Scholar] [CrossRef]
- Cramer, P.; Hallek, M. Prognostic factors in chronic lymphocytic leukemia—What do we need to know? Nat. Rev. Clin. Oncol. 2010, 8, 38–47. [Google Scholar] [CrossRef]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V Gene Mutation Status and CD38 Expression as Novel Prognostic Indicators in Chronic Lymphocytic Leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F.K. Unmutated Ig VH Genes Are Associated with a More Aggressive Form of Chronic Lymphocytic Leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, G.; Dalla-Favera, G.F.R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat. Rev. Cancer 2016, 16, 145–162. [Google Scholar] [CrossRef]
- Parikh, S.A.; Shanafelt, T.D. Risk Factors for Richter Syndrome in Chronic Lymphocytic Leukemia. Curr. Hematol. Malign-Rep. 2014, 9, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Crombie, J.; Davids, M.S. IGHVmutational status testing in chronic lymphocytic leukemia. Am. J. Hematol. 2017, 92, 1393–1397. [Google Scholar] [CrossRef]
- Thompson, P.A.; Tam, C.S.; O’Brien, S.M.; Wierda, W.G.; Stingo, F.; Plunkett, W.; Smith, S.C.; Kantarjian, H.M.; Freireich, E.J.; Keating, M.J. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood 2016, 127, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Bahlo, J.; Fink, A.M.; Goede, V.; Herling, C.D.; Cramer, P.; Langerbeins, P.; Von Tresckow, J.; Engelke, A.; Maurer, C.; et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: Updated results of the CLL8 trial. Blood 2016, 127, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, S.; Agathangelidis, A.; Schneider, C.; Bahlo, J.; Robrecht, S.; Tausch, E.; Bloehdorn, J.; Hoechstetter, M.; Fischer, K.; Eichhorst, B.; et al. Prognostic impact of prevalent chronic lymphocytic leukemia stereotyped subsets: Analysis within prospective clinical trials of the German CLL Study Group (GCLLSG). Haematologica 2019, 105, 2598–2607. [Google Scholar] [CrossRef]
- Rosenquist, R.; Ghia, P.; Hadzidimitriou, A.; Sutton, L.-A.; Agathangelidis, A.; Baliakas, P.; Darzentas, N.; Giudicelli, V.; Lefranc, M.-P.; Langerak, A.W.; et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: Updated ERIC recommendations. Leukemia 2017, 31, 1477–1481. [Google Scholar] [CrossRef] [PubMed]
- Rassenti, L.Z.; Huynh, L.; Toy, T.L.; Chen, L.; Keating, M.J.; Gribben, J.G.; Neuberg, D.S.; Flinn, I.W.; Rai, K.R.; Byrd, J.C.; et al. ZAP-70 Compared with Immunoglobulin Heavy-Chain Gene Mutation Status as a Predictor of Disease Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2004, 351, 893–901. [Google Scholar] [CrossRef]
- Falay, M.; Ceran, F.; Güneş, A.; Dagdas, S.; Ayli, M.; Özet, G. CD38 Expression and Variation as a Prognostic Factor Chronic Lymphocytic Leukemia. Clin. Lab. 2016, 62, 1287–1293. [Google Scholar] [CrossRef]
- Crespo, M.; Bosch, F.; Villamor, N.; Bellosillo, B.; Colomer, D.; Rozman, M.; Marcé, S.; López-Guillermo, A.; Campo, E.; Montserrat, E. ZAP-70 Expression as a Surrogate for Immunoglobulin-Variable-Region Mutations in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2003, 348, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Wiestner, A.; Rosenwald, A.; Barry, T.S.; Wright, G.; Davis, R.E.; Henrickson, S.E.; Zhao, H.; Ibbotson, R.E.; Orchard, J.A.; Davis, Z.; et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood 2003, 101, 4944–4951. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Zucchetto, A.; Rossi, F.M.; Capello, D.; Cerri, M.; Deambrogi, C.; Cresta, S.; Rasi, S.; De Paoli, L.; Bodoni, C.L.; et al. CD49d expression is an independent risk factor of progressive disease in early stage chronic lymphocytic leukemia. Haematologica 2008, 93, 1575–1579. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vosoughi, T.; Bagheri, M.; Hosseinzadeh, M.; Ehsanpour, A.; Davari, N.; Saki, N. CD markers variations in chronic lymphocytic leukemia: New insights into prognosis. J. Cell. Physiol. 2019, 234, 19420–19439. [Google Scholar] [CrossRef]
- Tissino, E.; Pozzo, F.; Benedetti, D.; Caldana, C.; Bittolo, T.; Rossi, F.M.; Bomben, R.; Nanni, P.; Chivilò, H.; Cattarossi, I.; et al. CD49d promotes disease progression in chronic lymphocytic leukemia: New insights from CD49d bimodal expression. Blood 2020, 135, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Juliusson, G.; Oscier, D.G.; Fitchett, M.; Ross, F.M.; Stockdill, G.; Mackie, M.J.; Parker, A.C.; Castoldi, G.L.; Cuneo, A.; Knuutila, S.; et al. Prognostic Subgroups in B-Cell Chronic Lymphocytic Leukemia Defined by Specific Chromosomal Abnormalities. N. Engl. J. Med. 1990, 323, 720–724. [Google Scholar] [CrossRef]
- Dierlamm, J.; Michaux, L.; Criel, A.; Wlodarska, I.; Berghe, H.V.D.; Hossfeld, D.K. Genetic abnormalities in chronic lymphocytic leukemia and their clinical and prognostic implications. Cancer Genet. Cytogenet. 1997, 94, 27–35. [Google Scholar] [CrossRef]
- Haferlach, C.; Bacher, U. Cytogenetic Methods in Chronic Lymphocytic Leukemia. Methods Mol. Biol. 2011, 730, 119–130. [Google Scholar] [CrossRef]
- Hernández, J.Á.; Rodríguez, A.E.; González, M.; Benito, R.; Fontanillo, C.; Sandoval, V.; Romero, M.; Martín-Núñez, G.; De Coca, A.G.; Fisac, R.; et al. A high number of losses in 13q14 chromosome band is associated with a worse outcome and biological differences in patients with B-cell chronic lymphoid leukemia. Haematologica 2009, 94, 364–371. [Google Scholar] [CrossRef]
- Hernández, J.A.; Hernández-Sánchez, M.; Rodríguez-Vicente, A.E.; Grossmann, V.; Collado, R.; Heras, C.; Puiggros, A.; Martín, A.Á.; Puig, N.; Benito, R.; et al. A Low Frequency of Losses in 11q Chromosome Is Associated with Better Outcome and Lower Rate of Genomic Mutations in Patients with Chronic Lymphocytic Leukemia. PLoS ONE 2015, 10, e0143073. [Google Scholar] [CrossRef] [PubMed]
- Marín, I.G.-G.Y.; Hernández-Sánchez, M.; Rodríguez-Vicente, A.-E.; Sanzo, C.; Aventín, A.; Puiggros, A.; Collado, R.; Heras, C.; Muñoz, C.; Delgado, J.; et al. A high proportion of cells carrying trisomy 12 is associated with a worse outcome in patients with chronic lymphocytic leukemia. Hematol. Oncol. 2015, 34, 84–92. [Google Scholar] [CrossRef]
- Van Dyke, D.L.; Werner, L.; Rassenti, L.Z.; Neuberg, D.; Ghia, E.; Heerema, N.A.; Cin, P.D.; Aquila, M.D.; Sreekantaiah, C.; Greaves, A.W.; et al. The Dohner fluorescence in situ hybridization prognostic classification of chronic lymphocytic leukaemia (CLL): The CLL Research Consortium experience. Br. J. Haematol. 2016, 173, 105–113. [Google Scholar] [CrossRef]
- Tam, C.S.; Shanafelt, T.D.; Wierda, W.G.; Abruzzo, L.V.; Van Dyke, D.L.; O’Brien, S.; Ferrajoli, A.; Lerner, S.A.; Lynn, A.; Kay, N.E.; et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: The M. D. Anderson and Mayo Clinic experience. Blood 2009, 114, 957–964. [Google Scholar] [CrossRef]
- Yuan, Y.-Y.; Zhu, H.-Y.; Wu, J.-Z.; Xia, Y.; Liang, J.-H.; Wu, W.; Cao, L.; Wang, L.; Fan, L.; Li, J.-Y.; et al. The percentage of cells with 17p deletion and the size of 17p deletion subclones show prognostic significance in chronic lymphocytic leukemia. Genes Chromosom. Cancer 2019, 58, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Cavazzini, F.; Hernandez, J.A.; Gozzetti, A.; Rossi, A.R.; De Angeli, C.; Tiseo, R.; Bardi, A.; Tammiso, E.; Crupi, R.; Lenoci, M.P.; et al. Chromosome 14q32 translocations involving the immunoglobulin heavy chain locus in chronic lymphocytic leukaemia identify a disease subset with poor prognosis. Br. J. Haematol. 2008, 142, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Kong, Y.; Yue, C. Genetic and immunophenotypic profile of IGH@ rearrangement detected by fluorescence in situ hybridization in 149 cases of B-cell chronic lymphocytic leukemia. Cancer Genet. Cytogenet. 2010, 196, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Carretero, C.; Hernández-Sánchez, M.; González, T.; Quijada-Álamo, M.; Martín-Izquierdo, M.; Hernández-Sánchez, J.; Vidal, M.; De Coca, A.G.; Aguilar, C.; Vargas-Pabón, M.; et al. Chronic lymphocytic leukemia patients with IGH translocations are characterized by a distinct genetic landscape with prognostic implications. Int. J. Cancer 2020, 147, 2780–2792. [Google Scholar] [CrossRef]
- Cuneo, A.; Rigolin, G.M.; Bigoni, R.; De Angeli, C.; Veronese, A.; Cavazzini, F.; Bardi, A.; Roberti, M.G.; Tammiso, E.; Agostini, P.; et al. Chronic lymphocytic leukemia with 6q−shows distinct hematological features and intermediate prognosis. Leukemia 2003, 18, 476–483. [Google Scholar] [CrossRef]
- Wang, D.-M.; Miao, K.-R.; Fan, L.; Qiu, H.-R.; Fang, C.; Zhu, D.-X.; Qiu, H.-X.; Xu, W.; Li, J.-Y. Intermediate prognosis of 6q deletion in chronic lymphocytic leukemia. Leuk. Lymphoma 2011, 52, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, D.; Pantic, M.; Skatulla, I.; Rawluk, J.; Kreutz, C.; Martens, U.M.; Fisch, P.; Timmer, J.; Veelken, H.; De Laat, B.; et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood 2006, 109, 1202–1210. [Google Scholar] [CrossRef]
- Gunn, S.R.; Mohammed, M.S.; Gorre, M.E.; Cotter, P.D.; Kim, J.; Bahler, D.W.; Preobrazhensky, S.N.; Higgins, R.A.; Bolla, A.R.; Ismail, S.H.; et al. Whole-Genome Scanning by Array Comparative Genomic Hybridization as a Clinical Tool for Risk Assessment in Chronic Lymphocytic Leukemia. J. Mol. Diagn. 2008, 10, 442–451. [Google Scholar] [CrossRef]
- Kujawski, L.; Ouillette, P.; Erba, H.; Saddler, C.; Jakubowiak, A.; Kaminski, M.; Shedden, K.; Malek, S.N. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood 2008, 112, 1993–2003. [Google Scholar] [CrossRef]
- Rodríguez, A.E.; Robledo, C.; García, J.L.; González, M.; Gutiérrez, N.C.; Hernández, J.A.; Sandoval, V.; De Coca, A.G.; Recio, I.; Risueño, A.; et al. Identification of a novel recurrent gain on 20q13 in chronic lymphocytic leukemia by array CGH and gene expression profiling. Ann. Oncol. 2012, 23, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Puiggros, A.; Xochelli, A.; Sutton, L.-A.; Nguyen-Khac, F.; Gardiner, A.; Plevova, K.; Minga, E.; Hadzidimitriou, A.; Walewska, R.; et al. Additional trisomies amongst patients with chronic lymphocytic leukemia carrying trisomy 12: The accompanying chromosome makes a difference. Haematology 2016, 101, e299–e302. [Google Scholar] [CrossRef] [PubMed]
- Leeksma, A.C.; Baliakas, P.; Moysiadis, T.; Puiggros, A.; Plevova, K.; Van Der Kevie-Kersemaekers, A.-M.; Posthuma, H.; Rodriguez-Vicente, A.E.; Tran, A.N.; Barbany, G.; et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: A multi-center study. Haematologica 2020, 106, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Rigolin, G.M.; Cibien, F.; Martinelli, S.; Formigaro, L.; Rizzotto, L.; Tammiso, E.; Saccenti, E.; Bardi, A.; Cavazzini, F.; Ciccone, M.; et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with “normal” FISH: Correlations with clinicobiologic parameters. Blood 2012, 119, 2310–2313. [Google Scholar] [CrossRef]
- Jaglowski, S.M.; Ruppert, A.S.; Heerema, N.A.; Bingman, A.; Flynn, J.M.; Grever, M.R.; Jones, J.A.; Elder, P.; Devine, S.M.; Byrd, J.C.; et al. Complex karyotype predicts for inferior outcomes following reduced-intensity conditioning allogeneic transplant for chronic lymphocytic leukaemia. Br. J. Haematol. 2012, 159, 82–87. [Google Scholar] [CrossRef]
- Puiggros, A.; Collado, R.; Calasanz, M.J.; Ortega, M.; Ruiz-Xivillé, N.; Rivas-Delgado, A.; Luño, E.; González, T.; Navarro, B.; García-Malo, M.; et al. Patients with chronic lymphocytic leukemia and complex karyotype show an adverse outcome even in absence of TP53/ATM FISH deletions. Oncotarget 2017, 8, 54297–54303. [Google Scholar] [CrossRef] [PubMed]
- Marín, I.G.-G.Y.; Hernández-Sanchez, M.; Rodríguez-Vicente, A.E.; Puiggros, A.; Collado, R.; Luño, E.; González, T.; Ruiz-Xivillé, N.; Ortega, M.; Gimeno, E.; et al. Characterizing patients with multiple chromosomal aberrations detected by FISH in chronic lymphocytic leukemia. Leuk. Lymphoma 2017, 59, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Herling, C.D.; Klaumünzer, M.; Rocha, C.K.; Altmüller, J.; Thiele, H.; Bahlo, J.; Kluth, S.; Crispatzu, G.; Herling, M.; Schiller, J.; et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood 2016, 128, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Le Bris, Y.; Struski, S.; Guièze, R.; Rouvellat, C.; Prade, N.; Troussard, X.; Tournilhac, O.; Béné, M.C.; Delabesse, E.; Ysebaert, L. Major prognostic value of complex karyotype in addition toTP53andIGHVmutational status in first-line chronic lymphocytic leukemia. Hematol. Oncol. 2016, 35, 664–670. [Google Scholar] [CrossRef]
- Thompson, P.A.; O’Brien, S.M.; Wierda, W.G.; Ferrajoli, A.; Stingo, F.; Smith, S.C.; Burger, J.A.; Estrov, Z.; Jain, N.; Kantarjian, H.M.; et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer 2015, 121, 3612–3621. [Google Scholar] [CrossRef]
- Anderson, M.A.; Tam, C.; Lew, T.E.; Juneja, S.; Juneja, M.; Westerman, D.; Wall, M.; Lade, S.; Gorelik, A.; Huang, D.C.S.; et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood 2017, 129, 3362–3370. [Google Scholar] [CrossRef]
- Zenz, T.; Vollmer, D.; Trbusek, M.; Smardova, J.; Benner, A.; Soussi, T.; Helfrich, H.; Heuberger, M.; Hoth, P.; Fuge, M.; et al. TP53 Mutation Profile in Chronic Lymphocytic Leukemia: Evidence for a Disease Specific Profile from a Comprehensive Analysis of 268 Mutations. Leukemia 2010, 24, 2072–2079. [Google Scholar] [CrossRef]
- Zenz, T.; Kröber, A.; Scherer, K.; Häbe, S.; Bühler, A.; Benner, A.; Denzel, T.; Winkler, D.; Edelmann, J.; Schwänen, C.; et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: Results from a detailed genetic characterization with long-term follow-up. Blood 2008, 112, 3322–3329. [Google Scholar] [CrossRef]
- Malcikova, J.; Smardova, J.; Rocnova, L.; Tichy, B.; Kuglik, P.; Vranova, V.; Cejkova, S.; Svitakova, M.; Francova, H.S.; Brychtova, Y.; et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: Selection, impact on survival, and response to DNA damage. Blood 2009, 114, 5307–5314. [Google Scholar] [CrossRef]
- Zenz, T.; Eichhorst, B.; Busch, R.; Denzel, T.; Häbe, S.; Winkler, D.; Bühler, A.; Edelmann, J.; Bergmann, M.; Hopfinger, G.; et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2010, 28, 4473–4479. [Google Scholar] [CrossRef]
- Rossi, D.; Cerri, M.; Deambrogi, C.; Sozzi, E.; Cresta, S.; Rasi, S.; De Paoli, L.; Spina, V.; Gattei, V.; Capello, D.; et al. The Prognostic Value of TP53 Mutations in Chronic Lymphocytic Leukemia Is Independent of Del17p13: Implications for Overall Survival and Chemorefractoriness. Clin. Cancer Res. 2009, 15, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Dicker, F.; Herholz, H.; Schnittger, S.; Nakao, A.; Patten, N.; Wu, L.; Kern, W.; Haferlach, T. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia 2008, 23, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.; Martinez, P.; Wade, R.; Hockley, S.; Oscier, D.; Matutes, E.; Dearden, C.E.; Richards, S.M.; Catovsky, D.; Morgan, G.J. Mutational Status of the TP53 Gene as a Predictor of Response and Survival in Patients with Chronic Lymphocytic Leukemia: Results From the LRF CLL4 Trial. J. Clin. Oncol. 2011, 29, 2223–2229. [Google Scholar] [CrossRef]
- Pospisilova, S.; Gonzalez, D.; Malcikova, J.; Trbusek, M.; Rossi, D.; Kater, A.P.; Cymbalista, F.; Eichhorst, B.; Hallek, M.; Döhner, H.; et al. ERIC Recommendations on TP53 Mutation Analysis in Chronic Lymphocytic Leukemia. Leukemia 2012, 26, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Malcikova, J.; Tausch, E.; Rossi, D.; Sutton, L.A.; Soussi, T.; Zenz, T.; Kater, A.P.; Niemann, C.U.; Gonzalez, D.; Davi, F.; et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia—Update on methodological approaches and results interpretation. Leukemia 2018, 32, 1070–1080. [Google Scholar] [CrossRef]
- Austen, B.; Powell, J.E.; Alvi, A.; Edwards, I.; Hooper, L.; Starczynski, J.; Taylor, A.M.R.; Fegan, C.; Moss, P.; Stankovic, T. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood 2005, 106, 3175–3182. [Google Scholar] [CrossRef]
- Austen, B.; Skowronska, A.; Baker, C.; Powell, J.E.; Gardiner, A.; Oscier, D.; Majid, A.; Dyer, M.; Siebert, R.; Taylor, A.M.; et al. Mutation Status of the Residual ATM Allele Is an Important Determinant of the Cellular Response to Chemotherapy and Survival in Patients with Chronic Lymphocytic Leukemia Containing an 11q Deletion. J. Clin. Oncol. 2007, 25, 5448–5457. [Google Scholar] [CrossRef]
- Skowronska, A.; Parker, A.; Ahmed, G.; Oldreive, C.; Davis, Z.; Richards, S.; Dyer, M.; Matutes, E.; González, D.; Taylor, A.M.R.; et al. Biallelic ATM Inactivation Significantly Reduces Survival in Patients Treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 Trial. J. Clin. Oncol. 2012, 30, 4524–4532. [Google Scholar] [CrossRef]
- Kipps, T.J.; Fraser, G.; Coutre, S.E.; Brown, J.R.; Barrientos, J.C.; Barr, P.M.; Byrd, J.C.; O’Brien, S.M.; Dilhuydy, M.-S.; Hillmen, P.; et al. Long-Term Studies Assessing Outcomes of Ibrutinib Therapy in Patients With Del(11q) Chronic Lymphocytic Leukemia. Clin. Lymphoma Myeloma Leuk. 2019, 19, 715–722.e6. [Google Scholar] [CrossRef]
- Barr, P.M.; Robak, T.; Owen, C.; Tedeschi, A.; Bairey, O.; Bartlett, N.L.; Burger, J.A.; Hillmen, P.; Coutre, S.; Devereux, S.; et al. Sustained efficacy and detailed clinical follow-up of first-line ibrutinib treatment in older patients with chronic lymphocytic leukemia: Extended phase 3 results from RESONATE-2. Haematologica 2018, 103, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Furman, R.R.; Coutre, S.E.; Flinn, I.W.; Burger, J.A.; Blum, K.A.; Sharman, J.P.; Wierda, W.; Zhao, W.; Heerema, N.A.; et al. Ibrutinib Treatment for First-Line and Relapsed/Refractory Chronic Lymphocytic Leukemia: Final Analysis of the Pivotal Phase Ib/II PCYC-1102 Study. Clin. Cancer Res. 2020, 26, 3918–3927. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.; Furman, R.R.; Coutre, S.; Flinn, I.W.; Burger, J.A.; Blum, K.; Sharman, J.; Wierda, W.; Jones, J.; Zhao, W.; et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: A 5-year experience. Blood 2018, 131, 1910–1919. [Google Scholar] [CrossRef]
- Ahn, I.E.; Underbayev, C.; Albitar, A.; Herman, S.E.M.; Tian, X.; Maric, I.; Arthur, D.C.; Wake, L.; Pittaluga, S.; Yuan, C.M.; et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood 2017, 129, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Quijada-Álamo, M.; Hernández-Sánchez, M.; Alonso-Pérez, V.; Rodríguez-Vicente, A.E.; García-Tuñón, I.; Martín-Izquierdo, M.; Hernández-Sánchez, J.M.; Herrero, A.B.; Bastida, J.M.; Segundo, L.S.; et al. CRISPR/Cas9-generated models uncover therapeutic vulnerabilities of del(11q) CLL cells to dual BCR and PARP inhibition. Leukemia 2020, 34, 1599–1612. [Google Scholar] [CrossRef]
- Quijada-Álamo, M.; Pérez-Carretero, C.; Hernández-Sánchez, M.; Rodríguez-Vicente, A.; Herrero, A.; Hernández-Sánchez, J.; Martín-Izquierdo, M.; Santos-Mínguez, S.; del Rey, M.; González, T.; et al. Dissecting the role of TP53 alterations in del(11q) chronic lymphocytic leukemia. Clin. Transl. Med. 2021, 11, e304. [Google Scholar] [CrossRef]
- Kwok, M.; Davies, N.; Agathanggelou, A.; Smith, E.; Oldreive, C.; Petermann, E.; Stewart, G.; Brown, J.; Lau, A.; Pratt, G.; et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016, 127, 582–595. [Google Scholar] [CrossRef]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nat. Cell Biol. 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Quesada, V.; Conde, L.; Villamor, N.; Ordóñez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Beà, S.; Pinyol, M.; Martínez-Trillos, A.; et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011, 44, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the Chronic Lymphocytic Leukemia Coding Genome: Role of NOTCH1 Mutational Activation. Available online: https://pubmed.ncbi.nlm.nih.gov/21670202/ (accessed on 8 February 2021).
- Stilgenbauer, S.; Schnaiter, A.; Paschka, P.; Zenz, T.; Rossi, M.; Döhner, K.; Bühler, A.; Böttcher, S.; Ritgen, M.; Kneba, M.; et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 2014, 123, 3247–3254. [Google Scholar] [CrossRef]
- Rodríguez-Vicente, A.E.; Bikos, V.; Hernández-Sánchez, M.; Malcikova, J.; Hernández-Rivas, J.-M.; Pospisilova, S. Next-generation sequencing in chronic lymphocytic leukemia: Recent findings and new horizons. Oncotarget 2017, 8, 71234–71248. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Rasi, S.; Fabbri, G.; Spina, V.; Fangazio, M.; Forconi, F.; Marasca, R.; Laurenti, L.; Bruscaggin, A.; Cerri, M.; et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012, 119, 521–529. [Google Scholar] [CrossRef]
- Lionetti, M.; Fabris, S.; Cutrona, G.; Agnelli, L.; Ciardullo, C.; Matis, S.; Ciceri, G.; Colombo, M.; Maura, F.; Mosca, L.; et al. High-throughput sequencing for the identification of NOTCH1mutations in early stage chronic lymphocytic leukaemia: Biological and clinical implications. Br. J. Haematol. 2014, 165, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1and Other Novel Cancer Genes in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Khiabanian, H.; Spina, V.; Ciardullo, C.; Bruscaggin, A.; Famà, R.; Rasi, S.; Monti, S.; Deambrogi, C.; De Paoli, L.; et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014, 123, 2139–2147. [Google Scholar] [CrossRef]
- Mansouri, L.; Noerenberg, D.; Young, E.; Mylonas, E.; Abdulla, M.; Frick, M.; Asmar, F.; Ljungström, V.; Schneider, M.; Yoshida, K.; et al. Frequent NFKBIE deletions are associated with poor outcome in primary mediastinal B-cell lymphoma. Blood 2016, 128, 2666–2670. [Google Scholar] [CrossRef]
- Young, E.; Noerenberg, D.; Mansouri, L.; Ljungström, V.; Frick, M.; Sutton, L.-A.; Blakemore, S.J.; Galan-Sousa, J.; Plevova, K.; Baliakas, P.; et al. EGR2 mutations define a new clinically aggressive subgroup of chronic lymphocytic leukemia. Leukemia 2017, 31, 1547–1554. [Google Scholar] [CrossRef]
- Rossi, D.; Fangazio, M.; Rasi, S.; Vaisitti, T.; Monti, S.; Cresta, S.; Chiaretti, S.; Del Giudice, I.; Fabbri, G.; Bruscaggin, A.; et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood 2012, 119, 2854–2862. [Google Scholar] [CrossRef]
- Nadeu, F.; Delgado, J.; Royo, C.; Baumann, T.; Stankovic, T.; Pinyol, M.; Jares, P.; Navarro, A.; Martín-García, D.; Beà, S.; et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 2016, 127, 2122–2130. [Google Scholar] [CrossRef]
- Rasi, S.; Khiabanian, H.; Ciardullo, C.; Terzi-Di-Bergamo, L.; Monti, S.; Spina, V.; Bruscaggin, A.; Cerri, M.; Deambrogi, C.; Martuscelli, L.; et al. Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica 2016, 101, e135–e138. [Google Scholar] [CrossRef]
- Ljungström, V.; Cortese, D.; Young, E.; Pandzic, T.; Mansouri, L.; Plevova, K.; Ntoufa, S.; Baliakas, P.; Clifford, R.; Sutton, L.-A.; et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: Clinical impact of recurrent RPS15 mutations. Blood 2016, 127, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Roos-Weil, D.; Nguyen-Khac, F.; Chevret, S.; Touzeau, C.; Roux, C.; Lejeune, J.; Cosson, A.; Mathis, S.; Feugier, P.; Leprêtre, S.; et al. Mutational and cytogenetic analyses of 188 CLL patients with trisomy 12: A retrospective study from the French Innovative Leukemia Organization (FILO) working group. Genes Chromosom. Cancer 2018, 57, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, S.J.; Clifford, R.; Parker, H.; Antoniou, P.; Stec-Dziedzic, E.; Larrayoz, M.; Davis, Z.; Kadalyayil, L.; Colins, A.; Robbe, P.; et al. Clinical significance of TP53, BIRC3, ATM and MAPK-ERK genes in chronic lymphocytic leukaemia: Data from the randomised UK LRF CLL4 trial. Leukemia 2020, 34, 1760–1774. [Google Scholar] [CrossRef]
- Hernández-Sánchez, M.; Rodríguez-Vicente, A.E.; Marín, I.G.-G.Y.; Quijada-Álamo, M.; Hernández-Sánchez, J.M.; Martín-Izquierdo, M.; Hernández-Rivas, J.Á.; Benito, R.; Hernández-Rivas, J.M. DNA damage response-related alterations define the genetic background of patients with chronic lymphocytic leukemia and chromosomal gains. Exp. Hematol. 2019, 72, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Guièze, R.; Robbe, P.; Clifford, R.; De Guibert, S.; Pereira, B.; Timbs, A.; Dilhuydy, M.-S.; Cabes, M.; Ysebaert, L.; Burns, A.; et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood 2015, 126, 2110–2117. [Google Scholar] [CrossRef] [PubMed]
- Brieghel, C.; Da Cunha-Bang, C.; Yde, C.W.; Schmidt, A.Y.; Kinalis, S.; Nadeu, F.; Andersen, M.A.; Jacobsen, L.O.; Andersen, M.K.; Pedersen, L.B.; et al. The Number of Signaling Pathways Altered by Driver Mutations in Chronic Lymphocytic Leukemia Impacts Disease Outcome. Clin. Cancer Res. 2020, 26, 1507–1515. [Google Scholar] [CrossRef]
- Del Giudice, I.; Rossi, D.; Chiaretti, S.; Marinelli, M.; Tavolaro, S.; Gabrielli, S.; Laurenti, L.; Marasca, R.; Rasi, S.; Fangazio, M.; et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica 2011, 97, 437–441. [Google Scholar] [CrossRef]
- Balatti, V.; Bottoni, A.; Palamarchuk, A.; Alder, H.; Rassenti, L.Z.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. NOTCH1 mutations in CLL associated with trisomy. Blood 2012, 119, 329–331. [Google Scholar] [CrossRef] [PubMed]
- López, C.; Delgado, J.; Costa, D.; Conde, L.; Ghita, G.; Villamor, N.; Navarro, A.; Cazorla, M.; Gómez, C.; Arias, A.; et al. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosom. Cancer 2012, 51, 881–889. [Google Scholar] [CrossRef]
- Jeromin, S.; Weissmann, S.; Haferlach, C.; Dicker, F.; Bayer, K.; Grossmann, V.; Alpermann, T.; Roller, A.; Kohlmann, A.; Kern, W.; et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014, 28, 108–117. [Google Scholar] [CrossRef]
- Rose-Zerilli, M.J.; Forster, J.; Parker, H.; Parker, A.; Rodríguez-Vicente, A.E.; Chaplin, T.; Gardiner, A.; Steele, A.J.; Collins, A.; Young, B.D.; et al. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: Data from the UK LRF CLL4 trial. Haematologica 2014, 99, 736–742. [Google Scholar] [CrossRef]
- Baliakas, P.; Hadzidimitriou, A.; Sutton, L.-A.; Rossi, D.; Minga, E.; Villamor, N.; Larrayoz, M.; Kminkova, J.; Agathangelidis, A.; Davis, Z.; et al. Recurrent Mutations Refine Prognosis in Chronic Lymphocytic Leukemia. Leukemia 2015, 29, 329–336. [Google Scholar] [CrossRef]
- Raponi, S.; Del Giudice, I.; Ilari, C.; Cafforio, L.; Messina, M.; Cappelli, L.V.; Bonina, S.; Piciocchi, A.; Marinelli, M.; Peragine, N.; et al. Biallelic BIRC 3 inactivation in chronic lymphocytic leukaemia patients with 11q deletion identifies a subgroup with very aggressive disease. Br. J. Haematol. 2018, 185, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Trillos, A.; Navarro, A.; Aymerich, M.; Delgado, J.; López-Guillermo, A.; Campo, E.; Villamor, N. Clinical impact of MYD88 mutations in chronic lymphocytic leukemia. Blood 2016, 127, 1611–1613. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C.; Wu, C.J. Clonal dynamics in chronic lymphocytic leukemia. Blood Adv. 2019, 3, 3759–3769. [Google Scholar] [CrossRef]
- Gruber, M.; Bozic, I.; Leshchiner, I.; Livitz, D.; Stevenson, K.; Rassenti, L.; Rosebrock, D.; Taylor-Weiner, A.; Olive, O.; Goyetche, R.; et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nat. Cell Biol. 2019, 570, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Schuh, A.; Becq, J.; Humphray, S.; Alexa, A.; Burns, A.; Clifford, R.; Feller, S.M.; Grocock, R.; Henderson, S.; Khrebtukova, I.; et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 2012, 120, 4191–4196. [Google Scholar] [CrossRef] [PubMed]
- Damm, F.; Mylonas, E.; Cosson, A.; Yoshida, K.; Della Valle, V.; Mouly, E.; Diop, M.; Scourzic, L.; Shiraishi, Y.; Chiba, K.; et al. Acquired Initiating Mutations in Early Hematopoietic Cells of CLL Patients. Cancer Discov. 2014, 4, 1088–1101. [Google Scholar] [CrossRef]
- Quijada-Álamo, M.; Hernández-Sánchez, M.; Robledo, C.; Hernández-Sánchez, J.-M.; Benito, R.; Montaño, A.; Rodríguez-Vicente, A.E.; Quwaider, D.; Martín, A.-Á.; García-Álvarez, M.; et al. Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia. J. Hematol. Oncol. 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Hernández-Sánchez, M.; Kotaskova, J.; Rodríguez, A.E.; Radova, L.; Tamborero, D.; Abáigar, M.; Plevova, K.; Benito, R.; Tom, N.; Quijada-Álamo, M.; et al. CLL cells cumulate genetic aberrations prior to the first therapy even in outwardly inactive disease phase. Leukemia 2018, 33, 518–558. [Google Scholar] [CrossRef]
- Leeksma, A.C.; Taylor, J.; Wu, B.; Gardner, J.R.; He, J.; Nahas, M.; Gonen, M.; Alemayehu, W.G.; Raa, D.T.; Walther, T.; et al. Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia 2019, 33, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Herling, C.D.; Abedpour, N.; Weiss, J.; Schmitt, A.; Jachimowicz, R.D.; Merkel, O.; Cartolano, M.; Oberbeck, S.; Mayer, P.; Berg, V.; et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Yu, L.; Kim, H.T.; Kasar, S.N.; Benien, P.; Du, W.; Hoang, K.; Aw, A.; Tesar, B.; Improgo, R.; Fernandes, S.M.; et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin. Cancer Res. 2017, 23, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Bo, M.D.; Del Principe, M.I.; Pozzo, F.; Ragusa, D.; Bulian, P.; Rossi, D.; Capelli, G.; Rossi, F.M.; Niscola, P.; Buccisano, F.; et al. NOTCH1 mutations identify a chronic lymphocytic leukemia patient subset with worse prognosis in the setting of a rituximab-based induction and consolidation treatment. Ann. Hematol. 2014, 93, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Hillmen, P.; O’Brien, S.; Barrientos, J.C.; Reddy, N.M.; Coutre, S.E.; Tam, C.S.; Mulligan, S.P.; Jaeger, U.; Barr, P.M.; et al. Extended follow-up and impact of high-risk prognostic factors from the phase 3 RESONATE study in patients with previously treated CLL/SLL. Leukemia 2018, 32, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Estenfelder, S.; Tausch, E.; Robrecht, S.; Bahlo, J.; Goede, V.; Ritgen, M.; Van Dongen, J.J.; Langerak, A.W.; Fingerle-Rowson, G.; Kneba, M.; et al. Gene Mutations and Treatment Outcome in the Context of Chlorambucil (Clb) without or with the Addition of Rituximab (R) or Obinutuzumab (GA-101, G)—Results of an Extensive Analysis of the Phase III Study CLL11 of the German CLL Study Group. Blood 2016, 128, 3227. [Google Scholar] [CrossRef]
- Rossi, D.; Rasi, S.; Spina, V.; Bruscaggin, A.; Monti, S.; Ciardullo, C.; Deambrogi, C.; Khiabanian, H.; Serra, R.; Bertoni, F.; et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013, 121, 1403–1412. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 Cluster Controls B Cell Proliferation and Its Deletion Leads to Chronic Lymphocytic Leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef]
- Hernández-Sánchez, M.; Rodríguez-Vicente, A.E.; Hernández, J.-Á.; Lumbreras, E.; Sarasquete, M.-E.; Martín, A.-Á.; Benito, R.; Vicente-Gutiérrez, C.; Robledo, C.; Heras, N.D.L.; et al. MiRNA expression profile of chronic lymphocytic leukemia patients with 13q deletion. Leuk. Res. 2016, 46, 30–36. [Google Scholar] [CrossRef]
- Visone, R.; Rassenti, L.Z.; Veronese, A.; Taccioli, C.; Costinean, S.; Aguda, B.D.; Volinia, S.; Ferracin, M.; Palatini, J.; Balatti, V.; et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009, 114, 3872–3879. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Bottoni, A.; Shimizu, M.; Spizzo, R.; Nicoloso, M.S.; Rossi, S.; Barbarotto, E.; Cimmino, A.; Adair, B.; Wojcik, S.E.; et al. Association of a MicroRNA/TP53 Feedback Circuitry with Pathogenesis and Outcome of B-Cell Chronic Lymphocytic Leukemia. JAMA 2011, 305, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mao, M.; Liu, H.; Wang, X.; Kou, Z.; Nie, Y.; Wang, Y.; Wang, Z.; Huang, Q.; Lang, T.; et al. miR-34a and miR-29b as indicators for prognosis of treatment-free survival of chronic lymphocytic leukemia patients in Chinese Uygur and Han populations. Mol. Cell. Probes 2019, 47, 101436. [Google Scholar] [CrossRef]
- Pepe, F.; Balatti, V. Role of Non-Coding RNAs in the Development of Targeted Therapy and Immunotherapy Approaches for Chronic Lymphocytic Leukemia. J. Clin. Med. 2020, 9, 593. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of Non-Coding Somatic Drivers in 2,658 Cancer Whole Genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Larrayoz, M.; Rose-Zerilli, M.J.J.; Kadalayil, L.; Parker, H.; Blakemore, S.J.; Forster, J.; Davis, Z.; Steele, A.J.; Collins, A.; Else, M.; et al. Non-coding NOTCH1 mutations in chronic lymphocytic leukemia; their clinical impact in the UK CLL4 trial. Leukemia 2016, 31, 510–514. [Google Scholar] [CrossRef]
- Kulis, M.; Heath, S.C.; Bibikova, M.; Queirós, A.C.; Navarro, A.; Clot, G.; Martínez-Trillos, A.; Castellano, G.; Brun-Heath, I.; Pinyol, M.; et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 1236–1242. [Google Scholar] [CrossRef]
- Oakes, C.C.; Seifert, M.; Assenov, Y.; Gu, L.; Przekopowitz, M.; Ruppert, A.S.; Wang, Q.; Imbusch, C.D.; Serva, A.; Koser, S.D.; et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 2016, 48, 253–264. [Google Scholar] [CrossRef]
- Mansouri, L.; Wierzbinska, J.A.; Plass, C.; Rosenquist, R. Epigenetic deregulation in chronic lymphocytic leukemia: Clinical and biological impact. Semin. Cancer Biol. 2018, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsagiopoulou, M.; Chapaprieta, V.; Duran-Ferrer, M.; Moysiadis, T.; Psomopoulos, F.; Kollia, P.; Papakonstantinou, N.; Campo, E.; Stamatopoulos, K.; Martin-Subero, J.I. Chronic lymphocytic leukemias with trisomy 12 show a distinct DNA methylation profile linked to altered chromatin activation. Haematologica 2020, 105, 2864–2867. [Google Scholar] [CrossRef] [PubMed]
- Wojdacz, T.K.; Amarasinghe, H.E.; Kadalayil, L.; Beattie, A.; Forster, J.; Blakemore, S.J.; Parker, H.; Bryant, D.; Larrayoz, M.; Clifford, R.; et al. Clinical significance of DNA methylation in chronic lymphocytic leukemia patients: Results from 3 UK clinical trials. Blood Adv. 2019, 3, 2474–2481. [Google Scholar] [CrossRef] [PubMed]
- Queirós, A.C.; Villamor, N.; Clot, G.; Martineztrillos, A.; Kulis, M.; Navarro, A.; Penas, E.M.M.; Jayne, S.; Majid, A.M.S.A.; Richter, J.A.; et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 2014, 29, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Beekman, R.; Chapaprieta, V.; Russiñol, N.; Vilarrasa-Blasi, R.; Verdaguer-Dot, N.; Martens, J.H.A.; Duran-Ferrer, M.; Kulis, M.; Serra, F.; Javierre, B.M.; et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018, 24, 868–880. [Google Scholar] [CrossRef]
- Yun, X.; Zhang, Y.; Wang, X. Recent progress of prognostic biomarkers and risk scoring systems in chronic lymphocytic leukemia. Biomark. Res. 2020, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- International CLL-IPI Working Group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016, 17, 779–790. [Google Scholar] [CrossRef]
- Delgado, J.; Doubek, M.; Baumann, T.; Kotaskova, J.; Molica, S.; Mozas, P.; Rivas-Delgado, A.; Morabito, F.; Pospisilova, S.; Montserrat, E. Chronic lymphocytic leukemia: A prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am. J. Hematol. 2017, 92, 375–380. [Google Scholar] [CrossRef]
- Muñoz-Novas, C.; Poza-Santaella, M.; Marín, I.G.-G.Y.; Hernández-Sánchez, M.; Rodríguez-Vicente, A.-E.; Infante, M.-S.; Heras, C.; Foncillas, M.-Á.; Marín, K.; Hernández-Rivas, J.-M.; et al. The International Prognostic Index for Patients with Chronic Lymphocytic Leukemia Has the Higher Value in Predicting Overall Outcome Compared with the Barcelona-Brno Biomarkers Only Prognostic Model and the MD Anderson Cancer Center Prognostic Index. BioMed Res. Int. 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- González-Gascón-Y-Marín, I.; Muñoz-Novas, C.; Figueroa, I.; Hernández-Sánchez, M.; Rodríguez-Vicente, A.-E.; Quijada-Álamo, M.; Pérez-Carretero, C.; Moreno, C.; Collado, R.; Espinet, B.; et al. Prognosis Assessment of Early-Stage Chronic Lymphocytic Leukemia: Are We Ready to Predict Clinical Evolution without a Crystal Ball? Clin. Lymphoma Myeloma Leuk. 2020, 20, 548–555.e4. [Google Scholar] [CrossRef]
- Condoluci, A.; Di Bergamo, L.T.; Langerbeins, P.; Hoechstetter, M.A.; Herling, C.D.; De Paoli, L.; Delgado, J.; Rabe, K.G.; Gentile, M.; Doubek, M.; et al. International prognostic score for asymptomatic early-stage chronic lymphocytic leukemia. Blood 2020, 135, 1859–1869. [Google Scholar] [CrossRef]
- Baliakas, P.; Moysiadis, T.; Hadzidimitriou, A.; Xochelli, A.; Jeromin, S.; Agathangelidis, A.; Mattsson, M.; Sutton, L.-A.; Minga, E.; Scarfò, L.; et al. Tailored approaches grounded on immunogenetic features for refined prognostication in chronic lymphocytic leukemia. Haematologica 2018, 104, 360–369. [Google Scholar] [CrossRef]
- Böttcher, S.; Ritgen, M.; Fischer, K.; Stilgenbauer, S.; Busch, R.M.; Fingerle-Rowson, G.; Fink, A.M.; Bühler, A.; Zenz, T.; Wenger, M.K.; et al. Minimal Residual Disease Quantification Is an Independent Predictor of Progression-Free and Overall Survival in Chronic Lymphocytic Leukemia: A Multivariate Analysis from the Randomized GCLLSG CLL8 Trial. J. Clin. Oncol. 2012, 30, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Brander, D.; Nabhan, C.; Mato, A. Minimal Residual Disease in Chronic Lymphocytic Leukemia in the Era of Novel Agents: A Review. JAMA Oncol. 2018, 4, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Yosifov, D.Y.; Wolf, C.; Stilgenbauer, S.; Mertens, D. From Biology to Therapy: The CLL Success Story. HemaSphere 2019, 3, e175. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Treatment of Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2020, 383, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Travade, P.; Chastang, C.; Dighiero, G.; Binet, J.L. New Trends in CLL Treatment. Blood Cells 1987, 12, 485–502. [Google Scholar]
- Jóhannsson, J.; Specht, L.; Mejer, J.; Jensen, B.A. Phase II study of palliative low-dose local radiotherapy in disseminated indolent non-Hodgkin’s lymphoma and chronic lymphocytic leukemia. Int. J. Radiat. Oncol. 2002, 54, 1466–1470. [Google Scholar] [CrossRef]
- Seymour, J.F.; Cusack, J.D.; Lerner, S.A.; Pollock, R.E.; Keating, M.J. Case/control study of the role of splenectomy in chronic lymphocytic leukemia. J. Clin. Oncol. 1997, 15, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Milne, K.; Sturrock, B.; Chevassut, T. Chronic Lymphocytic Leukaemia in 2020: The Future Has Arrived. Curr. Oncol. Rep. 2020, 22, 1–9. [Google Scholar] [CrossRef]
- CLL Trialists’ Collaborative Group. Chemotherapeutic Options in Chronic Lymphocytic Leukemia: A Meta-Analysis of the Randomized Trials. J. Natl. Cancer Inst. 1999, 91, 861–868. [Google Scholar] [CrossRef]
- Keating, M.J.; O’Brien, S.; Albitar, M.; Lerner, S.; Plunkett, W.; Giles, F.; Andreeff, M.; Cortes, J.; Faderl, S.; Thomas, D.; et al. Early Results of a Chemoimmunotherapy Regimen of Fludarabine, Cyclophosphamide, and Rituximab as Initial Therapy for Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2005, 23, 4079–4088. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Fischer, K.; Fingerle-Rowson, G.; Fink, A.; Busch, R.; Mayer, J.; Hensel, M.; Hopfinger, G.; Hess, G.; von Grünhagen, U.; et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet 2010, 376, 1164–1174. [Google Scholar] [CrossRef]
- Fischer, K.; Cramer, P.; Busch, R.; Stilgenbauer, S.; Bahlo, J.; Schweighofer, C.D.; Böttcher, S.; Staib, P.; Kiehl, M.; Eckart, M.J.; et al. Bendamustine Combined with Rituximab in Patients with Relapsed and/or Refractory Chronic Lymphocytic Leukemia: A Multicenter Phase II Trial of the German Chronic Lymphocytic Leukemia Study Group. J. Clin. Oncol. 2011, 29, 3559–3566. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Cramer, P.; Busch, R.; Böttcher, S.; Bahlo, J.; Schubert, J.; Pflüger, K.H.; Schott, S.; Goede, V.; Isfort, S.; et al. Bendamustine in Combination with Rituximab for Previously Untreated Patients With Chronic Lymphocytic Leukemia: A Multicenter Phase II Trial of the German Chronic Lymphocytic Leukemia Study Group. J. Clin. Oncol. 2012, 30, 3209–3216. [Google Scholar] [CrossRef]
- Eichhorst, B.; Fink, A.-M.; Bahlo, J.; Busch, R.; Kovacs, G.; Maurer, C.; Lange, E.; Köppler, H.; Kiehl, M.; Sökler, M.; et al. First-line chemoimmunotherapy with bendamustine and rituximab versus fludarabine, cyclophosphamide, and rituximab in patients with advanced chronic lymphocytic leukaemia (CLL10): An international, open-label, randomised, phase 3, non-inferiority trial. Lancet Oncol. 2016, 17, 928–942. [Google Scholar] [CrossRef]
- Goede, V.; Fischer, K.; Busch, R.; Engelke, A.; Eichhorst, B.; Wendtner, C.M.; Chagorova, T.; De La Serna, J.; Dilhuydy, M.-S.; Illmer, T.; et al. Obinutuzumab plus Chlorambucil in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2014, 370, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Hillmen, P.; Robak, T.; Janssens, A.; Babu, K.G.; Kloczko, J.; Grosicki, S.; Doubek, M.; Panagiotidis, P.; Kimby, E.; Schuh, A.; et al. Chlorambucil plus ofatumumab versus chlorambucil alone in previously untreated patients with chronic lymphocytic leukaemia (COMPLEMENT 1): A randomised, multicentre, open-label phase 3 trial. Lancet 2015, 385, 1873–1883. [Google Scholar] [CrossRef]
- Woyach, J.A.; Ruppert, A.S.; Heerema, N.A.; Zhao, W.; Booth, A.M.; Ding, W.; Bartlett, N.L.; Brander, D.M.; Barr, P.M.; Rogers, K.A.; et al. Ibrutinib Regimens versus Chemoimmunotherapy in Older Patients with Untreated CLL. N. Engl. J. Med. 2018, 379, 2517–2528. [Google Scholar] [CrossRef]
- Shanafelt, T.D.; Wang, X.V.; Kay, N.E.; Hanson, C.A.; O’Brien, S.; Barrientos, J.; Jelinek, D.F.; Braggio, E.; Leis, J.F.; Zhang, C.C.; et al. Ibrutinib–Rituximab or Chemoimmunotherapy for Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2019, 381, 432–443. [Google Scholar] [CrossRef]
- Wierda, W.G.; Byrd, J.C.; Abramson, J.S.; Bilgrami, S.F.; Bociek, G.; Brander, D.; Brown, J.; Chanan-Khan, A.A.; Chavez, J.C.; Coutre, S.E.; et al. Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma, Version 4.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2020, 18, 185–217. [Google Scholar] [CrossRef]
- Munir, T.; Brown, J.R.; O’Brien, S.; Barrientos, J.C.; Barr, P.M.; Reddy, N.M.; Coutre, S.; Tam, C.S.; Mulligan, S.P.; Jaeger, U.; et al. Final analysis from RESONATE: Up to six years of follow-up on ibrutinib in patients with previously treated chronic lymphocytic leukemia or small lymphocytic lymphoma. Am. J. Hematol. 2019, 94, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Barr, P.M.; Robak, T.; Owen, C.; Ghia, P.; Tedeschi, A.; Bairey, O.; Hillmen, P.; Coutre, S.E.; Devereux, S.; et al. Long-term efficacy and safety of first-line ibrutinib treatment for patients with CLL/SLL: 5 years of follow-up from the phase 3 RESONATE-2 study. Leukemia 2020, 34, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Brown, J.R.; O’Brien, S.; Barrientos, J.C.; Kay, N.E.; Reddy, N.M.; Coutre, S.; Tam, C.S.; Mulligan, S.P.; Jaeger, U.; et al. Ibrutinib versus Ofatumumab in Previously Treated Chronic Lymphoid Leukemia. N. Engl. J. Med. 2014, 371, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Sharman, J.P.; Egyed, M.; Jurczak, W.; Skarbnik, A.; Pagel, J.M.; Flinn, I.W.; Kamdar, M.; Munir, T.; Walewska, R.; Corbett, G.; et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzumab for treatment-naive chronic lymphocytic leukaemia (ELEVATE-TN): A randomised, controlled, phase 3 trial. Lancet 2020, 395, 1278–1291. [Google Scholar] [CrossRef]
- Ghia, P.; Pluta, A.; Wach, M.; Lysak, D.; Kozak, T.; Simkovic, M.; Kaplan, P.; Kraychok, I.; Illes, A.; De La Serna, J.; et al. ASCEND: Phase III, Randomized Trial of Acalabrutinib Versus Idelalisib Plus Rituximab or Bendamustine Plus Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2020, 38, 2849–2861. [Google Scholar] [CrossRef]
- Al-Sawaf, O.; Zhang, C.; Tandon, M.; Sinha, A.; Fink, A.-M.; Robrecht, S.; Samoylova, O.; Liberati, A.M.; Pinilla-Ibarz, J.; Opat, S.; et al. Venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab for previously untreated chronic lymphocytic leukaemia (CLL14): Follow-up results from a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2020, 21, 1188–1200. [Google Scholar] [CrossRef]
- Kater, A. Five-Year Analysis of Murano Study Demonstrates Enduring Undetectable Minimal Residual Disease (UMRD) in a Subset of Relapsed/Refractory Chronic Lymphocytic Leukemia (R/R CLL) Patients (Pts) Following Fixed-Duration Venetoclax-Rituximab (VenR) Therapy (Tx). In Proceedings of the 62nd Annual Meeting and Exposition of the American Society of Hematology, Online, 5–8 December 2020. [Google Scholar]
- Burger, J.A.; Tedeschi, A.; Barr, P.M.; Robak, T.; Owen, C.; Ghia, P.; Bairey, O.; Hillmen, P.; Bartlett, N.L.; Li, J.; et al. Ibrutinib as Initial Therapy for Patients with Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2015, 373, 2425–2437. [Google Scholar] [CrossRef]
- Mato, A.R.; Nabhan, C.; Thompson, M.C.; Lamanna, N.; Brander, D.M.; Hill, B.; Howlett, C.; Skarbnik, A.; Cheson, B.D.; Zent, C.; et al. Toxicities and outcomes of 616 ibrutinib-treated patients in the United States: A real-world analysis. Haematologica 2018, 103, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Beckwith, K.; Do, P.; Mundy, B.L.; Gordon, A.; Lehman, A.M.; Maddocks, K.J.; Cheney, C.; Jones, J.A.; Flynn, J.M.; et al. Ibrutinib treatment improves T cell number and function in CLL patients. J. Clin. Investig. 2017, 127, 3052–3064. [Google Scholar] [CrossRef]
- Tam, C.S.; Trotman, J.; Opat, S.; Burger, J.A.; Cull, G.; Gottlieb, D.; Harrup, R.; Johnston, P.B.; Marlton, P.; Munoz, J.; et al. Phase 1 study of the selective BTK inhibitor zanubrutinib in B-cell malignancies and safety and efficacy evaluation in CLL. Blood 2019, 134, 851–859. [Google Scholar] [CrossRef]
- Danilov, A.V.; Herbaux, C.; Walter, H.S.; Hillmen, P.; Rule, S.A.; Kio, E.A.; Karlin, L.; Dyer, M.J.; Mitra, S.S.; Yi, P.C.; et al. Phase Ib Study of Tirabrutinib in Combination with Idelalisib or Entospletinib in Previously Treated Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2020, 26, 2810–2818. [Google Scholar] [CrossRef]
- Iskierka-Jażdżewska, E.; Robak, T. Investigational treatments for chronic lymphocytic leukemia: A focus on phase 1 and 2 clinical trials. Expert Opin. Investig. Drugs 2020, 29, 709–722. [Google Scholar] [CrossRef]
- Roberts, A.W.; Davids, M.S.; Pagel, J.M.; Kahl, B.S.; Puvvada, S.D.; Gerecitano, J.F.; Kipps, T.J.; Anderson, M.A.; Brown, J.R.; Gressick, L.A.; et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2016, 374, 311–322. [Google Scholar] [CrossRef]
- Seymour, J.F.; Kipps, T.J.; Eichhorst, B.; Hillmen, P.; D’Rozario, J.; Assouline, S.; Owen, C.; Gerecitano, J.; Robak, T.; De La Serna, J.; et al. Venetoclax–Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2018, 378, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Kater, A.P.; Seymour, J.F.; Hillmen, P.; Eichhorst, B.; Langerak, A.W.; Owen, C.; Verdugo, M.; Wu, J.; Punnoose, E.A.; Jiang, Y.; et al. Fixed Duration of Venetoclax-Rituximab in Relapsed/Refractory Chronic Lymphocytic Leukemia Eradicates Minimal Residual Disease and Prolongs Survival: Post-Treatment Follow-Up of the MURANO Phase III Study. J. Clin. Oncol. 2019, 37, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Kater, A.P.; Wu, J.Q.; Kipps, T.; Eichhorst, B.; Hillmen, P.; D’Rozario, J.; Assouline, S.; Owen, C.; Robak, T.; De La Serna, J.; et al. Venetoclax Plus Rituximab in Relapsed Chronic Lymphocytic Leukemia: 4-Year Results and Evaluation of Impact of Genomic Complexity and Gene Mutations from the MURANO Phase III Study. J. Clin. Oncol. 2020, 38, 4042–4054. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.-M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Sharman, J.P.; Coutre, S.E.; Furman, R.R.; Cheson, B.D.; Pagel, J.M.; Hillmen, P.; Barrientos, J.C.; Zelenetz, A.D.; Kipps, T.J.; Flinn, I.W.; et al. Final Results of a Randomized, Phase III Study of Rituximab with or without Idelalisib Followed by Open-Label Idelalisib in Patients with Relapsed Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2019, 37, 1391–1402. [Google Scholar] [CrossRef]
- Lunning, M.; Vose, J.; Nastoupil, L.; Fowler, N.; Burger, J.A.; Wierda, W.G.; Schreeder, M.T.; Siddiqi, T.; Flowers, C.R.; Cohen, J.B.; et al. Ublituximab and umbralisib in relapsed/refractory B-cell non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood 2019, 134, 1811–1820. [Google Scholar] [CrossRef]
- Rai, K.R.; Freter, C.E.; Mercier, R.J.; Cooper, M.R.; Mitchell, B.S.; Stadtmauer, E.A.; Santábarbara, P.; Wacker, B.; Brettman, L. Alemtuzumab in Previously Treated Chronic Lymphocytic Leukemia Patients Who Also Had Received Fludarabine. J. Clin. Oncol. 2002, 20, 3891–3897. [Google Scholar] [CrossRef] [PubMed]
- Kreitman, R.J.; Pastan, I. Antibody Fusion Proteins: Anti-CD22 Recombinant Immunotoxin Moxetumomab Pasudotox. Clin. Cancer Res. 2011, 17, 6398–6405. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Keating, M.J.; Wierda, W.G.; Hartmann, E.; Hoellenriegel, J.; Rosin, N.Y.; de Weerdt, I.; Jeyakumar, G.; Ferrajoli, A.; Cardenas-Turanzas, M.; et al. Safety and activity of ibrutinib plus rituximab for patients with high-risk chronic lymphocytic leukaemia: A single-arm, phase 2 study. Lancet Oncol. 2014, 15, 1090–1099. [Google Scholar] [CrossRef]
- Burger, J.A.; Sivina, M.; Jain, N.; Kim, E.; Kadia, T.; Estrov, Z.; Nogueras-Gonzalez, G.M.; Huang, X.; Jorgensen, J.; Li, J.; et al. Randomized trial of ibrutinib vs ibrutinib plus rituximab in patients with chronic lymphocytic leukemia. Blood 2019, 133, 1011–1019. [Google Scholar] [CrossRef]
- Jain, N.; Keating, M.J.; Thompson, P.A.; Ferrajoli, A.; Burger, J.A.; Borthakur, G.M.; Takahashi, K.; Estrov, Z.E.; Fowler, N.H.; Kadia, T.M.; et al. Combined Ibrutinib and Venetoclax for First-Line Treatment for Patients with Chronic Lymphocytic Leukemia (CLL). Blood 2019, 134, 34. [Google Scholar] [CrossRef]
- Hillmen, P.; Rawstron, A.C.; Brock, K.; Muñoz-Vicente, S.; Yates, F.J.; Bishop, R.; Boucher, R.; Macdonald, D.; Fegan, C.; McCaig, A.; et al. Ibrutinib Plus Venetoclax in Relapsed/Refractory Chronic Lymphocytic Leukemia: The CLARITY Study. J. Clin. Oncol. 2019, 37, 2722–2729. [Google Scholar] [CrossRef]
- Rogers, K.A.; Huang, Y.; Ruppert, A.S.; Awan, F.T.; Heerema, N.A.; Hoffman, C.; Lozanski, G.; Maddocks, K.J.; Moran, M.E.; Reid, M.A.; et al. Phase 1b study of obinutuzumab, ibrutinib, and venetoclax in relapsed and refractory chronic lymphocytic leukemia. Blood 2018, 132, 1568–1572. [Google Scholar] [CrossRef]
- Von Tresckow, J.; Cramer, P.; Bahlo, J.; Robrecht, S.; Langerbeins, P.; Fink, A.-M.; Al-Sawaf, O.; Illmer, T.; Klaproth, H.; Estenfelder, S.; et al. CLL2-BIG: Sequential treatment with bendamustine, ibrutinib and obinutuzumab (GA101) in chronic lymphocytic leukemia. Leukemia 2018, 33, 1161–1172. [Google Scholar] [CrossRef]
- Woyach, J.A.; Blachly, J.S.; Rogers, K.A.; Bhat, S.A.; Jianfar, M.; Lozanski, G.; Weiss, D.M.; Andersen, B.L.; Gulrajani, M.; Frigault, M.M.; et al. Acalabrutinib plus Obinutuzumab in Treatment-Naïve and Relapsed/Refractory Chronic Lymphocytic Leukemia. Cancer Discov. 2020, 10, 394–405. [Google Scholar] [CrossRef]
- Davids, M.S.; Fisher, D.C.; Tyekucheva, S.; McDonough, M.; Hanna, J.; Lee, B.; Francoeur, K.; Montegaard, J.; Odejide, O.; Armand, P.; et al. A phase 1b/2 study of duvelisib in combination with FCR (DFCR) for frontline therapy for younger CLL patients. Leukemia 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Davids, M.S.; Brander, D.M.; Kim, H.T.; Tyekucheva, S.; Bsat, J.; Savell, A.; Hellman, J.M.; Bazemore, J.; Francoeur, K.; Alencar, A.; et al. Ibrutinib plus fludarabine, cyclophosphamide, and rituximab as initial treatment for younger patients with chronic lymphocytic leukaemia: A single-arm, multicentre, phase 2 trial. Lancet Haematol. 2019, 6, e419–e428. [Google Scholar] [CrossRef]
- Jain, N.; Keating, M.; Thompson, P.; Ferrajoli, A.; Burger, J.; Borthakur, G.; Takahashi, K.; Estrov, Z.; Fowler, N.; Kadia, T.; et al. Ibrutinib and Venetoclax for First-Line Treatment of CLL. N. Engl. J. Med. 2019, 380, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Roeker, L.E.; Dreger, P.; Brown, J.R.; Lahoud, O.B.; Eyre, T.A.; Brander, D.M.; Skarbnik, A.; Coombs, C.C.; Kim, H.T.; Davids, M.; et al. Allogeneic stem cell transplantation for chronic lymphocytic leukemia in the era of novel agents. Blood Adv. 2020, 4, 3977–3989. [Google Scholar] [CrossRef] [PubMed]
- Kharfan-Dabaja, M.A.; Kumar, A.; Hamadani, M.; Stilgenbauer, S.; Ghia, P.; Anasetti, C.; Dreger, P.; Montserrat, E.; Perales, M.-A.; Alyea, E.P.; et al. Clinical Practice Recommendations for Use of Allogeneic Hematopoietic Cell Transplantation in Chronic Lymphocytic Leukemia on Behalf of the Guidelines Committee of the American Society for Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2016, 22, 2117–2125. [Google Scholar] [CrossRef]
- Porter, D.L.; Grupp, S.A.; Kalos, M.; Loren, A.W.; Lledo, L.; Gilmore, J.; Milone, M.C.; Chew, A.; Levine, B.L.; June, C.H. Chimeric Antigen Receptor T Cells Directed Against CD19 Induce Durable Responses and Transient Cytokine Release Syndrome in Relapsed, Refractory CLL and ALL. Blood 2012, 120, 717. [Google Scholar] [CrossRef]
- Porter, D.L.; Hwang, W.-T.; Frey, N.V.; Lacey, S.F.; Shaw, P.A.; Loren, A.W.; Bagg, A.; Marcucci, K.T.; Shen, A.; Gonzalez, V.; et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 2015, 7, 303ra139. [Google Scholar] [CrossRef]
- Gauthier, J.; Hirayama, A.V.; Purushe, J.; Hay, K.A.; Lymp, J.; Li, D.H.; Yeung, C.C.S.; Sheih, A.; Pender, B.S.; Hawkins, R.M.; et al. Feasibility and efficacy of CD19-targeted CAR T cells with concurrent ibrutinib for CLL after ibrutinib failure. Blood 2020, 135, 1650–1660. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Kerbauy, L.N.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef]
- Langerbeins, P.; Bahlo, J.; Rhein, C.; Cramer, P.; Pflug, N.; Fischer, K.; Stilgenbauer, S.; Kreuzer, K.A.; Wendtner, C.-M.; Eichhorst, B.; et al. The CLL12 trial protocol: A placebo-controlled double-blind Phase III study of ibrutinib in the treatment of early-stage chronic lymphocytic leukemia patients with risk of early disease progression. Future Oncol. 2015, 11, 1895–1903. [Google Scholar] [CrossRef]
- Sedlarikova, L.; Petrackova, A.; Papajik, T.; Turcsanyi, P.; Kriegova, E. Resistance-Associated Mutations in Chronic Lymphocytic Leukemia Patients Treated with Novel Agents. Front. Oncol. 2020, 10, 894. [Google Scholar] [CrossRef]
- Woyach, J.A.; Furman, R.R.; Liu, T.-M.; Ozer, H.G.; Zapatka, M.; Ruppert, A.S.; Xue, L.; Li, D.H.-H.; Steggerda, S.M.; Versele, M.; et al. Resistance Mechanisms for the Bruton’s Tyrosine Kinase Inhibitor Ibrutinib. N. Engl. J. Med. 2014, 370, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Ruppert, A.S.; Guinn, D.; Lehman, A.; Blachly, J.S.; Lozanski, A.; Heerema, N.A.; Zhao, W.; Coleman, J.; Jones, D.; et al. BTKC481S-Mediated Resistance to Ibrutinib in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2017, 35, 1437–1443. [Google Scholar] [CrossRef]
- Kadri, S.; Lee, J.; Fitzpatrick, C.; Galanina, N.; Sukhanova, M.; Venkataraman, G.; Sharma, S.; Long, B.; Petras, K.; Theissen, M.; et al. Clonal evolution underlying leukemia progression and Richter transformation in patients with ibrutinib-relapsed CLL. Blood Adv. 2017, 1, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Guo, A.; Lu, P.; Ma, J.; Coleman, M.P.; Wang, Y.L. Functional characterization of BTKC481S mutation that confers ibrutinib resistance: Exploration of alternative kinase inhibitors. Leukemia 2014, 29, 895–900. [Google Scholar] [CrossRef]
- Hamasy, A.; Wang, Q.; Blomberg, K.E.M.; Mohammad, D.K.; Yu, L.; Vihinen, M.; Berglöf, A.; Smith, C.I.E. Substitution scanning identifies a novel, catalytically active ibrutinib-resistant BTK cysteine 481 to threonine (C481T) variant. Leukemia 2017, 31, 177–185. [Google Scholar] [CrossRef]
- Woyach, J.A.; Johnson, A.J. Targeted therapies in CLL: Mechanisms of resistance and strategies for management. Blood 2015, 126, 471–477. [Google Scholar] [CrossRef]
- Liu, T.-M.; Woyach, J.A.; Zhong, Y.; Lozanski, A.; Lozanski, G.; Dong, S.; Strattan, E.; Lehman, A.; Zhang, X.; Jones, J.A.; et al. Hypermorphic mutation of phospholipase C, γ2 acquired in ibrutinib-resistant CLL confers BTK independency upon B-cell receptor activation. Blood 2015, 126, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, K.J.; Ruppert, A.S.; Lozanski, G.; Heerema, N.A.; Zhao, W.; Abruzzo, L.V.; Lozanski, A.; Davis, M.; Gordon, A.L.; Smith, L.L.; et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients with Chronic Lymphocytic Leukemia. JAMA Oncol. 2015, 1, 80–87. [Google Scholar] [CrossRef]
- Woyach, J.A. Patterns of resistance to B cell–receptor pathway antagonists in chronic lymphocytic leukemia and strategies for management. Hematology 2015, 2015, 355–360. [Google Scholar] [CrossRef][Green Version]
- Landau, D.A.; Sun, C.; Rosebrock, D.; Herman, S.E.M.; Fein, J.; Sivina, M.; Underbayev, C.; Liu, D.; Hoellenriegel, J.; Ravichandran, S.; et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Lama, T.G.; Kyung, D.; O’Brien, S. Mechanisms of ibrutinib resistance in chronic lymphocytic leukemia and alternative treatment strategies. Expert Rev. Hematol. 2020, 13, 1–13. [Google Scholar] [CrossRef]
- Tausch, E.; Close, W.; Dolnik, A.; Bloehdorn, J.; Chyla, B.; Bullinger, L.; Döhner, H.; Mertens, D.; Stilgenbauer, S. Venetoclax resistance and acquired BCL2 mutations in chronic lymphocytic leukemia. Haematologica 2019, 104, e434–e437. [Google Scholar] [CrossRef] [PubMed]
- Blombery, P.; Thompson, E.R.; Nguyen, T.; Birkinshaw, R.W.; Gong, J.-N.; Chen, X.; McBean, M.; Thijssen, R.; Conway, T.; Anderson, M.A.; et al. Multiple BCL2 mutations cooccurring with Gly101Val emerge in chronic lymphocytic leukemia progression on venetoclax. Blood 2020, 135, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Blombery, P.; Anderson, M.A.; Gong, J.-N.; Thijssen, R.; Birkinshaw, R.W.; Thompson, E.R.; Teh, C.E.; Nguyen, T.; Xu, Z.; Flensburg, C.; et al. Acquisition of the Recurrent Gly101Val Mutation in BCL2 Confers Resistance to Venetoclax in Patients with Progressive Chronic Lymphocytic Leukemia. Cancer Discov. 2019, 9, 342–353. [Google Scholar] [CrossRef]
- Guièze, R.; Liu, V.M.; Rosebrock, D.; Jourdain, A.A.; Hernández-Sánchez, M.; Zurita, A.M.; Sun, J.; Hacken, E.T.; Baranowski, K.; Thompson, P.A.; et al. Mitochondrial Reprogramming Underlies Resistance to BCL-2 Inhibition in Lymphoid Malignancies. Cancer Cell 2019, 36, 369.e13–384.e13. [Google Scholar] [CrossRef] [PubMed]
- Mato, A.R.; Roeker, L.E.; Lamanna, N.; Allan, J.N.; Leslie, L.; Pagel, J.M.; Patel, K.; Osterborg, A.; Wojenski, D.; Kamdar, M.; et al. Outcomes of COVID-19 in patients with CLL: A multicenter international experience. Blood 2020, 136, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Scarfò, L.; Chatzikonstantinou, T.; Rigolin, G.M.; Quaresmini, G.; Motta, M.; Vitale, C.; Garcia-Marco, J.A.; Hernández-Rivas, J.Á.; Mirás, F.; Baile, M.; et al. COVID-19 severity and mortality in patients with chronic lymphocytic leukemia: A joint study by ERIC, the European Research Initiative on CLL, and CLL Campus. Leukemia 2020, 34, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- Muntañola, A.; Villacampa, G.; Hernández-Rivas, J.Á.; Alonso, R.; Mirás, F.; Osorio, S.; Baile, M.; Baltasar, P.; López Jiménez, J.; Hernandez-Rodriguez, I.; et al. Clinical Characteristics and Outcome of SARS-CoV-2 Infection in Admitted Patients with Chronic Lymphocytic Leukemia from a Single European Country. Exp. Hematol. Oncol. 2020, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Montserrat, E. When CLL meets COVID. Blood 2020, 136, 1115–1116. [Google Scholar] [CrossRef]
- Treon, S.P.; Castillo, J.J.; Skarbnik, A.P.; Soumerai, J.D.; Ghobrial, I.M.; Guerrera, M.L.; Meid, K.E.; Yang, G. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19-infected patients. Blood 2020, 135, 1912–1915. [Google Scholar] [CrossRef] [PubMed]
- Thibaud, S.; Tremblay, D.; Bhalla, S.; Zimmerman, B.; Sigel, K.; Gabrilove, J. Protective role of Bruton tyrosine kinase inhibitors in patients with chronic lymphocytic leukaemia and COVID-19. Br. J. Haematol. 2020, 190, e73–e76. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.Y.; Cuttica, M.J.; Ison, M.G.; Gordon, L.I. Ibrutinib for chronic lymphocytic leukemia in the setting of respiratory failure from severe COVID-19 infection: Case report and literature review. eJHaem 2020, 1, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Lionakis, M.S.; Sharman, J.P.; Roswarski, J.; Goy, A.; Monticelli, M.A.; Roshon, M.; Wrzesinski, S.H.; Desai, J.V.; Zarakas, M.A.; et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020, 5, eabd0110. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Shadman, M.; Condoluci, A.; Brown, J.R.; Byrd, J.C.; Gaidano, G.; Hallek, M.; Hillmen, P.; Mato, A.; Montserrat, E.; et al. How We Manage Patients with Chronic Lymphocytic Leukemia during the SARS-CoV-2 Pandemic. HemaSphere 2020, 4, e432. [Google Scholar] [CrossRef] [PubMed]
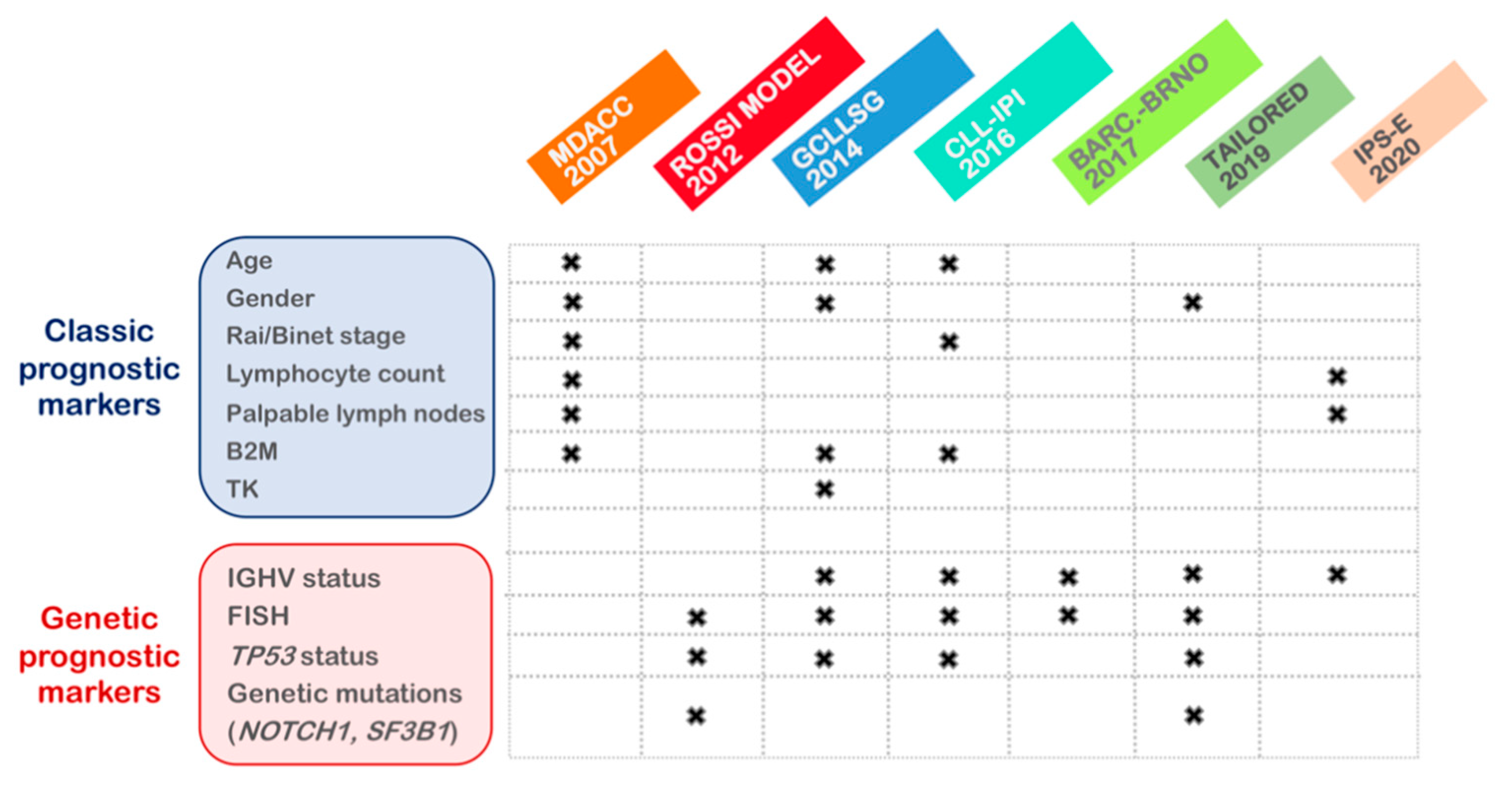
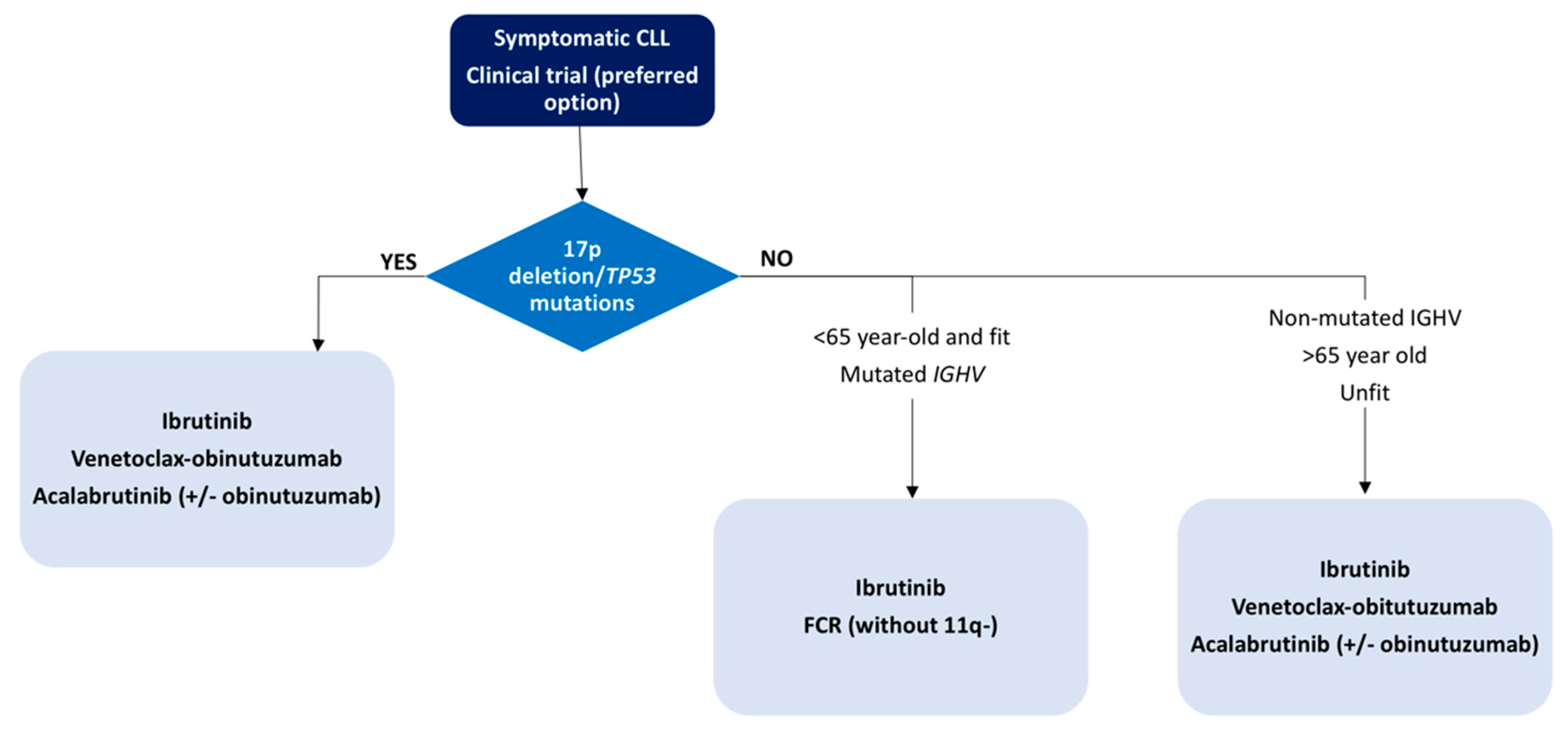
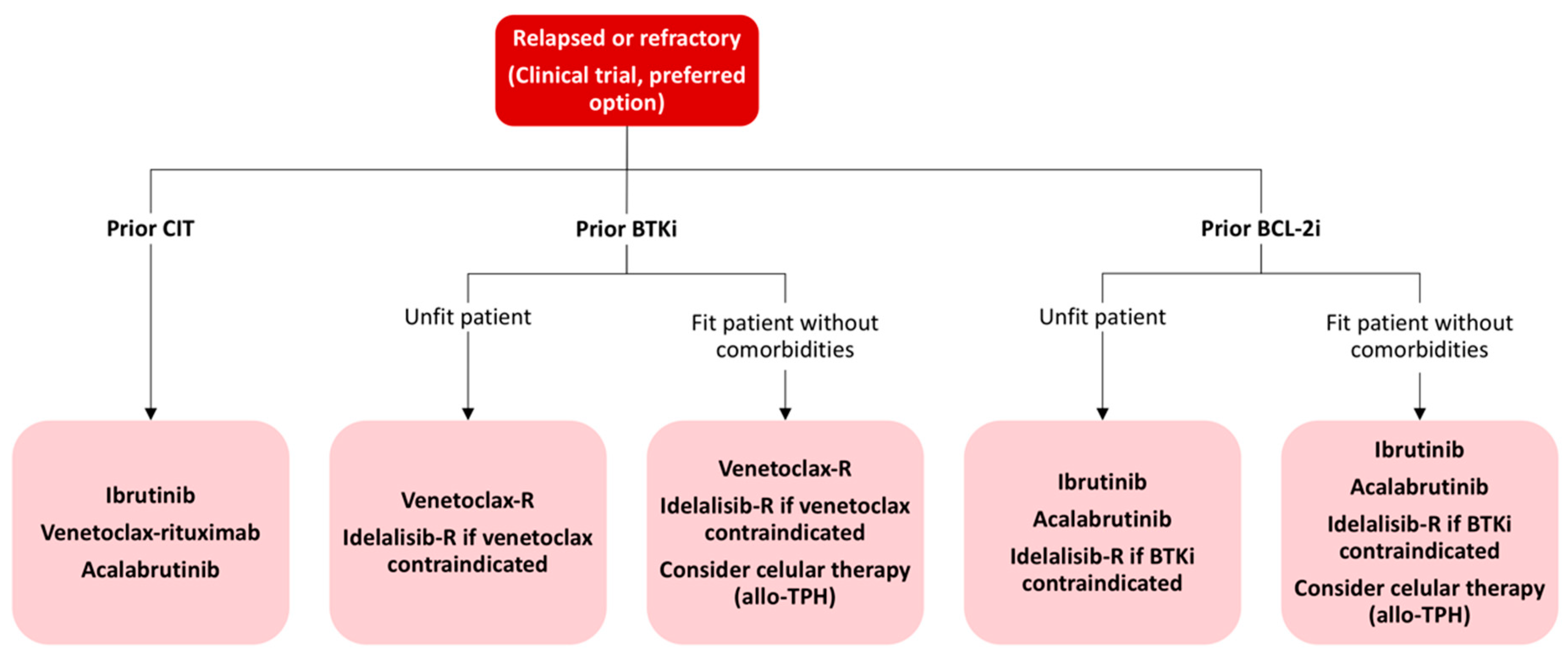
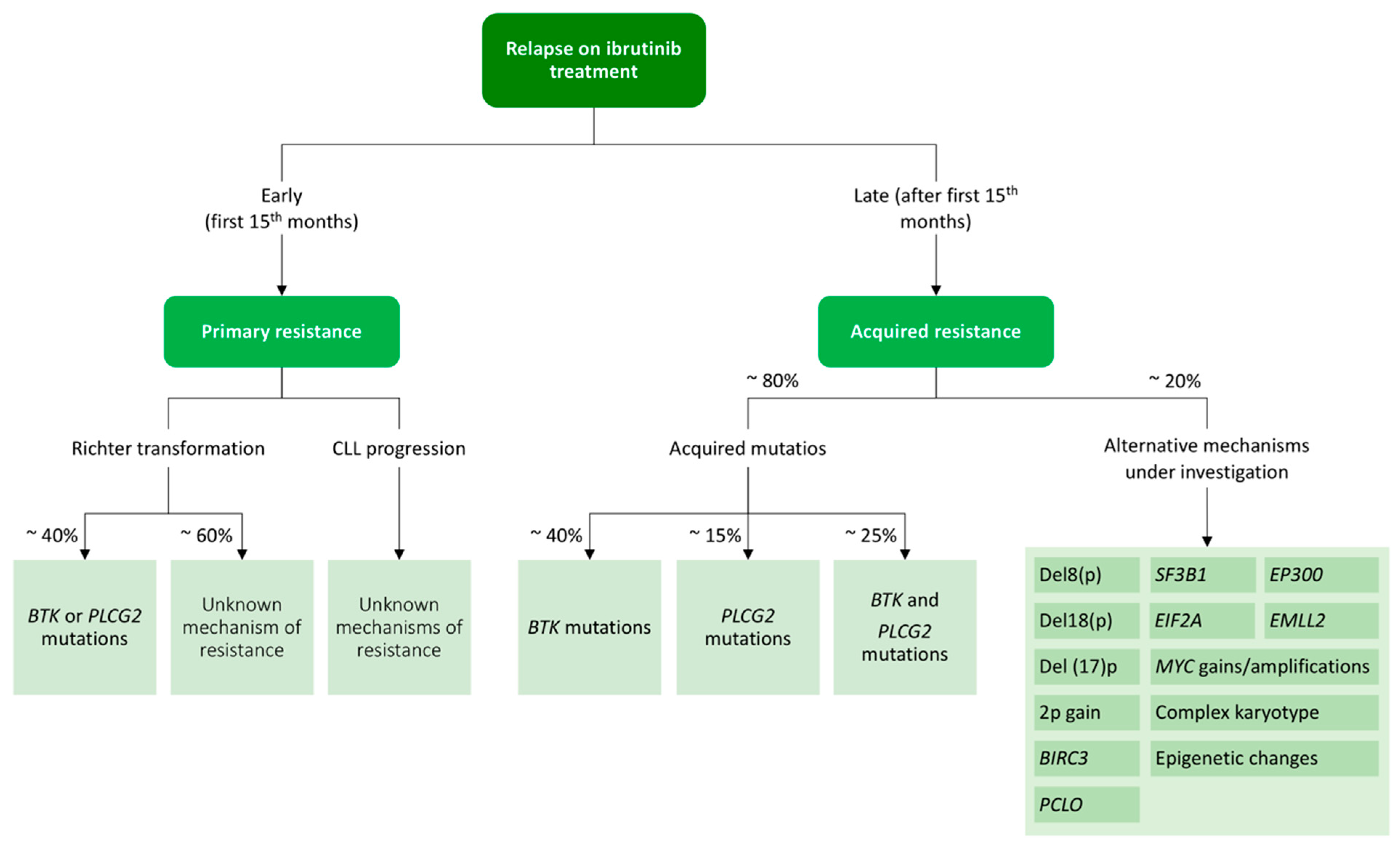
| Biomarkers | Clinical Significance in Prognosis |
|---|---|
| Rai/Binet advance stage | Associated with unfavorable disease course. Not enough to predict disease progression. |
| β2M high (>3.5 mg/L) | Predicts worse outcome and short-term remission after fludarabine-based CIT. Included in different risk scoring systems. |
| CD49d expression | Predicts shorter survival and remains valid for predicting treatment-free survival after ibrutinib treatment 1. |
| IGHV unmutated | Associated with a shorter time to first treatment and poorer response to CIT. Its assessment is highly recommended in pre-treatment evaluation and only once since its status remains stable during disease course. |
| Del(11q)/ATM mutation | Associated with a shorter time to first treatment but better response to BTK inhibitors in the presence of del(11q) 1. |
| Del(17p)/TP53 mutation | Confers resistance to CIT and predicts rapid disease progression. Its assessment is mandatory in pre-treatment evaluation. |
| Complex karyotype | Predicts unfavorable outcome after CIT independently of TP53 alterations. Its role is controversial after novel targeted agents 1. |
| NOTCH1 mutation | Refines cytogenetic-risk stratification and is associated with worse outcome and poor response to rituximab treatment 1. |
| SF3B1 mutation | Refines cytogenetic-risk stratification and has been associated with poor prognostis 1. |
| BTK/PCLG2 mutation | Confers resistance to BTK inhibitors. |
| BCL2 mutation | Confers resistance to venetoclax. |
| MRD positive | Predicts shorter progression free-survival for CIT. Remains valid for venetoclax-based regimens 1. |
| Drug | Line | Trial | Treatment (N) | ORR | PFS | OS | AE ≥ G3 | Follow Up | References |
|---|---|---|---|---|---|---|---|---|---|
| Ibru | 1 | Resonate-2 | Ibru (136) | 92% | NR | NR | Neutropenia (13%), pneumonia (12%), major hemorrhage (11%), hypertension (8%), anemia (7%), atrial fibrillation (5%), diarrhea (4%) | 57 m | [204] |
| Chl (133) | 37% | 15 m | NR | ||||||
| R/R | Resonate | Ibru (195) | 91% | 44.1 m | 67.7 | Neutropenia (25%), pneumonia (21%), hypertension (9%), major hemorrhage (10%) anemia (9%), atrial fibrillation (6%), diarrhea (7%) | 65 m | [203] | |
| Ofatumumab (196) | 24% | 8.1 m | 65.1 | ||||||
| Acala | 1 | Elevate-TN | Acala-Obi (179) | 94% | NR | NR | Neutropenia (30%), pneumonia (6%), anemia (6%), atrial fibrillation (4%), diarrhea (4%), hypertension (3%), major hemorrhage (2%), infusion reaction (2.2%) | 28 m | [206] |
| Acala (179) | 86% | NR | NR | Neutropenia (9%), anemia (7%), atrial fibrillation (3%), pneumonia (2%), hypertension (2%), major hemorrhage (2%), diarrhea (1%) | |||||
| Chl-Obi (177) | 79% | 22.6 m | NR | Neutropenia (41.4%), thrombocytopenia (11.8%), anemia (7.1%), infusion reaction (5.6%), pneumonia (1.8%) | |||||
| R/R | ASCEND | Acala (155) | 81% | NR | NR | Neutropenia (15%), anemia (11%), pneumonia (5%), atrial fibrillation (5%), hypertension (2%), diarrhea (1%), major hemorrhage (1%) | 16.1 m | [207] | |
| I.C.: Idela-R (119) | 75% | 15.8 m | NR | Neutropenia (39%), diarrhea (24%), pneumonia (8%), anemia (7%) | |||||
| I.C.: BR (36) | 16.9 m | NR | Neutropenia (31%), anemia (9%) pneumonia (3%) | ||||||
| Ven | 1 | CLL-14 | Ven-Obi (216) | 85% | NR | NR | Neutropenia (53%), infusion reaction (9%), thrombocytopenia (9%), anemia (9%), pneumonia (7%), tumor lysis syndrome (2%) | 39.6 m | [208] |
| Chl-Obi (216) | 71% | 35.6 m | NR | Neutropenia (46%), thrombocytopenia (15%), infusion reaction (11%), anemia (7%), pneumonia (5%), tumor lysis syndrome (3%) | |||||
| R/R | Murano | Ven-R (195) | 92.3% | 53.6 m | 83.5% 1 | Neutropenia (57.5%), infections (17.5%), anemia (10.8%), pneumonia (5.2%), tumor lysis syndrome (3.1%) | 59.2 m | [209] | |
| BR (194) | 72.3% | 17 m | 66.8% 1 | Neutropenia (38.8%), infections (21.8%), anemia (13.8%), pneumonia (8%) |
| Therapeutic Approach | Treatment | Phase | N R/R | N TN | Duration of Treatment | Response Rate | % uMRD (BM) | References |
|---|---|---|---|---|---|---|---|---|
| TA + anti CD20 | Ibru-R | II | 208 | 27 | Indefinite | 92.3% | BM, 24 m: 19.8% | [226] |
| Ibru | 92.3% | BM, 24 m: 12.2% | ||||||
| Ibru-O (benda) | II | 30 | 31 | Possible if MRD-neg | 100% | PB, 12 m: 48% | [230] | |
| Acala-O | Ib/II | 26 | 19 | Indefinite | 92% (R/R) 95% (TN) | 12 m: 15% (R/R), 26% (TN) | [231] | |
| Ven-R | III | 389 | 24 months | 92.3% | PB, 24 m: 62.4% | [217] | ||
| Benda-R | 72.3% | PB, 24 m: 13.3% | ||||||
| Ven-O | III | 432 | 12 cycles | 85% | BM, 12 m: 57% | [220] | ||
| Chl-O | 71% | BM, 12 m: 17% | ||||||
| TA + CIT | FCR-ibru | II | 85 | Possible if MRD-neg | 96% | BM, 24 m: 78% | [233] | |
| FCO-ibru | II | 45 | Possible if MRD-neg | 73% | BM, 12 m: 100% | [227] | ||
| FCR-duvelisib | Ib/II | 32 | 24 months | 88% | BM, 66% at best response | [232] | ||
| TA + TA | Ibru-ven | II | 53 | Possible if MRD-neg | 89% | BM, 12 Mm 36% | [228] | |
| Ibru-ven | II | 80 | Possible if MRD-neg | 88% | BM, 12 m: 61% | [234] | ||
| Ibru-ven-O | Ib | 25 | 25 | 14 cycles | 88% (R/R) 84% TN | BM and PB, 7 m: 70% | [229] |
| Ibrutinib | Venetoclax | ||
|---|---|---|---|
| Mutation Type | BTK | PLCG2 | BCL-2 |
| Prevalence in relapsed patients | 57% | 13% | 47% |
| Mechanism | Loss of covalent binding of ibrutinib to BTK | Activating BCR signaling independent of BTK | Disruption of the bond of venetoclax to BCL-2 |
| Variants | |||
| More frequent | C481S | Different subclones coexist with low allelic burden | G101V (subclonal) |
| Others | C481R, C281F, C481Y, R28S, G164D, T316A, T474I/S, R490H, Q516K, L528W, V537I | F82S, P664S, R665W, S707Y, S707P, S707F, L845F, L845V, L845G, L848R, D993Y, D993H, D1140N, M1141K, M1141R, S1192G | D103Y, A103T, A103G, A103V, A113G, A129L, V156A |
| Median time since drug exposure | 34.3 months (14–76.8) | 35.1 months (17.4–64.6) | 36 months (6.5–73) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Carretero, C.; González-Gascón-y-Marín, I.; Rodríguez-Vicente, A.E.; Quijada-Álamo, M.; Hernández-Rivas, J.-Á.; Hernández-Sánchez, M.; Hernández-Rivas, J.M. The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment. Diagnostics 2021, 11, 853. https://doi.org/10.3390/diagnostics11050853
Pérez-Carretero C, González-Gascón-y-Marín I, Rodríguez-Vicente AE, Quijada-Álamo M, Hernández-Rivas J-Á, Hernández-Sánchez M, Hernández-Rivas JM. The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment. Diagnostics. 2021; 11(5):853. https://doi.org/10.3390/diagnostics11050853
Chicago/Turabian StylePérez-Carretero, Claudia, Isabel González-Gascón-y-Marín, Ana E. Rodríguez-Vicente, Miguel Quijada-Álamo, José-Ángel Hernández-Rivas, María Hernández-Sánchez, and Jesús María Hernández-Rivas. 2021. "The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment" Diagnostics 11, no. 5: 853. https://doi.org/10.3390/diagnostics11050853
APA StylePérez-Carretero, C., González-Gascón-y-Marín, I., Rodríguez-Vicente, A. E., Quijada-Álamo, M., Hernández-Rivas, J.-Á., Hernández-Sánchez, M., & Hernández-Rivas, J. M. (2021). The Evolving Landscape of Chronic Lymphocytic Leukemia on Diagnosis, Prognosis and Treatment. Diagnostics, 11(5), 853. https://doi.org/10.3390/diagnostics11050853








