Detection of Autophagy-Related Gene Expression by Conjunctival Impression Cytology in Age-Related Macular Degeneration
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture of ARPE Cells
2.2. Real-Time PCR
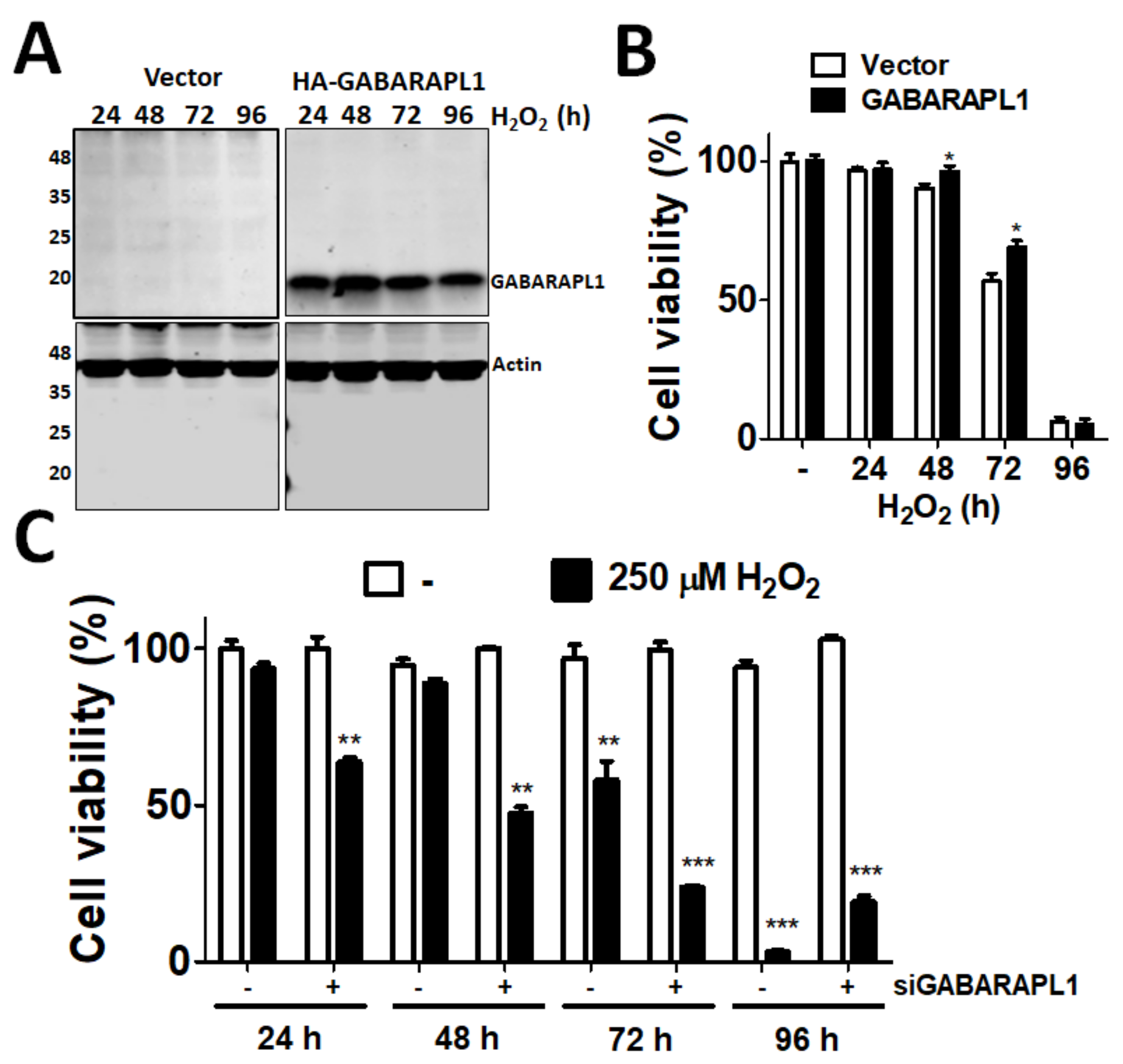
2.3. Cell Viability Assay
2.4. Transfection
2.5. Autophagic Flux Measurement and Immunoblotting
2.6. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| PCV | polypoidal choroidal vasculopathy |
| ERBB2 | erb-b2 receptor tyrosine-protein kinase 2 |
| GABARAPL1 | Gamma-aminobutyric acid receptor-associated protein-like 1 |
| AMD | age-related macular degeneration |
| ATG | autophagy-related gene |
| SQSTM1/p62 | Sequestosome 1 |
| MAP1LC3B | Microtubule-associated proteins 1A/1B light chain 3B |
References
- Solomon, S.D.; Lindsley, K.; Vedula, S.S.; Krzystolik, M.G.; Hawkins, B.S. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. Cochrane Database Syst. Rev. 2019, 3, CD005139. [Google Scholar] [CrossRef]
- Van Lookeren Campagne, M.; LeCouter, J.; Yaspan, B.L.; Ye, W. Mechanisms of age-related macular degeneration and therapeutic opportunities. J. Pathol. 2014, 232, 151–164. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Wong, C.W.; Yanagi, Y.; Lee, W.K.; Ogura, Y.; Yeo, I.; Wong, T.Y.; Cheung, C.M.G. Age-related macular degeneration and polypoidal choroidal vasculopathy in Asians. Prog. Retin. Eye Res. 2016, 53, 107–139. [Google Scholar] [CrossRef]
- Ambati, J.; Fowler, B.J. Mechanisms of age-related macular degeneration. Neuron 2012, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Plafker, S.M.; O’Mealey, G.B.; Szweda, L.I. Mechanisms for countering oxidative stress and damage in retinal pigment epithelium. Int. Rev. Cell Mol. Biol. 2012, 298, 135–177. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef]
- Boya, P.; Esteban-Martinez, L.; Serrano-Puebla, A.; Gomez-Sintes, R.; Villarejo-Zori, B. Autophagy in the eye: Development, degeneration, and aging. Prog. Retin. Eye Res. 2016, 55, 206–245. [Google Scholar] [CrossRef] [PubMed]
- Terluk, M.R.; Ebeling, M.C.; Fisher, C.R.; Kapphahn, R.J.; Yuan, C.; Kartha, R.V.; Montezuma, S.R.; Ferrington, D.A. n-Acetyl-l-cysteine protects human retinal pigment epithelial cells from oxidative damage: Implications for age-related macular degeneration. Oxid. Med. Cell Longev. 2019, 2019, 5174957. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Tsao, Y.N.; Shu, C.W. Autophagy modulation as a potential targeted cancer therapy: From drug repurposing to new drug development. Kaohsiung J. Med. Sci. 2021. [Google Scholar] [CrossRef]
- Liu, P.F.; Farooqi, A.A.; Peng, S.Y.; Yu, T.J.; Dahms, H.U.; Lee, C.H.; Tang, J.Y.; Wang, S.C.; Shu, C.W.; Chang, H.W. Regulatory effects of noncoding RNAs on the interplay of oxidative stress and autophagy in cancer malignancy and therapy. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; Nucci, C.; Corasaniti, M.T.; Bagetta, G.; Morrone, L.A. Autophagy dysregulation and the fate of retinal ganglion cells in glaucomatous optic neuropathy. Prog. Brain Res. 2015, 220, 87–105. [Google Scholar] [CrossRef]
- Sheu, S.J.; Chen, J.L.; Bee, Y.S.; Lin, S.H.; Shu, C.W. ERBB2-modulated ATG4B and autophagic cell death in human ARPE19 during oxidative stress. PLoS ONE 2019, 14, e0213932. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Qiu, Y.; Frontera, E.; Jia, L.; Khan, N.W.; Klionsky, D.J.; Ferguson, T.A.; Thompson, D.A.; Zacks, D.N. Inhibiting autophagy reduces retinal degeneration caused by protein misfolding. Autophagy 2018, 14, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Filek, R.; Wang, J.M.; Wu, C.H.; Liu, H.; Hutnik, C.M. Impression cytology implicates cell autophagy in aqueous deficiency dry eye. Clin. Ophthalmol. 2017, 11, 773–779. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hagan, S. Biomarkers of ocular surface disease using impression cytology. Biomark. Med. 2017, 11, 1135–1147. [Google Scholar] [CrossRef]
- Chang, H.W.; Liu, P.F.; Tsai, W.L.; Hu, W.H.; Hu, Y.C.; Yang, H.C.; Lin, W.Y.; Weng, J.R.; Shu, C.W. Xanthium strumarium fruit extract inhibits ATG4B and diminishes the proliferation and metastatic characteristics of colorectal cancer cells. Toxins 2019, 11, 313. [Google Scholar] [CrossRef] [PubMed]
- Kaarniranta, K.; Tokarz, P.; Koskela, A.; Paterno, J.; Blasiak, J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell Biol. Toxicol. 2017, 33, 113–128. [Google Scholar] [CrossRef]
- Kivinen, N. The role of autophagy in age-related macular degeneration (AMD)—Studies into the pathogenesis of AMD. Acta Ophthalmol. 2018, 96, 531–532. [Google Scholar] [CrossRef]
- Mitter, S.K.; Song, C.; Qi, X.; Mao, H.; Rao, H.; Akin, D.; Lewin, A.; Grant, M.; Dunn, W., Jr.; Ding, J.; et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 2014, 10, 1989–2005. [Google Scholar] [CrossRef]
- Ci, Y.; Shi, K.; An, J.; Yang, Y.; Hui, K.; Wu, P.; Shi, L.; Xu, C. ROS inhibit autophagy by downregulating ULK1 mediated by the phosphorylation of p53 in selenite-treated NB4 cells. Cell Death Dis. 2014, 5, e1542. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Qin, Z.H. The divergent roles of autophagy in ischemia and preconditioning. Acta Pharmacol. Sin. 2015, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Celik Buyuktepe, T.; Ozmert, E.; Demirel, S.; Batioglu, F. Role of inflammation in retinal microcirculation in diabetic eyes: Correlation between aqueous flare and microvascular findings. Ophthalmologica 2020, 243, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Rozing, M.P.; Durhuus, J.A.; Krogh Nielsen, M.; Subhi, Y.; Kirkwood, T.B.; Westendorp, R.G.; Sorensen, T.L. Age-related macular degeneration: A two-level model hypothesis. Prog. Retin. Eye Res. 2020, 76, 100825. [Google Scholar] [CrossRef]
- Ang, W.J.; Zunaina, E.; Norfadzillah, A.J.; Raja-Norliza, R.O.; Julieana, M.; Ab-Hamid, S.A.; Mahaneem, M. Evaluation of vascular endothelial growth factor levels in tears and serum among diabetic patients. PLoS ONE 2019, 14, e0221481. [Google Scholar] [CrossRef]



| Gene Symbol | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| ULK1 | TGCCCCTGGTTGAATGTTCT | ACACCAGCCCAACAATTCCA |
| ATG 2A | GCCTGGCGCCTAGAGAAG | ACCCGCTCTTTCACACAGTT |
| ATG 2B | CGTTGTCAGTTCAGCCAAGA | AGATGTGGCAAACCACGGAT |
| ATG 3 | TCCCATGTGTTCAGTTCACCC | AACAGCCATTTTGCCACTAATC |
| ATG 4A | AGTCAGAGTAAGGGCACCTCT | CCCCTAAAGACTGTGGCATCT |
| ATG 4B | TCCATCGCTGTGGGGTTTTT | AGAATCTAGGGACAGGTTCAGGA |
| ATG 4C | TTACTACGGTGGCCGGGGT | AAAAATGTGCAGGAGCCACCAA |
| ATG 4D | AACGTCAAGTACGGTTGGGT | ACACAAAGTCCCGCTGGAAA |
| ATG 5 | AGACCTTCTGCACTGTCCATC | GCAATCCCATCCAGAGTTGCT |
| BECN1 | AACCAGATGCGTTATGCCCA | TCCATTCCACGGGAACACTG |
| ATG 7 | AGCGGCGGCAAGAAATAATG | GTCCTTGGGAGCTTCATCCA |
| GABARAP | GAGGGCGAGAAGATCCGAAA | AGCTCGGAGATGAATTCGCTT |
| GABARAPL1 | TGAGACCTGAGGACGCCTT | GGGCTTCCAACCACTCATTTC |
| GABARAPL2 | GCGAAATATCCCGACAGGGT | CCACAAACAGGAAGATCGCC |
| MAP1LC3B | AAGGCTTTCAGAGAGACCCTG | CCGTTTACCCTGCGTTTGTG |
| ATG 9A | GCTGTTCCTGAGGTGGTCAA | TATGGTGCCAAGGTGACTTGC |
| ATG 9B | CTCTAGCCCCAGACAACAGTG | TACTCCACCCTCCAATCTCCT |
| ATG 10 | CGAGCGGAGAGGGTTATCATT | GCACATGTAGCCATCAGAACAG |
| ATG 12 | CGGATGTCTCCCCAGAAACAA | CCTTGGATGGTTCGTGTTCG |
| ATG 13 | CCGAAAAGTGGGGGCTTTTG | TCGGTATCCTCCAGCTCCAA |
| ATG 14 | GCTCTCCTCTCAGGCCATCAT | TCTGAACGCATTTGGCGCA |
| ATG 16 L1 | TCGAGGAGATCATCCTGCAATAAC | TCAGTTGGGCCATTTCTTGTAGC |
| ATG 16 L2 | GCCAAACAGTGTCACTCCCA | AAGCAAGCTCACCACAGACC |
| RB1CC1 | CATCCTAGACGAACGCCATGA | TGGTTTGAGATCCAGGGCAG |
| WIPI1 | TCCACGGAAGCAATGAAATCC | TAGGCAAACCAGCAGCCTTT |
| SNX 30 | TGGAGAGGTGGCAGAACAAC | GAATAATCGACTCCCACGCCA |
| SNX 4 | GTTTTCAGTGAATGGAGTGCC | TTCATGTTTCCTGCACACAGC |
| ATG 101 | TGTTTTAACCGTGTGCCCCCT | ACAAGAGACCAGCTCCACAGT |
| SQSTM1 | CTGCCCAGACTACGACTTGTGT | TCAACTTCAATGCCCAGAGG |
| GAPDH | TGCACCACCA ACTGCTTAGC | GGCATGGACT GTGGTCATGA G |
| Variable | % | Control (n = 27) | % | AMD (n = 42) | p Value * | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | p Value * | Mean ± SD | Median | p Value * | ||||
| Total | 0.0850 ± 0.0495 | 0.0773 | 0.3057 ± 0.4457 | 0.0972 | 0.013 | ||||
| Sex | |||||||||
| Female | 66.67% | 0.0923 ± 0.0478 | 0.0808 | 0.284 | 45.20% | 0.2845 ± 0.3971 | 0.0813 | 0.781 | |
| Male | 33.33% | 0.0702 ± 0.0525 | 0.0829 | 54.80% | 0.3241±0.4924 | 0.0983 | |||
| Age, y | |||||||||
| ≦60 | 25.93% | 0.0794 ± 0.0503 | 0.0864 | 0.739 | 14.30% | 0.7504 ± 0.5594 | 0.6178 | 0.007 | |
| >60 | 74.07% | 0.0868 ± 0.0504 | 0.0773 | 85.70% | 0.2295 ± 0.3832 | 0.0886 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, C.-W.; Bee, Y.-S.; Chen, J.-L.; Tsen, C.-L.; Tsai, W.-L.; Sheu, S.-J. Detection of Autophagy-Related Gene Expression by Conjunctival Impression Cytology in Age-Related Macular Degeneration. Diagnostics 2021, 11, 296. https://doi.org/10.3390/diagnostics11020296
Shu C-W, Bee Y-S, Chen J-L, Tsen C-L, Tsai W-L, Sheu S-J. Detection of Autophagy-Related Gene Expression by Conjunctival Impression Cytology in Age-Related Macular Degeneration. Diagnostics. 2021; 11(2):296. https://doi.org/10.3390/diagnostics11020296
Chicago/Turabian StyleShu, Chih-Wen, Youn-Shen Bee, Jiunn-Liang Chen, Chui-Lien Tsen, Wei-Lun Tsai, and Shwu-Jiuan Sheu. 2021. "Detection of Autophagy-Related Gene Expression by Conjunctival Impression Cytology in Age-Related Macular Degeneration" Diagnostics 11, no. 2: 296. https://doi.org/10.3390/diagnostics11020296
APA StyleShu, C.-W., Bee, Y.-S., Chen, J.-L., Tsen, C.-L., Tsai, W.-L., & Sheu, S.-J. (2021). Detection of Autophagy-Related Gene Expression by Conjunctival Impression Cytology in Age-Related Macular Degeneration. Diagnostics, 11(2), 296. https://doi.org/10.3390/diagnostics11020296






