Abstract
Plasmodium vivax malaria is one of the most lethal infectious diseases, with 7 million infections annually. One of the roadblocks to global malaria elimination is the lack of highly sensitive, specific, and accurate diagnostic tools. The absence of diagnostic tools in particular has led to poor differentiation among parasite species, poor prognosis, and delayed treatment. The improvement necessary in diagnostic tools can be broadly grouped into two categories: technologies-driven and omics-driven progress over time. This article discusses the recent advancement in omics-based malaria for identifying the next generation biomarkers for a highly sensitive and specific assay with a rapid and antecedent prognosis of the disease. We summarize the state-of-the-art diagnostic technologies, the key challenges, opportunities, and emerging prospects of multi-omics-based sensors.
1. Introduction
1.1. Grounding and Current Status of Malaria Diagnostics
Malaria is a significant public health concern that continues to claim the lives of more than 435,000 people each year. The antimalarial drug resistance and detection of low parasitemia form a primary barrier to achieving the United Nations Sustainable Development Goals of ending malaria epidemics by 2030 [1]. This aim can only be achieved when all cases are accurately diagnosed and treated appropriately.
The advancement in diagnostics can be grouped into two categories: technologies-driven and omics-driven (Figure 1). The advancements in technology-driven malaria diagnosis devices include microfluidics-based devices for cell-based diagnosis targeting hemozoin crystals in red blood cells (RBCs) [2,3,4,5,6], immuno-chromatographic tests (ICT) for the quantitative analysis of parasite protein (e.g., pfHRPII [7] or pLDH [8] {P. falciparum specific}, pLDH {P. vivax, P. ovale, and P. malariae-pan} [9], aldolase {pan} [8]) in the blood sample, 2-D paper matrix prototypes with dried reagents for quantitative analysis of parasite protein in the blood sample [10], and the DxBox 3-D plastic device [11] for differential diagnosis purposes. Spectrometry-based micro-Nuclear Magnetic Resonance (micro-NMR) [2,3,6,12,13,14,15,16] for diagnosing malaria, and mass spectrometry-based proteomics for host proteins [17,18,19] and parasite proteins [19,20,21] have also shown promising results in the laboratory setup (Figure 1a). In parallel to the advances on the technology front, the development of different omics approaches has been explored to achieve ideal biomarker candidates [22,23,24,25,26]. Omics is a robust tool in terms of looking at a biological problem with varying points of reference, but to understand the bigger picture, one needs to combine the findings of various omics. Multi-omics (e.g., genomics, transcriptomics, proteomics, metabolomics, or phenomics) or integrated omics reveal substantially novel insights into the pathobiology of chronic diseases [27,28,29], but remains a challenge in infectious diseases (e.g., malaria). While novel technologies are being developed, light microscopy remains the gold standard due to various challenges imposed by limited resources in rural areas (Table 1).

Figure 1.
(a) Malaria diagnosis and differentiation of Plasmodium spp. in the intra-erythrocyte circle of the host system. (b) Schematic representation exhibits the workflow of understanding the “black box” of the biological system using a systemic sample type. The unraveling of the biomolecules and their interactions allows pathways to be decoded using technology and integrated omics-based approaches.

Table 1.
Comparison of the gold standard and widely used malaria diagnostic tools across the world.
In short, we will discuss the recent development of omics-based malaria biomarkers [31,32,33,34] (Figure 1, Table 2) and the emergence of malaria diagnosis technologies targeting the inorganic biocrystal (hemozoin) as a marker. Hemozoin is a by-product of heme utilisation through various redox transitioning for the parasite’s survival during the intra-erythrocyte cycle [2,3,12,14,15]. Omics-based comprehensive analysis and exploration provide a panel of biomolecules with high throughput techniques to answer various questions related to diagnostics, parasite species differentiation, the antimalarial status of the infecting parasite, and prognosis. We will summarise the state-of-the-art diagnostic technologies and the current challenges and discuss the emerging prospects for multi-omics-based sensors.

Table 2.
List of tools for malaria diagnosis.
1.2. Multi-Omics Approaches in Malaria Diagnosis
Diagnosis of Plasmodium pathogenic species can be performed in clinical settings using phenome assays such as light microscopy-based diagnosis and differentiation of Plasmodium species [49]. The infected red blood cells (iRBCs) exhibit alterations in the morphology and physiology, exploited for enriching iRBCs using microfluidics [5,50], flow cytometry [51,52], magnetic beads [53], or detergent [54] gradients for reliable diagnosis of malaria pathogen, for studying the parasite, or for understanding the pathobiology of the parasite. The phenotypic changes include the loss of flexibility of the RBC (more in P. falciparum as compared to in P. vivax) [55], loss of discoidal shape, change of surface proteins making the P. falciparum-iRBCs sticky leading to severe clinical complications such as anemia and cerebral malaria [55], and enhanced permeability to selective ionic entities through a new permeability pathway (NPP) [56] creating an ideal microenvironment for flourishing parasitic growth [57]. These deformities were studied and exploited by researchers and clinicians to diagnose malaria using light microscopy [49] or enrichment of iRBCs [50,51,52,53,58,59] for enhanced chances of differentiation and diagnosis of the malaria parasite. There is a certain amount of human error in microscopy-based manual phenome analysis. To overcome this, Poostchi et al. and Fuhad et al. have successfully reported using captured microscopic images of the infected RBC stages for automation of malaria diagnosis using machine learning [60,61].
Chew et al., Malpartida-Cardenas et al., and Hede et al. have reported a nucleic acid-based mode of diagnosis of Plasmodium species with high accuracy and sensitivity by nested-PCR targeting of ssrRNA [62,63,64]. Nucleic acid-based diagnosis and differentiation studies have also exhibited the identification of polymorphs in different parasite genes leading to resistance against antimalarials, accurate diagnosis, and efficient medication [65,66,67,68,69], and create a mode of disguise from known RDTs such as deletions of PfHRP2/3 [70,71,72,73,74,75]. There are various reports on specific gene sequencing for understanding the polymorphism [76,77,78] of satellite genes leading to multidrug resistance and predicting the lineage of distribution across the globe [79]. In many cases, it may also predict or indicate the cases of malaria relapse [80].
Proteomics of parasite pellet and parasite secreted proteins in plasma give abundant parasite proteins used for diagnostic application [19,20,21]. Additionally, the host protein profile indicates the prognosis of the disease by using a panel of host proteins differentially regulated in malaria compared to febrile control or healthy volunteers’ serum profiles [17,18,19,81]. The differentially altered host proteins also indicate the affected pathways in severe and non-severe malaria conditions such as lipid metabolism, complement, platelet degranulation, and homeostasis [19]. Hence, the parasite [19,20,21] or host proteome profile [17,18,19,81,82] can be explored for the most recurring and stable peptides, proteins, or metabolites unique to parasite response compared to febrile control. Furthermore, the characteristic of metabolites being highly dynamic and involved in regulating multiple pathways may help in the prognosis of disease beforehand [31,47,48,83,84,85,86]. In the case of malaria, metabolomics has facilitated finding the altered pathways and significantly altered metabolites as compared to the apt controls such as retinol metabolism and glucose metabolism in P. vivax and P. falciparum, respectively [47]. Metabolomics may help predict the chloroquine resistance in the infected patients [85] or erythrocyte metabolism may facilitate understanding of parasite survival in the RBCs for P. falciparum [86]. The tools to perform integrated omics require an annotated genomics database, protein database, and post-translation modifications with major proteins’ functions structure [87]. This information is lacking for P. vivax due to the lack of continuous in vitro culturing property. Swearingen and Lindner, 2018, Swearingen et al., 2017, and Lindner et al., 2019 have reported a few studies to attempt proteogenomics of P. vivax and P. falciparum to compare the salivary stage of the respective parasites [33,87,88]. Another group, Gardinassi et al., has integrated the metabolomics and transcriptomic results of parasites, reporting T-cell activation and activation of innate immunity [31]. The omics field has been driven largely by technological advances that have made cost-efficient, high-throughput analysis of biomolecules possible (Figure 1b). Integration of different omics data types is often used to elucidate potential causative changes that lead to disease or treatment targets, which can then be tested in further molecular studies [88].
However, the omics-based approaches have their limitations (Table 3) and hence provide a limited understanding of the pathogenesis and progression; for example, on a genetic level, there are reports suggesting polymorphism in many of the malaria parasites, P. falciparum [89], that hinders the screening of the antimalarial driving genes and the role of the mutants based on sole proteomics, highlighting the need for a comprehensive proteogenomic study to understand the parasite. At present, due to the lack of knowledge about P. vivax, one cannot categorize the malaria infection as severe or non-severe [90].

Table 3.
SWOT analysis of omics-based malaria diagnostic methodologies.
2. Challenges in the Development of Diagnostic Tools for P. vivax and P. falciparum
Both parasites, P. vivax and P. falciparum belong to the same genus with considerable differences in their G + C content in genetic material and their capability to infect hepatic cells to form hypnozoites, adaptability towards antimalarial drugs, and ability to grow in vitro continuously.
Plasmodium vivax (Pv) exhibits low parasitemia in the host and has an uncharacterized genome and proteome due to a lack of knowledge of how to culture P. vivax in vitro. For several decades, the hepatocytic stage of P. vivax has failed to grow in in vitro conditions. Sangiamsuntorn K et al. [91] and Yongyut Pewkliang et al. [92] successfully produced immortalized mesenchymal stem cells (MSCs) in a stable immortal hepatocytes (imHC) cell culture supporting liver-stage culturing when infected with sporozoites in vitro. However, continuous in vitro P. vivax culture remains a roadblock in understanding its pathobiology [92,93]. Additionally, P. vivax undergoes the quiescence known as hypnozoites in the host liver, which is responsible for disease relapse. The major impediment is that we lack a method or instrument capable of detecting hypnozoites to preclude the relapse. Primaquine and Tafenoquine (in clinical trials) are the most effective drugs against hypnozoites of P. vivax. However, it leads to severe-hemolytic conditions leading to death in classes 1 and 2 G-6-PD deficient patients. This makes the detection of G6PD deficiency in the patient an essential prerequisite in vivax malaria treatment.
On the other hand, P. falciparum is very well studied and has a parasite protein for its diagnosis and differentiation. However, P. falciparum has started deleting HRP2/3 genes, differentiation biomarkers, from the genome, suggesting that the role of the gene might not be essential for the pathogen’s survival [72]. It has engendered the urgent need to explore biomolecules that are not disposable for the parasite. There is a high demand to explore ways to diagnose and differentiate the parasite species and antimalarial drugs for effective treatment (Figure 2).
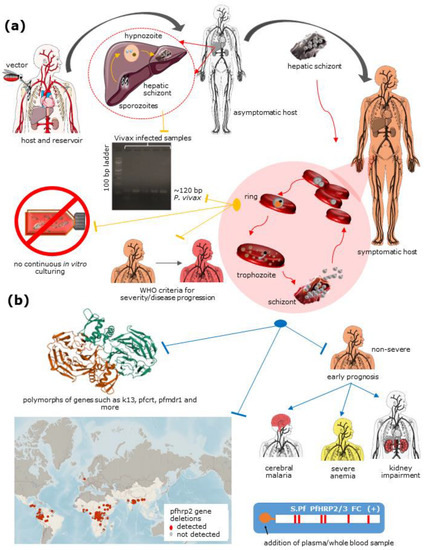
Figure 2.
Challenges related to P. vivax understanding and eradication include: (a) A lack of diagnostic aid for hypnozoites, differentiation of P. vivax from other Plasmodium species, lack of continuous culture in vitro conditions, and a lack of information about P. vivax to document severity criteria for the species; and (b) P. falciparum includes polymorphs in particular genes resulting in antimalarial drug resistance, HRP2/3 deletion in populations across the globe (source: Malaria threat map, WHO) leading to false negatives using RDTs and early prognosis of non-severe falciparum malaria to cerebral malaria, severe anemia, and kidney impairment. The red arrow represents parasite stage transition common to both the species, the yellow arrow represents stage-specific to P. vivax, and the blue arrow represents stage-specific to P. falciparum. This figure was created using Servier medical art templates, licensed under a creative commons attribution 3.0 unreported license; https://smart.servier.com (accessed on 20 March 2021).
3. Recent Advancements in Multi-Omics Based Malaria Diagnosis
Recent advancements in diagnosis techniques have also enabled the in-depth exploration of the biomolecules including a higher resolution and increased accuracy and sensitivity (Box 1). The other major cause of advancement in the diagnostics field is omics-based diagnostic assays that include (1) phenomics-based assays, (2) nucleic acid-based assays, (3) protein-based assays, (4) metabolite-based assays, and (5) non-protein based assays. The various categories of these assays target different stages of the parasite life cycle.
Box 1. Highlights.
- Malaria parasites show a vast difference on their molecular level resulting in unique life cycle stages such as hypnozoites in P. vivax and P. ovale.
- There is a paucity of accurate, reliable, rapid, and pathogen species differentiating diagnostic assays in endemic regions of the world.
- Delayed diagnosis contributes to the augmentation of resistant strains and antimalarial drugs facilitate the selection of mutants, aggravating the condition.
- Advancements in malaria diagnosis are propelled by advancements in technology and omics fields.
- The dearth of understanding about the pathogen results in a scarcity of promising diagnostic or therapeutic targets.
- Omics-based understanding of the pathogen or host response may facilitate understanding of the pathogenesis.
- Integration of multiple omics facilitates a holistic view of pathobiology and highlights crucial biomolecules for diagnostic and therapeutic purposes.
Phenome-based diagnostics have advanced by using whole blood cells on a microarray chip to stain iRBCs using nuclear staining dye and correlating it with microscopy findings. The method is fast, easy to use, and has shown reliable outcomes [94].
The nucleic acid-based diagnosis uses small subunit ribosomal RNA (ssrRNA) to amplify the genus and species-specific stretch to identify Plasmodium species [39]. Loop-mediated isothermal amplification (LAMP) of 18S rRNA and mitochondrial DNA has efficiently detected P. falciparum [63,64]. LAMP coupled with complementary metal-oxide-semiconductor (CMOS) technology provides the diagnosis of P. falciparum along with Artemisinin resistance status in the infected pathogen strain [63]. Hede et al. [64] also demonstrated that the rolling circle enhanced enzyme activity detection (REEAD) assay might be favourable under clinical settings with scarce resources. Genomics-based diagnosis has recently observed a spike in the usage of clustered regularly interspaced short palindromic repeats (CRISPR) for targeted and efficient diagnostics due to its highly sensitive and specific nature [95]. CRISPR has shown a high success rate in the diagnosis of viral infectious diseases such as Zika, Dengue [96], and HPV [97], and is being tested for SARS-CoV2 [98] diagnosis. Similarly, potential CRISPR-based panels are being explored to diagnose and differentiate symptomatic from asymptomatic patients [99], differentiate Plasmodium species, and genotype antimalarial drug resistance [100] in malaria patients.
Proteomics-based assays, such as serum proteomics, have shown steady growth in the field of diagnostic applications for the detection of cancer and other human diseases with high sensitivity [101]. It can also facilitate the understanding of host-parasite interaction by studying the alteration in the host proteome. Ray et al. 2012, 2016, and 2017 have reported various differentially expressed proteins in malaria-infected patient sera under various severity conditions and different time points [17,18,80]. A recent publication from the same group has also exhibited the use of machine learning to predict a biomarker panel of host proteins to diagnose and differentiate P. falciparum malaria from P. vivax malaria [19]. Protein-based diagnosis includes antibody antigen-based interacting assays such as rapid diagnostics tests (RDTs) [102,103,104,105]. Additionally, parasite proteomics exhibits the range of parasite proteins to obtain consistently expressing proteins. These proteins can be sorted to obtain a list of proteins that may serve as potential biomarker candidates [20,21].
Metabolome profiling of serum/plasma [47,81,106] or urine [107] of P. falciparum-infected patients compared to healthy volunteers can also be used for a diagnostic marker search [48]. A P. vivax infected host [47] exhibits significant alteration in glucose metabolism of gluconeogenesis and glycolysis in P. falciparum, whereas there is a significant alteration of retinol metabolism in P. vivax. Metabolites have also been reported to show the prediction of the antimalarial mechanism of action [106]. In addition, the byproduct of hemoglobin, hemozoin, has stood out due to its unique property of storing iron ions in their oxidized state, making it capable of rotating polarized light [108] and providing a magnetic field [2,3,5,30,109,110] that can be used for diagnostic purposes. Morang’a et al. [111] have reported predicting malaria infection using a machine learning approach for hematological parameters [16].
4. Approaches to Integrative Multi-Omics
The integrated omics approach helps unravel the intricacies of pathobiology, given that the required databases and tools are available to the researcher. Of P. falciparum and P. vivax, P. falciparum is well studied, resulting in integrated omics, transcriptomics, and proteomics analysis to understand different stages in infected hosts [112]. Proteogenomics of haptoglobin in infected host blood aided understanding of the correlation between the gene and protein of polymorph in haptoglobin co-dominant alleles to understand the malaria progression [32]. Transcriptomic and proteomic study of the sporozoites and oocytes of P. falciparum extracted from the salivary gland of mosquitos exhibited the role of a set of genes that help in parasite maturation and are capable of infecting the liver in humans [33]. However, not much has been done on the multi-omics integration of P. vivax due to limited information. Swearingen et al. reported using a proteogenomic tool to prepare a customized database of P. vivax isolated from the Thai population. The study explains the similarity and differences between P. vivax and P. falciparum proteomes [109]. The metabolomic and transcriptomic integration of the patients’ semi-immune and naïve plasma samples has exhibited the activation of T cells and innate immunity [31]. However, due to the lack of vivax information, many of the crucial aspects of malaria pathology remains an unsolved impediment.
5. Advantage of Multi-Omics Approaches
The infected human exhibits symptoms on exposure to a disease that results from the alteration of the biochemical, physiological, or combination of both. The understanding of the disease is first looked at based on physiological alterations such as RBCs morphology change in the presence of the Plasmodium parasite, which can be confirmed by using microscopy [49]. The altered shape can be further researched for molecular reasons to have the altered morphology using advanced technologies such as proteomics, genomics, and metabolomics tools [110]. Understanding altered biomolecules lead to the following question: which pathways are affected due to infection? Pathway enrichment with significantly altered biomolecules facilitates the identification of the driving molecules in the affected pathways and affected biomolecules, resulting in potential candidates for therapeutic purposes [110]. This increases the demand for an integrated omics approach in search of a potential target for drugs or for creating vaccines with minimal disturbance to the microenvironment of the target cells. All the organisms are similar to a great extent, yet each human exhibits vast differences in terms of their environment and how individuals may react to similar exposures. Similarly, the same cell in different organisms of the same species has its microenvironment and genetic polymorphs/mutants that result in a personalized response to a similar stimulus or exposure, engendering the need for precision medicines. However, the biological system is so vastly variant [113,114] and dynamic that the key to designing an effective treatment or cure is knowledge [115]. Knowing all of the possible omics and their integration is key to understanding the organism as per its biochemical and genetic makeup, but with vast knowledge, the biggest hurdle is handling the enormous variants involved in making sense out of data without introducing false discoveries.
6. Limitation in Multi-Omics Approaches
Multiple omics enhance perceptiveness towards an unknown organism such as P. vivax; however, each omics individually has its limitations. For example, the phenome of the infected cells is altered in terms of formability, rigidity, and adhesive character. However, these phenotypes can be due to other factors and micro-environmental variations such as sickle cell anemia [116], other intracellular parasites, or even parasites contributing to the host RBCs’ cytoskeleton [117] leading to the rigidity of the RBCs. Hence, the phenotype may not provide clear evidence of parasite species infecting the RBC. Therefore, understanding molecular-level information is crucial. On the one hand, proteomics has been developed to successfully identify the peptide using hybrid mass spectrometry [118]. Diagnostic biomarkers and prognostic biomarker exploration for making RDTs have been possible because of the proteomics field, as differentially expressed genes lead to diseased conditions in autonomous diseases. However, proteins are dynamic compared to nucleic acids, which may result in an increased rate of false positives or false negatives [119].
On the other hand, genomic research has led us from predicting genotype by studying phenotype [120] to target point mutation in the genome [121]. Providing the knowledge about one’s heritage and vulnerability to specific defects and diseases will facilitate a prolonged lifespan, but includes highly interlinked proteins, leading to a false prediction of a gene defect and related disease(s). Hence, it raises the need for an integrated tool to compare and correlate the gene defect, protein expression, and its effect on the whole system rather than studying the individual proteins or genes [87]. Philipp Mertins et al. included the correlation of genes with their mRNA expression and mRNA expression for the selected proteins. They reported that all the genes, mRNA, and proteins cannot be correlated similarly [122]. Hence, understanding the clinical conditions and factors responsible for it requires a thorough study of the proteome and genome of the same subject. Still, sample availability is the limiting step.
In contrast, metabolomics is easy to extract but is highly dynamic, unlike proteins/antibodies, which have a higher self-life resulting in false positives even after the infection is clear from the system. However, handling metabolites is a challenging task; due to their highly dynamic nature, they tend to convert to their byproducts if not ceased properly. There are tools such as Qemistree that look promising in retracing the molecular signature back to predict the precursor metabolite [123], but it might impose a challenge for diagnostic and prognostic purposes.
7. Current Challenges and Future Opportunities
Diagnostic and prognostic biomarkers are required with higher accuracy, longer shelf-life, and faster turnover rate. It is necessary to have a panel of biomolecules from host and parasite to accurately detect the causative agent and its antimalarial profile, such as geneXpert in Mycobacterium tuberculosis diagnosis [124]. The possibility of curating and integrating knowledge can be achieved by an online consortium of published data such as the ICPC [125] and CPTAC [126,127,128] in case of cancer. A unified SOP data sharing for meta-analysis of the multination population might help us to better understand the pathobiology.
The Foundation for Innovative New Diagnostics (FIND), along with WHO as a collaborating centre, has focused on laboratory strengthening and diagnostic technology evaluation since 2003. Utilization of funding to translate the findings to the clinics where the diagnosis, differentiation of parasite species, prognosis, and antimalarial profile can be screened in the minimum time with the highest accuracy and sensitivity is crucial to provide a better quality of life in a malaria-prone population. The dormant stage of vivax diagnosis coupled with a quantitative assay for G6PD testing or polymorph confirmation is necessary to explore the distribution of classes of G6PD deficiency in the population followed by Primaquine treatment. Identifying dormant vivax carriers and the asymptomatic patients may lead to a targeted approach to eliminate the vivax reservoirs. Proper classification of non-severe and severe vivax malaria is crucial in a timely prognosis for effective treatment (Box 2). These are the current challenges in reducing malaria up to 90% by 2030, as per WHO’s global technical strategy for malaria.
Box 2. Unresolved Questions.
- A biomarker or tool is necessary for the differentiation of malaria-causing pathogen species in humans with high sensitivity and specificity.
- A biomarker or methodology is necessary for the diagnosis of asymptomatic malaria patients and hypnozoite carriers for eliminating the parasite reservoir from the host.
- There is inaccessibility of the P. vivax parasite for comprehensive analysis of the pathogen due to low parasitemia in clinical samples.
- There is an inability to grow P. vivax continuously in the in vitro cell culture condition.
- The paucity of complete proteomic and metabolomic databases of P. vivax is one of the major roadblocks in the quest for a diagnostic, differentiation, and prognostic biomarker search.
- There is a lack of annotation of the genome of P. vivax. Most of the genes are hypothetical, uncharacterized, or unannotated.
- There is a lack of clinical information with heterogeneity information such as gender, race, age, and geographic location to understand the vivax malaria severity.
- There is a lack of information on the correlation between parasitemia and the severity of vivax malaria.
- The coupled diagnosis of G-6-PD deficiency class along with P. vivax for the efficient treatment and elimination of the parasite reservoir from the host system.
8. Concluding Remarks and Future Perspective
The advancements in the field of diagnosis concerning the development of technology on the omics-driven front have facilitated highly sensitive and specific diagnostic prototypes [34,114], but a solid political will (e.g., human capital, logistics) has to follow, as many of these malaria-endemic regions are in developing countries. P. vivax information is the source of extrapolation for the understanding of P. falciparum. Hence, the need has peaked for a short-culture-based multi-omic for the patient. Multi-omic information may help diagnostics and therapeutic modalities on a mass screening of vivax malaria from mixed or other parasite species in endemic regions. Additionally, the biosensor development towards the ultra-low parasitemia detection to check the correlation of parasitemia and severity for prognostics purposes, along with a global repository for the curated data of the clinical conditions, might help in understanding the heterogeneity involved in providing global/customized statistically significant biomarker candidates using artificial intelligence. This will help achieve the goal of timely diagnosis and efficient treatment towards the UN mission of global malaria elimination.
Author Contributions
The article is conceptualized and written by W.K.P., S.S. and S.A. Original draft preparation by S.A. Figures and tables prepared by S.A. Review and editing by W.K.P. and S.S. Supervision by W.K.P. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This review received no external funding. The funding agencies involved in this research are MHRD-UAY (IITB001) RD/0120-MHRDUP3-001 and BT/INF/22/SP23026/2017, Infrastructure Facility for Advanced Research and Education in Diagnostics. This research was supported by the MYRG2020-00132-IME, Songshan Lake Materials Laboratory Innovation Grant.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The figure was created using Servier Medical Art templates, licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com (accessed on 20 March 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. WHO World Malaria Report 2019. Available online: https://www.who.int/malaria/publications/world-malaria-report-2019/en/ (accessed on 25 May 2020).
- Peng, W.K.; Chen, L.; Han, J. Development of Miniaturized, Portable Magnetic Resonance Relaxometry System for Point-of-Care Medical Diagnosis. Rev. Sci. Instrum. 2012, 83, 095115. [Google Scholar] [CrossRef]
- Peng, W.K.; Kong, T.F.; Ng, C.S.; Chen, L.; Huang, Y.; Bhagat, A.A.S.; Nguyen, N.-T.; Preiser, P.R.; Han, J. Micromagnetic Resonance Relaxometry for Rapid Label-Free Malaria Diagnosis. Nat. Med. 2014, 20, 1069–1073. [Google Scholar] [CrossRef]
- Toh, R.J.; Peng, W.K.; Han, J.; Pumera, M. Direct In Vivo Electrochemical Detection of Haemoglobin in Red Blood Cells. Sci. Rep. 2014, 4, 6209. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.F.; Ye, W.; Peng, W.K.; Hou, H.W.; Preiser, P.R.; Nguyen, N.T.; Han, J. Enhancing Malaria Diagnosis through Microfluidic Cell Enrichment and Magnetic Resonance Relaxometry Detection. Sci. Rep. 2015, 5, 11425. [Google Scholar] [CrossRef]
- Dupré, A.; Lei, K.-M.; Mak, P.-I.; Martins, R.P.; Peng, W.K. Micro- and Nanofabrication NMR Technologies for Point-of-Care Medical Applications—A Review. Microelectron. Eng. 2019, 209, 66–74. [Google Scholar] [CrossRef]
- Choi, J.; Cho, S.-J.; Kim, Y.T.; Shin, H. Development of a Film-Based Immunochromatographic Microfluidic Device for Malaria Diagnosis. Biomed Microdevices 2019, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.; Jones, S.; Gatton, M.L.; Barnwell, J.W.; Cheng, Q.; Chiodini, P.L.; Glenn, J.; Incardona, S.; Kosack, C.; Luchavez, J.; et al. A Review of the WHO Malaria Rapid Diagnostic Test Product Testing Programme (2008–2018): Performance, Procurement and Policy. Malar. J. 2019, 18, 387. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Malaria Diagnostic Tests. Available online: https://www.cdc.gov/malaria/diagnosis_treatment/diagnostic_tools.html (accessed on 7 March 2021).
- Fridley, G.E.; Le, H.; Yager, P. Highly Sensitive Immunoassay Based on Controlled Rehydration of Patterned Reagents in a 2-Dimensional Paper Network. Anal. Chem. 2014, 86, 6447–6453. [Google Scholar] [CrossRef]
- Lafleur, L.; Stevens, D.; McKenzie, K.; Ramachandran, S.; Spicar-Mihalic, P.; Singhal, M.; Arjyal, A.; Osborn, J.; Kauffman, P.; Yager, P.; et al. Progress toward Multiplexed Sample-to-Result Detection in Low Resource Settings Using Microfluidic Immunoassay Cards. Lab Chip 2012, 12, 1119–1127. [Google Scholar] [CrossRef]
- Gregorio, E.D.; Ferrauto, G.; Schwarzer, E.; Gianolio, E.; Valente, E.; Ulliers, D.; Aime, S.; Skorokhod, O. Relaxometric Studies of Erythrocyte Suspensions Infected by Plasmodium Falciparum: A Tool for Staging Infection and Testing Anti-Malarial Drugs. Magn. Reson. Med. 2020, 84, 3366–3378. [Google Scholar] [CrossRef]
- Thamarath, S.S.; Xiong, A.; Lin, P.-H.; Preiser, P.R.; Han, J. Enhancing the Sensitivity of Micro Magnetic Resonance Relaxometry Detection of Low Parasitemia Plasmodium Falciparum in Human Blood. Sci. Rep. 2019, 9, 2555. [Google Scholar] [CrossRef]
- Gupta, M.; Singh, K.; Lobiyal, D.K.; Safvan, C.P.; Sahu, B.K.; Yadav, P.; Singh, S. A Sensitive On-Chip Probe–Based Portable Nuclear Magnetic Resonance for Detecting Low Parasitaemia Plasmodium Falciparum in Human Blood. Med. Devices Sens. 2020, 3, e10098. [Google Scholar] [CrossRef]
- Han, J.; Peng, W.K. Reply to “Considerations Regarding the Micromagnetic Resonance Relaxometry Technique for Rapid Label-Free Malaria Diagnosis”. Nat. Med. 2015, 21, 1387–1389. [Google Scholar] [CrossRef]
- Peng, W.K. Clustering Nuclear Magnetic Resonance: Machine Learning Assistive Rapid Two-Dimensional Relaxometry Mapping. Eng. Rep. 2021, 3, e12383. [Google Scholar] [CrossRef]
- Ray, S.; Patel, S.K.; Venkatesh, A.; Chatterjee, G.; Ansari, N.N.; Gogtay, N.J.; Thatte, U.M.; Gandhe, P.; Varma, S.G.; Patankar, S.; et al. Quantitative Proteomics Analysis of Plasmodium Vivax Induced Alterations in Human Serum during the Acute and Convalescent Phases of Infection. Sci. Rep. 2017, 7, 4400. [Google Scholar] [CrossRef]
- Ray, S.; Patel, S.K.; Venkatesh, A.; Bhave, A.; Kumar, V.; Singh, V.; Chatterjee, G.; Shah, V.G.; Sharma, S.; Renu, D.; et al. Clinicopathological Analysis and Multipronged Quantitative Proteomics Reveal Oxidative Stress and Cytoskeletal Proteins as Possible Markers for Severe Vivax Malaria. Sci. Rep. 2016, 6, 24557. [Google Scholar] [CrossRef]
- Kumar, V.; Ray, S.; Aggarwal, S.; Biswas, D.; Jadhav, M.; Yadav, R.; Sabnis, S.V.; Banerjee, S.; Talukdar, A.; Kochar, S.K.; et al. Multiplexed Quantitative Proteomics Provides Mechanistic Cues for Malaria Severity and Complexity. Commun. Biol. 2020, 3, 683. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, A.; Lahiri, A.; Reddy, P.J.; Shastri, J.; Bankar, S.; Patankar, S.; Srivastava, S. Identification of Highly Expressed Plasmodium Vivax Proteins from Clinical Isolates Using Proteomics. Proteom. Clin. Appl. 2018, 12, e1700046. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, A.; Aggarwal, S.; Kumar, S.; Rajyaguru, S.; Kumar, V.; Bankar, S.; Shastri, J.; Patankar, S.; Srivastava, S. Comprehensive Proteomics Investigation of P. Vivax-Infected Human Plasma and Parasite Isolates. BMC Infect. Dis. 2020, 20, 188. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Jalandra, R.; Sharma, M.; Prakash, H.; Makharia, G.K.; Solanki, P.R.; Singh, R.; Kumar, A. Omics Technologies for Improved Diagnosis and Treatment of Colorectal Cancer: Technical Advancement and Major Perspectives. Biomed. Pharmacother. 2020, 131, 110648. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Carini, C. Are Innovation and New Technologies in Precision Medicine Paving a New Era in Patients Centric Care? J. Transl. Med. 2019, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Raja, K.; Patrick, M.; Gao, Y.; Madu, D.; Yang, Y.; Tsoi, L.C. A Review of Recent Advancement in Integrating Omics Data with Literature Mining towards Biomedical Discoveries. Int. J. Genom. 2017, 2017, e6213474. [Google Scholar] [CrossRef]
- Debnath, M.; Prasad, G.; Bisen, P. Omics Technology. In Molecular Diagnostics: Promises and Possibilities; Springer: Dordrecht, The Netherlands, 2010; pp. 11–31. ISBN 978-90-481-3260-7. [Google Scholar]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances and Future Approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and Metabolomic Characterization of Human Glioblastoma. Cancer Cell 2021, 39, 509–528. [Google Scholar] [CrossRef]
- Petralia, F.; Tignor, N.; Reva, B.; Koptyra, M.; Chowdhury, S.; Rykunov, D.; Krek, A.; Ma, W.; Zhu, Y.; Ji, J.; et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020, 183, 1962–1985.e31. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef]
- Chen, K.; Yuen, C.; Aniweh, Y.; Preiser, P.; Liu, Q. Towards Ultrasensitive Malaria Diagnosis Using Surface Enhanced Raman Spectroscopy. Sci. Rep. 2016, 6, 20177. [Google Scholar] [CrossRef] [PubMed]
- Gardinassi, L.G.; Arévalo-Herrera, M.; Herrera, S.; Cordy, R.J.; Tran, V.; Smith, M.R.; Johnson, M.S.; Chacko, B.; Liu, K.H.; Darley-Usmar, V.M.; et al. Integrative Metabolomics and Transcriptomics Signatures of Clinical Tolerance to Plasmodium Vivax Reveal Activation of Innate Cell Immunity and T Cell Signaling. Redox Biol. 2018, 17, 158–170. [Google Scholar] [CrossRef]
- Awasthi, G.; Tyagi, S.; Kumar, V.; Patel, S.K.; Rojh, D.; Sakrappanavar, V.; Kochar, S.K.; Talukdar, A.; Samanta, B.; Das, A.; et al. A Proteogenomic Analysis of Haptoglobin in Malaria. Proteom. Clin. Appl. 2018, 12, e1700077. [Google Scholar] [CrossRef]
- Lindner, S.E.; Swearingen, K.E.; Shears, M.J.; Walker, M.P.; Vrana, E.N.; Hart, K.J.; Minns, A.M.; Sinnis, P.; Moritz, R.L.; Kappe, S.H.I. Transcriptomics and Proteomics Reveal Two Waves of Translational Repression during the Maturation of Malaria Parasite Sporozoites. Nat. Commun. 2019, 10, 4964. [Google Scholar] [CrossRef]
- Peng, W.K.; Paesani, D. Omics Meeting Onics: Towards the Next Generation of Spectroscopic-Based Technologies in Personalized Medicine. J. Pers. Med. 2019, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Ngasala, B.; Bushukatale, S. Evaluation of Malaria Microscopy Diagnostic Performance at Private Health Facilities in Tanzania. Malar. J. 2019, 18, 375. [Google Scholar] [CrossRef]
- Ghindilis, A.L.; Chesnokov, O.; Ngasala, B.; Smith, M.W.; Smith, K.; Mårtensson, A.; Oleinikov, A.V. Detection of Sub-Microscopic Blood Levels of Plasmodium Falciparum Using Tandem Oligonucleotide Repeat Cascade Amplification (TORCA) Assay with an Attomolar Detection Limit. Sci. Rep. 2019, 9, 2901. [Google Scholar] [CrossRef]
- Heraud, P.; Chatchawal, P.; Wongwattanakul, M.; Tippayawat, P.; Doerig, C.; Jearanaikoon, P.; Perez-Guaita, D.; Wood, B.R. Infrared Spectroscopy Coupled to Cloud-Based Data Management as a Tool to Diagnose Malaria: A Pilot Study in a Malaria-Endemic Country. Malar. J. 2019, 18, 348. [Google Scholar] [CrossRef]
- Adeoye, G.O.; Nga, I.C. Comparison of Quantitative Buffy Coat Technique (QBC) with Giemsa-Stained Thick Film (GTF) for Diagnosis of Malaria. Parasitol. Int. 2007, 56, 308–312. [Google Scholar] [CrossRef]
- Pinheirob, V.E.; Thaithongc, S.; Browna, K.N. High Sensitivity of Detection of Human Malaria Parasites by the Use of Nested Polymerase Chain Reaction. Mol. Biochem. Parasitol. 1993, 61, 315–320. [Google Scholar] [CrossRef]
- Han, E.-T.; Watanabe, R.; Sattabongkot, J.; Khuntirat, B.; Sirichaisinthop, J.; Iriko, H.; Jin, L.; Takeo, S.; Tsuboi, T. Detection of Four Plasmodium Species by Genus- and Species-Specific Loop-Mediated Isothermal Amplification for Clinical Diagnosis. J. Clin. Microbiol. 2007, 45, 2521–2528. [Google Scholar] [CrossRef] [PubMed]
- Sirichaisinthop, J.; Buates, S.; Watanabe, R.; Han, E.-T.; Suktawonjaroenpon, W.; Krasaesub, S.; Takeo, S.; Tsuboi, T.; Sattabongkot, J. Evaluation of Loop-Mediated Isothermal Amplification (LAMP) for Malaria Diagnosis in a Field Setting. Am. J. Trop. Med. Hyg. 2011, 85, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Kikuchi, A.; Kaneko, A.; Isozumi, R.; Teramoto, I.; Kimura, M.; Hirasawa, N.; Hiratsuka, M. Rapid and Sensitive Multiplex Single-Tube Nested PCR for the Identification of Five Human Plasmodium Species. Parasitol. Int. 2018, 67, 277–283. [Google Scholar] [CrossRef]
- Jang, J.W.; Cho, C.H.; Han, E.T.; An, S.S.A.; Lim, C.S. PLDH Level of Clinically Isolated Plasmodium Vivax and Detection Limit of PLDH Based Malaria Rapid Diagnostic Test. Malar. J. 2013, 12, 181. [Google Scholar] [CrossRef]
- Wongsrichanalai, C.; Barcus, M.J.; Muth, S.; Sutamihardja, A.; Wernsdorfer, W.H. A Review of Malaria Diagnostic Tools: Microscopy and Rapid Diagnostic Test (RDT); American Society of Tropical Medicine and Hygiene: Arlington, VA, USA, 2007. [Google Scholar]
- Mens, P.F.; Matelon, R.J.; Nour, B.Y.; Newman, D.M.; Schallig, H.D. Laboratory Evaluation on the Sensitivity and Specificity of a Novel and Rapid Detection Method for Malaria Diagnosis Based on Magneto-Optical Technology (MOT). Malar. J. 2010, 9, 207. [Google Scholar] [CrossRef]
- Langen, A.J.D.; Dillen, J.V.; Witte, P.; de Mucheto, S.; Nagelkerke, N.; Kager, P. Automated Detection of Malaria Pigment: Feasibility for Malaria Diagnosing in an Area with Seasonal Malaria in Northern Namibia. Trop. Med. Int. Health 2006, 11, 809–816. [Google Scholar] [CrossRef]
- Na, J.; Khan, A.; Kim, J.K.; Wadood, A.; Choe, Y.L.; Walker, D.I.; Jones, D.P.; Lim, C.S.; Park, Y.H. Discovery of Metabolic Alterations in the Serum of Patients Infected with Plasmodium Spp. by High-Resolution Metabolomics. Metabolomics 2019, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Leopold, S.J.; Ghose, A.; Allman, E.L.; Kingston, H.W.F.; Hossain, A.; Dutta, A.K.; Plewes, K.; Chotivanich, K.; Day, N.P.J.; Tarning, J.; et al. Identifying the Components of Acidosis in Patients with Severe Plasmodium Falciparum Malaria Using Metabolomics. J. Infect. Dis. 2019, 219, 1766–1776. [Google Scholar] [CrossRef]
- Payne, D. Use and Limitations of Light Microscopy for Diagnosing Malaria at the Primary Health Care Level. Bull. World Health Organ. 1988, 66, 621–626. [Google Scholar]
- Geislinger, T.M.; Chan, S.; Moll, K.; Wixforth, A.; Wahlgren, M.; Franke, T. Label-Free Microfluidic Enrichment of Ring-Stage Plasmodium Falciparum-Infected Red Blood Cells Using Non-Inertial Hydrodynamic Lift. Malar. J. 2014, 13, 375. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Philipp, S.; Oberg, H.-H.; Janssen, O.; Leippe, M.; Gelhaus, C. Isolation of Erythrocytes Infected with Viable Early Stages of Plasmodium Falciparum by Flow Cytometry. Cytom. A 2012, 81, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.W.; Kim, J.Y.; Yoon, J.; Yoon, S.Y.; Cho, C.H.; Han, E.T.; An, S.S.A.; Lim, C.S. Flow Cytometric Enumeration of Parasitemia in Cultures of Plasmodium Falciparum Stained with SYBR Green I and CD235A. Sci. World J. 2014, 2014, 536723. [Google Scholar] [CrossRef]
- Mata-Cantero, L.; Lafuente, M.J.; Sanz, L.; Rodriguez, M.S. Magnetic Isolation of Plasmodium Falciparum Schizonts IRBCs to Generate a High Parasitaemia and Synchronized in Vitro Culture. Malar. J. 2014, 13, 112. [Google Scholar] [CrossRef]
- Brown, A.C.; Moore, C.C.; Guler, J.L. Cholesterol-Dependent Enrichment of Understudied Erythrocytic Stages of Human Plasmodium Parasites. Sci. Rep. 2020, 10, 4591. [Google Scholar] [CrossRef] [PubMed]
- Suwanarusk, R.; Cooke, B.M.; Dondorp, A.M.; Silamut, K.; Sattabongkot, J.; White, N.J.; Udomsangpetch, R. The Deformability of Red Blood Cells Parasitized by Plasmodium Falciparum and P. Vivax. J. Infect. Dis. 2004, 189, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Staines, H.M.; Ashmore, S.; Felgate, H.; Moore, J.; Powell, T.; Ellory, J.C. Solute Transport via the New Permeability Pathways in Plasmodium Falciparum–Infected Human Red Blood Cells Is Not Consistent with a Simple Single-Channel Model. Blood 2006, 108, 3187–3194. [Google Scholar] [CrossRef][Green Version]
- Mohandas, N.; An, X. Malaria and Human Red Blood Cells. Med. Microbiol. Immunol. 2012, 201, 593–598. [Google Scholar] [CrossRef]
- Brown, A.C.; Guler, J.L. SLOPE: A Two-Part Method for the Enrichment of Ring Stage Plasmodium falciparum Parasites. bioRxiv 2018, 474338. [Google Scholar] [CrossRef]
- Trang, D.T.X.; Huy, N.T.; Kariu, T.; Tajima, K.; Kamei, K. One-Step Concentration of Malarial Parasite-Infected Red Blood Cells and Removal of Contaminating White Blood Cells. Malar. J. 2004, 3, 7. [Google Scholar] [CrossRef][Green Version]
- Poostchi, M.; Silamut, K.; Maude, R.J.; Jaeger, S.; Thoma, G. Image Analysis and Machine Learning for Detecting Malaria. Transl. Res. 2018, 194, 36–55. [Google Scholar] [CrossRef]
- Fuhad, K.M.; Tuba, J.F.; Sarker, M.; Ali, R.; Momen, S.; Mohammed, N.; Rahman, T. Deep Learning Based Automatic Malaria Parasite Detection from Blood Smear and Its Smartphone Based Application. Diagnostics 2020, 10, 329. [Google Scholar] [CrossRef]
- Chew, C.H.; Lim, Y.A.L.; Lee, P.C.; Mahmud, R.; Chua, K.H. Hexaplex PCR Detection System for Identification of Five Human Plasmodium Species with an Internal Control. J. Clin. Microbiol. 2012, 50, 4012–4019. [Google Scholar] [CrossRef]
- Malpartida-Cardenas, K.; Miscourides, N.; Rodriguez-Manzano, J.; Yu, L.-S.; Moser, N.; Baum, J.; Georgiou, P. Quantitative and Rapid Plasmodium Falciparum Malaria Diagnosis and Artemisinin-Resistance Detection Using a CMOS Lab-on-Chip Platform. Biosens. Bioelectron. 2019, 145, 111678. [Google Scholar] [CrossRef] [PubMed]
- DNA Sensors for Malaria Diagnosis. Nano LIFE. Available online: https://www.worldscientific.com/doi/abs/10.1142/S1793984415410032 (accessed on 25 February 2021).
- Patgiri, S.J.; Sarma, K.; Sarmah, N.; Bhattacharyya, N.; Sarma, D.K.; Nirmolia, T.; Bhattacharyya, D.R.; Mohapatra, P.K.; Bansal, D.; Bharti, P.K.; et al. Characterization of Drug Resistance and Genetic Diversity of Plasmodium Falciparum Parasites from Tripura, Northeast India. Sci. Rep. 2019, 9, 13704. [Google Scholar] [CrossRef]
- Nayebare, P.; Asua, V.; Conrad, M.D.; Kajubi, R.; Kakuru, A.; Nankabirwa, J.I.; Muhanguzi, D.; Dorsey, G.; Kamya, M.R.; Nsobya, S.; et al. Associations between Malaria-Preventive Regimens and Plasmodium Falciparum Drug Resistance-Mediating Polymorphisms in Ugandan Pregnant Women. Antimicrob. Agents Chemother. 2020, 64, 12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, S.P.; Bhatt, R.; Singh, V. Genetic Profiling of the Plasmodium Falciparum Parasite Population in Uncomplicated Malaria from India. Malar. J. 2019, 18, 385. [Google Scholar] [CrossRef] [PubMed]
- Joy, S.; Mukhi, B.; Ghosh, S.K.; Achur, R.N.; Gowda, D.C.; Surolia, N. Drug Resistance Genes: Pvcrt-o and Pvmdr-1 Polymorphism in Patients from Malaria Endemic South Western Coastal Region of India. Malar. J. 2018, 17, 40. [Google Scholar] [CrossRef]
- Anantabotla, V.M.; Antony, H.A.; Parija, S.C.; Rajkumari, N.; Kini, J.R.; Manipura, R.; Nag, V.L.; Gadepalli, R.; Chayani, N.; Patro, S. Polymorphisms in Genes Associated with Drug Resistance of Plasmodium Vivax in India. Parasitol. Int. 2019, 70, 92–97. [Google Scholar] [CrossRef]
- Sepúlveda, N.; Phelan, J.; Diez-Benavente, E.; Campino, S.; Clark, T.G.; Hopkins, H.; Sutherland, C.; Drakeley, C.J.; Beshir, K.B. Global Analysis of Plasmodium Falciparum Histidine-Rich Protein-2 (Pfhrp2) and Pfhrp3 Gene Deletions Using Whole-Genome Sequencing Data and Meta-Analysis. Infect. Genet. Evol. 2018, 62, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.; Beshir, K.B.; Cunningham, J.; Baiden, F.; Bharmal, J.; Bruxvoort, K.J.; Maiteki-Sebuguzi, C.; Owusu-Agyei, S.; Staedke, S.G.; Hopkins, H. Pfhrp2 and Pfhrp3 Gene Deletions That Affect Malaria Rapid Diagnostic Tests for Plasmodium Falciparum: Analysis of Archived Blood Samples From 3 African Countries. J. Infect. Dis. 2019, 220, 1444–1452. [Google Scholar] [CrossRef]
- Agaba, B.B.; Yeka, A.; Nsobya, S.; Arinaitwe, E.; Nankabirwa, J.; Opigo, J.; Mbaka, P.; Lim, C.S.; Kalyango, J.N.; Karamagi, C.; et al. Systematic Review of the Status of Pfhrp2 and Pfhrp3 Gene Deletion, Approaches and Methods Used for Its Estimation and Reporting in Plasmodium Falciparum Populations in Africa: Review of Published Studies 2010–2019. Malar. J. 2019, 18, 355. [Google Scholar] [CrossRef]
- Bharti, P.K.; Chandel, H.S.; Ahmad, A.; Krishna, S.; Udhayakumar, V.; Singh, N. Prevalence of Pfhrp2 and/or Pfhrp3 Gene Deletion in Plasmodium Falciparum Population in Eight Highly Endemic States in India. PLoS ONE 2016, 11, e0157949. [Google Scholar] [CrossRef]
- Singh, V.; Kojom, L.P. Deletions in the Plasmodium Falciparum Histidine-Rich Protein 2 Gene: An Emerging Threat to the Elimination of Malaria in India. J. Vector Borne Dis. 2019, 56, 85. [Google Scholar] [CrossRef]
- Kumar, N.; Pande, V.; Bhatt, R.M.; Shah, N.K.; Mishra, N.; Srivastava, B.; Valecha, N.; Anvikar, A.R. Genetic Deletion of HRP2 and HRP3 in Indian Plasmodium Falciparum Population and False Negative Malaria Rapid Diagnostic Test. Acta Trop 2013, 125, 119–121. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Mbenda, H.G.N.; Soe, M.T.; Yu, C.; Feng, H.; Kyaw, M.P.; Cui, L.; Zhu, X.; Cao, Y. Genetic Diversity, Natural Selection and Haplotype Grouping of Plasmodium Vivax Duffy-Binding Protein Genes from Eastern and Western Myanmar Borders. Parasit. Vectors 2019, 12, 546. [Google Scholar] [CrossRef]
- Hailemeskel, E.; Menberu, T.; Shumie, G.; Behaksra, S.; Chali, W.; Keffale, M.; Belachew, M.; Shitaye, G.; Mohammed, H.; Abebe, D.; et al. Prevalence of Plasmodium Falciparum Pfcrt and Pfmdr1 Alleles in Settings with Different Levels of Plasmodium Vivax Co-Endemicity in Ethiopia. Int. J. Parasito.l Drugs Drug Resist 2019, 11, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Spotin, A.; Mahami-Oskouei, M.; Ahmadpour, E.; Parsaei, M.; Rostami, A.; Emami, S.; Gholipour, S.; Farmani, M. Global Assessment of Genetic Paradigms of Pvmdr1 Mutations in Chloroquine-Resistant Plasmodium Vivax Isolates. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 339–345. [Google Scholar] [CrossRef]
- Prajapati, S.K.; Joshi, H.; Shalini, S.; Patarroyo, M.A.; Suwanarusk, R.; Kumar, A.; Sharma, S.K.; Eapen, A.; Dev, V.; Bhatt, R.M.; et al. Plasmodium Vivax Lineages: Geographical Distribution, Tandem Repeat Polymorphism, and Phylogenetic Relationship. Malar. J. 2011, 10, 374. [Google Scholar] [CrossRef]
- Groger, M.; Veletzky, L.; Lalremruata, A.; Cattaneo, C.; Mischlinger, J.; Manego Zoleko, R.; Kim, J.; Klicpera, A.; Meyer, E.L.; Blessborn, D.; et al. Prospective Clinical and Molecular Evaluation of Potential Plasmodium Ovale Curtisi and Wallikeri Relapses in a High-Transmission Setting. Clin. Infect. Dis. 2019, 69, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Renu, D.; Srivastava, R.; Gollapalli, K.; Taur, S.; Jhaveri, T.; Dhali, S.; Chennareddy, S.; Potla, A.; Dikshit, J.B.; et al. Proteomic Investigation of Falciparum and Vivax Malaria for Identification of Surrogate Protein Markers. PLoS ONE 2012, 7, e41751. [Google Scholar] [CrossRef]
- Patel, S.K.; Rajora, N.; Kumar, S.; Sahu, A.; Kochar, S.K.; Krishna, C.M.; Srivastava, S. Rapid Discrimination of Malaria- and Dengue-Infected Patients Sera Using Raman Spectroscopy. Anal. Chem. 2019, 91, 7054–7062. [Google Scholar] [CrossRef] [PubMed]
- Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine. Metabolites 2020, 10, 51. [Google Scholar] [CrossRef]
- Gardinassi, L.G.; Cordy, R.J.; Lacerda, M.V.G.; Salinas, J.L.; Monteiro, W.M.; Melo, G.C.; Siqueira, A.M.; Val, F.F.; Tran, V.; Jones, D.P.; et al. Metabolome-Wide Association Study of Peripheral Parasitemia in Plasmodium Vivax Malaria. Int. J. Med. Microbiol. 2017, 307, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Salinas, J.L.; Monteiro, W.M.; Val, F.; Cordy, R.J.; Liu, K.; Melo, G.C.; Siqueira, A.M.; Magalhaes, B.; Galinski, M.R.; et al. Plasma Metabolomics Reveals Membrane Lipids, Aspartate/Asparagine and Nucleotide Metabolism Pathway Differences Associated with Chloroquine Resistance in Plasmodium Vivax Malaria. PLoS ONE 2017, 12, e0182819. [Google Scholar] [CrossRef]
- Srivastava, A.; Creek, D.J.; Evans, K.J.; De Souza, D.; Schofield, L.; Müller, S.; Barrett, M.P.; McConville, M.J.; Waters, A.P. Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites. PLoS Pathog. 2015, 11, e1004882. [Google Scholar] [CrossRef]
- Ruggles, K.V.; Krug, K.; Wang, X.; Clauser, K.R.; Wang, J.; Payne, S.H.; Fenyö, D.; Zhang, B.; Mani, D.R. Methods, Tools and Current Perspectives in Proteogenomics. Mol. Cell Proteom. 2017, 16, 959–981. [Google Scholar] [CrossRef]
- Badhwar, A.; McFall, G.P.; Sapkota, S.; Black, S.E.; Chertkow, H.; Duchesne, S.; Masellis, M.; Li, L.; Dixon, R.A.; Bellec, P. A Multiomics Approach to Heterogeneity in Alzheimer’s Disease: Focused Review and Roadmap. Brain 2020, 143, 1315–1331. [Google Scholar] [CrossRef]
- Fairhurst, R.M.; Dondorp, A.M. Artemisinin-Resistant Plasmodium Falciparum Malaria. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Severe Malaria. Trop. Med. Int. Health 2014, 19, 7–131. [CrossRef]
- Sa-ngiamsuntorn, K.; Wongkajornsilp, A.; Kasetsinsombat, K.; Duangsa-ard, S.; Nuntakarn, L.; Borwornpinyo, S.; Akarasereenont, P.; Limsrichamrern, S.; Hongeng, S. Upregulation of CYP 450s Expression of Immortalized Hepatocyte-like Cells Derived from Mesenchymal Stem Cells by Enzyme Inducers. BMC Biotechnol. 2011, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Pewkliang, Y.; Rungin, S.; Lerdpanyangam, K.; Duangmanee, A.; Kanjanasirirat, P.; Suthivanich, P.; Sa-ngiamsuntorn, K.; Borwornpinyo, S.; Sattabongkot, J.; Patrapuvich, R.; et al. A Novel Immortalized Hepatocyte-like Cell Line (ImHC) Supports in Vitro Liver Stage Development of the Human Malarial Parasite Plasmodium Vivax. Malar. J. 2018, 17, 50. [Google Scholar] [CrossRef]
- Bermúdez, M.; Moreno-Pérez, D.A.; Arévalo-Pinzón, G.; Curtidor, H.; Patarroyo, M.A. Plasmodium Vivax in Vitro Continuous Culture: The Spoke in the Wheel. Malar. J. 2018, 17, 301. [Google Scholar] [CrossRef]
- Yatsushiro, S.; Yamamoto, T.; Yamamura, S.; Abe, K.; Obana, E.; Nogami, T.; Hayashi, T.; Sesei, T.; Oka, H.; Okello-Onen, J.; et al. Application of a Cell Microarray Chip System for Accurate, Highly Sensitive, and Rapid Diagnosis for Malaria in Uganda. Sci. Rep. 2016, 6, 30136. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Mukama, O.; Yuan, T.; He, Z.; Li, Z.; de Dieu Habimana, J.; Hussain, M.; Li, W.; Yi, Z.; Liang, Q.; Zeng, L. A High Fidelity CRISPR/Cas12a Based Lateral Flow Biosensor for the Detection of HPV16 and HPV18. Sens. Actuators B Chem. 2020, 316, 128119. [Google Scholar] [CrossRef]
- Full Article: CRISPR-Cas Systems Based Molecular Diagnostic Tool for Infectious Diseases and Emerging 2019 Novel Coronavirus (COVID-19) Pneumonia. Available online: https://www.tandfonline.com/doi/full/10.1080/1061186X.2020.1769637 (accessed on 11 November 2021).
- Lee, R.A.; Puig, H.D.; Nguyen, P.Q.; Angenent-Mari, N.M.; Donghia, N.M.; McGee, J.P.; Dvorin, J.D.; Klapperich, C.M.; Pollock, N.R.; Collins, J.J. Ultrasensitive CRISPR-Based Diagnostic for Field-Applicable Detection of Plasmodium Species in Symptomatic and Asymptomatic Malaria. PNAS 2020, 117, 25722–25731. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.H.; Hennelly, C.M.; Lin, J.T.; Ubalee, R.; Boyce, R.M.; Mulogo, E.M.; Hathaway, N.; Thwai, K.L.; Phanzu, F.; Kalonji, A.; et al. A Novel CRISPR-Based Malaria Diagnostic Capable of Plasmodium Detection, Species Differentiation, and Drug-Resistance Genotyping. EBioMedicine 2021, 68, 103415. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Peeling, R.W.; WHO-Regional Office for the Western Pacific/TDR. Evaluation of Rapid Diagnostic Tests: Malaria. Nat. Rev. Microbiol. 2006, 4, S34–S38. [Google Scholar] [CrossRef]
- Moody, A. Rapid Diagnostic Tests for Malaria Parasites. Clin. Microbiol. Rev. 2002, 15, 66–78. [Google Scholar] [CrossRef]
- Das, S.; Peck, R.B.; Barney, R.; Jang, I.K.; Kahn, M.; Zhu, M.; Domingo, G.J. Performance of an Ultra-Sensitive Plasmodium Falciparum HRP2-Based Rapid Diagnostic Test with Recombinant HRP2, Culture Parasites, and Archived Whole Blood Samples. Malar. J. 2018, 17, 118. [Google Scholar] [CrossRef]
- Murray, C.K.; Gasser, R.A.; Magill, A.J.; Miller, R.S. Update on Rapid Diagnostic Testing for Malaria. CMR 2008, 21, 97–110. [Google Scholar] [CrossRef]
- Creek, D.J.; Chua, H.H.; Cobbold, S.A.; Nijagal, B.; MacRae, J.I.; Dickerman, B.K.; Gilson, P.R.; Ralph, S.A.; McConville, M.J. Metabolomics-Based Screening of the Malaria Box Reveals Both Novel and Established Mechanisms of Action. Antimicrob. Agents Chemother. 2016, 60, 6650–6663. [Google Scholar] [CrossRef] [PubMed]
- Abdelrazig, S.; Ortori, C.A.; Davey, G.; Deressa, W.; Mulleta, D.; Barrett, D.A.; Amberbir, A.; Fogarty, A.W. A Metabolomic Analytical Approach Permits Identification of Urinary Biomarkers for Plasmodium Falciparum Infection: A Case-Control Study. Malar. J. 2017, 16, 229. [Google Scholar] [CrossRef]
- Newman, D.M.; Heptinstall, J.; Matelon, R.J.; Savage, L.; Wears, M.L.; Beddow, J.; Cox, M.; Schallig, H.D.F.H.; Mens, P.F. A Magneto-Optic Route toward the In Vivo Diagnosis of Malaria: Preliminary Results and Preclinical Trial Data. Biophys. J. 2008, 95, 994–1000. [Google Scholar] [CrossRef][Green Version]
- Swearingen, K.E.; Lindner, S.E.; Flannery, E.L.; Vaughan, A.M.; Morrison, R.D.; Patrapuvich, R.; Koepfli, C.; Muller, I.; Jex, A.; Moritz, R.L.; et al. Proteogenomic Analysis of the Total and Surface-Exposed Proteomes of Plasmodium Vivax Salivary Gland Sporozoites. PLoS Negl. Trop. Dis. 2017, 11, e0005791. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef]
- Morang’a, C.M.; Amenga–Etego, L.; Bah, S.Y.; Appiah, V.; Amuzu, D.S.Y.; Amoako, N.; Abugri, J.; Oduro, A.R.; Cunnington, A.J.; Awandare, G.A.; et al. Machine Learning Approaches Classify Clinical Malaria Outcomes Based on Haematological Parameters. BMC Med. 2020, 18, 375. [Google Scholar] [CrossRef]
- Tomescu, O.A.; Mattanovich, D.; Thallinger, G.G. Integrative Omics Analysis. A Study Based on Plasmodium Falciparum MRNA and Protein Data. BMC Syst. Biol. 2014, 8 (Suppl. 2), S4. [Google Scholar] [CrossRef] [PubMed]
- Loh, T.P.; Peng, W.K.; Chen, L.; Sethi, S.K. Application of Smoothed Continuous Labile Haemoglobin A1c Reference Intervals for Identification of Potentially Spurious HbA1c Results. J. Clin. Pathol. 2014, 67, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Veiga, M.I.; Peng, W.K. Rapid Phenotyping towards Personalized Malaria Medicine. Malar. J. 2020, 19, 68. [Google Scholar] [CrossRef]
- Das, T.; Andrieux, G.; Ahmed, M.; Chakraborty, S. Integration of Online Omics-Data Resources for Cancer Research. Front. Genet. 2020, 11, 578345. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.; Shamoun, M.; Tanski, T.; Eniola-Adefeso, L. Characterization of Bulk Rigidity of Rigid Red Blood Cell Populations in Sickle Cell Patients Using a Parameterization Model of Ektacytometry. Blood 2019, 134, 3572. [Google Scholar] [CrossRef]
- Prajapati, S.K. Alteration of the Red Blood Cell Membrane. In Encyclopedia of Malaria; Hommel, M., Kremsner, P.G., Eds.; Springer: New York, NY, USA, 2021; pp. 1–12. ISBN 978-1-4614-8757-9. [Google Scholar]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and Comprehensive Peptide Identification in Mass Spectrometry-Based Proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Alareqi, L.M.Q.; Mahdy, M.A.K.; Lau, Y.-L.; Fong, M.-Y.; Abdul-Ghani, R.; Ali, A.A.; Cheong, F.-W.; Tawfek, R.; Mahmud, R. Field Evaluation of a PfHRP-2/PLDH Rapid Diagnostic Test and Light Microscopy for Diagnosis and Screening of Falciparum Malaria during the Peak Seasonal Transmission in an Endemic Area in Yemen. Malar. J. 2016, 15, 49. [Google Scholar] [CrossRef]
- Lange, K.; Papp, J.C.; Sinsheimer, J.S.; Sripracha, R.; Zhou, H.; Sobel, E.M. Mendel: The Swiss Army Knife of Genetic Analysis Programs. Bioinformatics 2013, 29, 1568–1570. [Google Scholar] [CrossRef] [PubMed]
- Akcakaya, P.; Bobbin, M.L.; Guo, J.A.; Lopez, J.M.; Clement, M.K.; Garcia, S.P.; Fellows, M.D.; Porritt, M.J.; Firth, M.A.; Carreras, A.; et al. In Vivo CRISPR-Cas Gene Editing with No Detectable Genome-Wide off-Target Mutations. bioRxiv 2018, 272724. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics Connects Somatic Mutations to Signalling in Breast Cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef]
- Tripathi, A.; Vázquez-Baeza, Y.; Gauglitz, J.M.; Wang, M.; Dührkop, K.; Nothias-Esposito, M.; Acharya, D.D.; Ernst, M.; van der Hooft, J.J.J.; Zhu, Q.; et al. Chemically Informed Analyses of Metabolomics Mass Spectrometry Data with Qemistree. Nat. Chem. Biol. 2021, 17, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Ahmad, M.; Iram, S.; Riaz, S.; Akhtar, M.; Aslam, M. GeneXpert Technology. Saudi. Med. J. 2017, 38, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Office of Cancer Clinical Proteomics Research. International Cancer Proteogenome Consortium. Available online: https://proteomics.cancer.gov/programs/international-cancer-proteogenome-consortium (accessed on 22 March 2021).
- Office of Cancer Clinical Proteomics Research. CPTAC. Available online: https://proteomics.cancer.gov/programs/cptac (accessed on 22 March 2021).
- NCI Genomic Data Commons. Clinical Proteomic Tumor Analysis Consortium (CPTAC). Available online: https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/clinical-proteomic-tumor-analysis-consortium-cptac (accessed on 22 March 2021).
- CPTAC-DCC. CPTAC Data Exchange Sandbox. Available online: https://cptac-data-portal.georgetown.edu/cptac/ (accessed on 22 March 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).