Development of a Multiplex Loop-Mediated Isothermal Amplification Assay for Diagnosis of Plasmodium spp., Plasmodium falciparum and Plasmodium vivax
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples and DNA Extraction
2.2. Primer Design
2.3. Multiplex Malaria Pan/Pf/Pv/IC LAMP Assay
2.4. Real-Time Reverse Transcription (RT)-PCR
2.5. Limit of Detection (LOD) Tests of the Multiplex Malaria Pan/Pf/Pv/IC LAMP Assay
3. Results
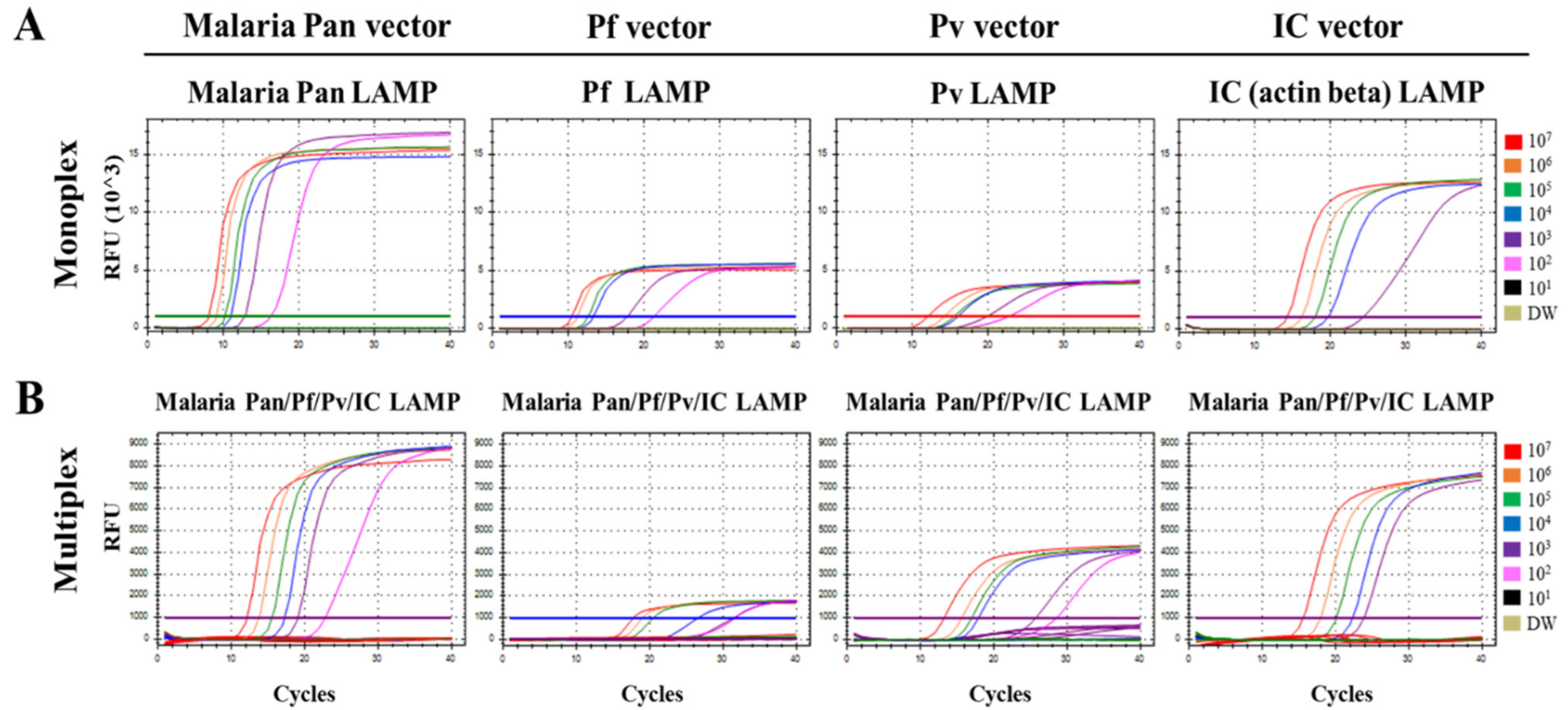
3.1. Optimization of the Multiplex Malaria Pan/Pf/Pv/IC LAMP Primer Set
3.2. Limit of Detection (LOD) of the Multiplex Malaria Pan/Pf/Pv/IC LAMP Assay
3.3. Comparison of Clinical Performance between the Multiplex Malaria Pan/Pf/Pv/IC LAMP and RealStar® Malaria Screen & Type PCR Kit 1.0 Using Clinical Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tuteja, R. Malaria—An overview. FEBS J. 2007, 274, 4670–4679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbacham, W.F.; Ayong, L.; Guewo-Fokeng, M.; Makoge, V. Current Situation of Malaria in Africa. Methods Mol. Biol. 2019, 2013, 29–44. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Malaria Report 2020: 20 Years of Global Progress and Challenges; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Nishimoto, Y.; Arisue, N.; Kawai, S.; Escalante, A.A.; Horii, T.; Tanabe, K.; Hashimoto, T. Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium. Mol. Phylogenetics Evol. 2008, 47, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Cohee, L.M.; Laufer, M.K. Malaria in children. Pediatric Clin. N. Am. 2017, 64, 851. [Google Scholar] [CrossRef] [PubMed]
- Bloland, P.B.; World Health Organization. Drug Resistance in Malaria; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- World Health Organization. Guidelines for the Treatment of Malaria; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Bousema, T.; Drakeley, C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 2011, 24, 377–410. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.L. Laboratory diagnosis of malaria: Conventional and rapid diagnostic methods. Arch. Pathol. Lab. Med. 2013, 137, 805–811. [Google Scholar] [CrossRef] [Green Version]
- Warhurst, D.C.; Williams, J.E. ACP Broadsheet no 148. July 1996. Laboratory diagnosis of malaria. J. Clin. Pathol. 1996, 49, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Poostchi, M.; Silamut, K.; Maude, R.J.; Jaeger, S.; Thoma, G. Image analysis and machine learning for detecting malaria. Transl. Res. 2018, 194, 36–55. [Google Scholar] [CrossRef] [Green Version]
- Ohrt, C.; Purnomo; Sutamihardja, M.A.; Tang, D.; Kain, K.C. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J. Infect. Dis. 2002, 186, 540–546. [Google Scholar] [CrossRef]
- Berzosa, P.; de Lucio, A.; Romay-Barja, M.; Herrador, Z.; González, V.; García, L.; Fernández-Martínez, A.; Santana-Morales, M.; Ncogo, P.; Valladares, B.; et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar. J. 2018, 17, 333. [Google Scholar] [CrossRef]
- Lee, M.A.; Tan, C.H.; Aw, L.T.; Tang, C.S.; Singh, M.; Lee, S.H.; Chia, H.P.; Yap, E.P. Real-time fluorescence-based PCR for detection of malaria parasites. J. Clin. Microbiol. 2002, 40, 4343–4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pöschl, B.; Waneesorn, J.; Thekisoe, O.; Chutipongvivate, S.; Karanis, P. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am. J. Trop. Med. Hyg. 2010, 83, 56–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, L.L.; Wong, B.W.; Ma, E.H.; Chan, K.H.; Chow, L.M.; Abeyewickreme, W.; Tangpukdee, N.; Yuen, K.Y.; Guan, Y.; Looareesuwan, S.; et al. Sensitive and inexpensive molecular test for falciparum malaria: Detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 2006, 52, 303–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersting, S.; Rausch, V.; Bier, F.F.; von Nickisch-Rosenegk, M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar. J. 2014, 13, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mens, P.F.; Schoone, G.J.; Kager, P.A.; Schallig, H.D. Detection and identification of human Plasmodium species with real-time quantitative nucleic acid sequence-based amplification. Malar. J. 2006, 5, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paris, D.H.; Imwong, M.; Faiz, A.M.; Hasan, M.; Yunus, E.B.; Silamut, K.; Lee, S.J.; Day, N.P.; Dondorp, A.M. Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am. J. Trop. Med. Hyg. 2007, 77, 972–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picot, S.; Cucherat, M.; Bienvenu, A.L. Systematic review and meta-analysis of diagnostic accuracy of loop-mediated isothermal amplification (LAMP) methods compared with microscopy, polymerase chain reaction and rapid diagnostic tests for malaria diagnosis. Int. J. Infect. Dis. 2020, 98, 408–419. [Google Scholar] [CrossRef]
- Sirichaisinthop, J.; Buates, S.; Watanabe, R.; Han, E.T.; Suktawonjaroenpon, W.; Krasaesub, S.; Takeo, S.; Tsuboi, T.; Sattabongkot, J. Evaluation of loop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am. J. Trop. Med. Hyg. 2011, 85, 594–596. [Google Scholar] [CrossRef] [Green Version]
- Surabattula, R.; Vejandla, M.P.; Mallepaddi, P.C.; Faulstich, K.; Polavarapu, R. Simple, rapid, inexpensive platform for the diagnosis of malaria by loop mediated isothermal amplification (LAMP). Exp. Parasitol. 2013, 134, 333–340. [Google Scholar] [CrossRef]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef] [Green Version]
- Ball, C.S.; Light, Y.K.; Koh, C.Y.; Wheeler, S.S.; Coffey, L.L.; Meagher, R.J. Quenching of Unincorporated Amplification Signal Reporters in Reverse-Transcription Loop-Mediated Isothermal Amplification Enabling Bright, Single-Step, Closed-Tube, and Multiplexed Detection of RNA Viruses. Anal. Chem. 2016, 88, 3562–3568. [Google Scholar] [CrossRef]
- Gadkar, V.J.; Goldfarb, D.M.; Gantt, S.; Tilley, P.A.G. Real-time Detection and Monitoring of Loop Mediated Amplification (LAMP) Reaction Using Self-quenching and De-quenching Fluorogenic Probes. Sci. Rep. 2018, 8, 5548. [Google Scholar] [CrossRef] [Green Version]
- Kubota, R.; Alvarez, A.; Su, W.; Jenkins, D. FRET-based assimilating probe for sequence-specific real-time monitoring of loop-mediated isothermal amplification (LAMP). Biol. Eng. Trans. 2011, 4, 81–100. [Google Scholar] [CrossRef]
- Liu, W.; Huang, S.; Liu, N.; Dong, D.; Yang, Z.; Tang, Y.; Ma, W.; He, X.; Ao, D.; Xu, Y.; et al. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci. Rep. 2017, 7, 40125. [Google Scholar] [CrossRef]
- Tanner, N.A.; Zhang, Y.; Evans, T.C., Jr. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. BioTechniques 2012, 53, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Zerilli, F.; Bonanno, C.; Shehi, E.; Amicarelli, G.; Adlerstein, D.; Makrigiorgos, G.M. Methylation-specific loop-mediated isothermal amplification for detecting hypermethylated DNA in simplex and multiplex formats. Clin. Chem. 2010, 56, 1287–1296. [Google Scholar] [CrossRef]
- Chen, J.H.; Lu, F.; Lim, C.S.; Kim, J.Y.; Ahn, H.J.; Suh, I.B.; Takeo, S.; Tsuboi, T.; Sattabongkot, J.; Han, E.T. Detection of Plasmodium vivax infection in the Republic of Korea by loop-mediated isothermal amplification (LAMP). Acta Trop. 2010, 113, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Mohon, A.N.; Getie, S.; Jahan, N.; Alam, M.S.; Pillai, D.R. Ultrasensitive loop mediated isothermal amplification (US-LAMP) to detect malaria for elimination. Malar. J. 2019, 18, 350. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.S.; Lim, D.H.; Nam, J.; Mihn, D.C.; Sung, H.W.; Lim, C.S.; Kim, J. Development of a multiplex isothermal amplification molecular diagnosis method for on-site diagnosis of influenza. PLoS ONE 2020, 15, e0238615. [Google Scholar] [CrossRef] [PubMed]
- Pousibet-Puerto, J.; Salas-Coronas, J.; Sánchez-Crespo, A.; Molina-Arrebola, M.A.; Soriano-Pérez, M.J.; Giménez-López, M.J.; Vázquez-Villegas, J.; Cabezas-Fernández, M.T. Impact of using artemisinin-based combination therapy (ACT) in the treatment of uncomplicated malaria from Plasmodium falciparum in a non-endemic zone. Malar. J. 2016, 15, 339. [Google Scholar] [CrossRef] [Green Version]
- Fransisca, L.; Kusnanto, J.H.; Satoto, T.B.; Sebayang, B.; Supriyanto; Andriyan, E.; Bangs, M.J. Comparison of rapid diagnostic test Plasmotec Malaria-3, microscopy, and quantitative real-time PCR for diagnoses of Plasmodium falciparum and Plasmodium vivax infections in Mimika Regency, Papua, Indonesia. Malar. J. 2015, 14, 103. [Google Scholar] [CrossRef] [Green Version]
- Payne, D. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull. World Health Organ. 1988, 66, 621–626. [Google Scholar]
- Curtis, K.A.; Rudolph, D.L.; Owen, S.M. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 2008, 151, 264–270. [Google Scholar] [CrossRef]
- Miyamoto, S.; Sano, S.; Takahashi, K.; Jikihara, T. Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal. Biochem. 2015, 473, 28–33. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, X.; Wu, B.; Qian, C.; Zhang, F.; Wang, L.; Ye, Z.; Wu, J. Rapid on-site detection of genetically modified soybean products by real-time loop-mediated isothermal amplification coupled with a designed portable amplifier. Food Chem. 2020, 323, 126819. [Google Scholar] [CrossRef] [PubMed]

| Target | Name | Sequence (5′-3′) | Length (mer) | Reference |
|---|---|---|---|---|
| Malaria pan (18S rRNA gene) | M. pan F3 | GTA TCA ATC GAG TTT CTG ACC | 21 | [31] |
| M. pan B3 | CTT GTC ACT ACC TCT CTT CT | 20 | ||
| M. pan FIP | TCG AAC TCT AAT TCC CCG TTA CCT ATC AGC TTT TGA TGT TAG GGT | 45 | ||
| M. pan BIP | CGG AGA GGG AGC CTG AGA AAT AGA ATT GGG TAA TTT ACG CG | 41 | ||
| M. pan FLP | CGT CAT AGC CAT GTT AGG CC | 20 | ||
| M. pan BLP | AGC TAC CAC ATC TAA GGA AGG CAG | 24 | ||
| M. pan BLP probe | [FAM]-CGG GCC CGT ACA AAG GGA ACA CCC ACA CTC CGA GCT ACC ACA TCT AAG GAA GGC AG | 56 | ||
| Plasmodium falciparum (LDH gene) | P. f F3 | AAT TGT AAA CTT ACA TGC ATC AC | 23 | Present study |
| P. f B3 | TCA AAT TTA GCT TTT TCC TCA CT | 23 | ||
| P. f FIP | GCC CTT CTA ACA AGG TTG AGC AGC TGC TGC TAT TAT CGA AAT | 42 | ||
| P. f BIP | TAT GGA CAC TCC GAT ATA TTC GGT GTA ATT CGA TAA CTT GTT CAA CAC | 48 | ||
| P. f FLP | CAA ATC TTT AAG TAG GAT TCA GCC | 24 | ||
| P. f BLP | GGT ACA CCT GTT GTT TTA GGT GCT | 24 | ||
| P. f BLP probe | [HEX]-CGG GCC CGT ACA AAG GGA ACA CCC ACA CTC CGG GTA CAC CTG TTG TTT TAG GTG CT | 56 | ||
| Plasmodium vivax (16S rRNA gene) | P. v F3 | GGA ATG ATG GGA ATT TAA AAC CT | 23 | [30] |
| P. v B3 | ACG AAG TAT CAG TTA TGT GGA T | 22 | ||
| P. v FIP | CTA TTG GAG CTG GAA TTA CCG CTC CCA AAA CTC AAT TGG AGG | 42 | ||
| P. v BIP | AAT TGT TGC AGT TAA AAC GCT CGT AAG CTA GAA GCG TTG CT | 41 | ||
| P. v FLP | GCT GCT GGC ACC AGA CTT | 18 | ||
| P. v BLP | AGT TGA ATT TCA AAG AAT CG | 20 | ||
| P. v BLP probe | [Texas red]-CGG GCC CGT ACA AAG GGA ACA CCC ACA CTC CGA GTT GAA TTT CAA AGA ATC G | 52 | ||
| Human (actin gene) | IC F3 | AGT ACC CCA TCG AGC ACG | 18 | [32] |
| IC B3 | AGC CTG GAT AGC AAC GTA CA | 20 | ||
| IC FIP | GAG CCA CAC GCA GCT CAT TGT ATC ACC AAC TGG GAC GAC A | 40 | ||
| IC BIP | CTG AAC CCC AAG GCC AAC CGG CTG GGG TGT TGA AGG TC | 38 | ||
| IC FLP | TGT GGT GCC AGA TTT TCT CCA | 21 | ||
| IC BLP | CGA GAA GAT GAC CCA GAT CAT GT | 23 | ||
| IC BLP probe | [Cy5]-CGG GCC CGT ACA AAG GGA ACA CCC ACA CTC CGC GAG AAG ATG ACC CAG ATC ATG T | 55 | ||
| Quencher probe 1 Quencher probe 2 | GAG TGT GGG TGT TCC CTT TGT ACG GGC CCG -BHQ1 | 30 | ||
| CCT ACC CTC GTC CTA ACA CGG GAG CCT GCA CTG AC -BHQ2 | 35 |
| Ct Values | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vector Dilution (Copies/µL) | |||||||||
| Primer set | Vector | 107 | 106 | 105 | 104 | 103 | 102 | 101 | |
| Monoplex LAMP | Pan (FAM) | Malaria Pan | 8.08 | 9.10 | 10.21 | 11.04 | 12.82 | 16.26 | Neg |
| Pf (HEX) | P. falciparum | 10.44 | 11.21 | 12.59 | 13.30 | 17.73 | 21.26 | Neg | |
| Pv (Tex) | P. vivax | 12.05 | 14.22 | 15.48 | 16.00 | 19.77 | 22.60 | Neg | |
| IC (Cy5) | IC | 14.26 | 16.18 | 18.00 | 19.62 | 24.28 | Neg | Neg | |
| Multiplex LAMP | Pan (FAM) + Pf (HEX) + Pv (Tex) + IC (Cy5) | Malaria Pan | 12.15 | 13.78 | 15.55 | 17.28 | 19.10 | 22.81 | Neg |
| P. falciparum | 17.79 | 19.00 | 20.53 | 26.65 | 31.48 | 31.67 | Neg | ||
| P. vivax | 13.26 | 15.65 | 17.03 | 17.93 | 25.73 | 28.58 | Neg | ||
| IC | 15.58 | 18.01 | 20.14 | 22.33 | 23.91 | Neg | Neg | ||
| Clinical Samples | Whole Blood Clinical Sample | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution * | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | ||||||||
| CT | RFU | CT | RFU | CT | RFU | CT | RFU | CT | RFU | CT | RFU | |||
| P. falciparum | Monoplex LAMP | Pan (FAM) | 9.86 | 15,145 | 10.83 | 16,032 | 12.68 | 17,075 | 14.43 | 17,203 | Neg | - | Neg | - |
| Pf (HEX) | 9.71 | 16,795 | 10.82 | 16,581 | 13.73 | 17,218 | Neg | - | Neg | - | Neg | - | ||
| Multiplex LAMP | Pan (FAM) | 16.70 | 7543 | 18.24 | 7629 | 20.70 | 8057 | 30.85 | 8118 | Neg | - | Neg | - | |
| Pf (HEX) | 21.16 | 3928 | 22.88 | 3969 | 24.72 | 4108 | 38.28 | 4020 | Neg | - | Neg | - | ||
| Pv (Tex) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| IC (Cy5) | 24.71 | 4603 | 37.16 | 2616 | Neg | - | Neg | - | Neg | - | Neg | - | ||
| RealStar® Malaria S&T PCR Kit 1.0 | Pf (FAM) | 22.61 | 11,985 | 27.09 | 12,807 | 30.82 | 12,548 | 35.41 | 11,329 | 39.26 | 7558 | Neg | - | |
| Pv (Cy5) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| P. vivax | Monoplex LAMP | Pan (FAM) | 16.12 | 13,226 | 18.05 | 14125 | 32.51 | 13,617 | 37.25 | 12,230 | Neg | - | Neg | - |
| Pv (Tex) | 12.80 | 9561 | 14.59 | 9454 | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Multiplex LAMP | Pan (FAM) | 19.36 | 7347 | 20.90 | 7527 | 25.27 | 7546 | Neg | - | Neg | - | Neg | - | |
| Pf (HEX) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Pv (Tex) | 18.10 | 5398 | 20.08 | 5360 | 21.99 | 5541 | Neg | - | Neg | - | Neg | - | ||
| IC (Cy5) | 54.86 | 1138 | 55.03 | 1124 | Neg | - | Neg | - | Neg | - | Neg | - | ||
| RealStar® Malaria S&T PCR Kit 1.0 | Pf (FAM) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | |
| Pv (Cy5) | 29.37 | 5712 | 33.71 | 5585 | 36.90 | 5049 | Neg | - | Neg | - | Neg | - | ||
| P. malariae | Monoplex LAMP | Pan (FAM) | 11.11 | 15,757 | 12.61 | 17,887 | 14.87 | 16,582 | 15.80 | 17,883 | Neg | - | Neg | - |
| Multiplex LAMP | Pan (FAM) | 20.05 | 7706 | 22.29 | 7741 | 26.23 | 7531 | Neg | - | Neg | - | Neg | - | |
| Pf (HEX) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Pv (Tex) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| IC (Cy5) | 32.60 | 6815 | 37.41 | 6486 | 38.92 | 6449 | Neg | - | Neg | - | Neg | - | ||
| RealStar® Malaria S&T PCR Kit 1.0 | Pm (FAM) | 24.56 | 13,326 | 28.16 | 12,145 | 31.22 | 1875 | 34.67 | 11,985 | 36.91 | 5500 | Neg | - | |
| Po (Cy5) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Pk (Tex) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| P. ovale | Monoplex LAMP | Pan (FAM) | 16.60 | 15,880 | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - |
| Multiplex LAMP | Pan (FAM) | 22.20 | 7885 | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | |
| Pf (HEX) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Pv (Tex) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| IC (Cy5) | 26.27 | 6097 | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| RealStar® Malaria S&T PCR Kit 1.0 | Pm (FAM) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | |
| Po (Cy5) | 34.12 | 2516 | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Pk (Tex) | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | Neg | - | ||
| Clinical Samples | Multiplex Malaria Pan/Pf/Pv/IC LAMP Assay | RealStar® Malaria Screen & Type PCR Kit 1.0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pan (FAM) | Pf (HEX) | Pv (Tex) | IC (Cy5) | Pf (FAM) | Pv (Cy5) | IC (HEX) | Pm (FAM) | Po (Cy5) | IC (HEX) | ||
| P. falciparum (n = 61) | P/N | 61/0 | 61/0 | 0/61 | 46/15 | 61/0 | 1/60 | 58/3 | - | - | - |
| Sensitivity | 100% | 100% | - | 75.41% | 100% | - | 95.08% | - | - | - | |
| Specificity | - | - | 100% | - | - | 98.36% | - | - | - | - | |
| P. vivax (n = 54) | P/N | 54/0 | 0/54 | 54/0 | 44/10 | 0/54 | 54/0 | 54/0 | - | - | - |
| Sensitivity | 100% | - | 100% | 81.48% | - | 100% | 100% | - | - | - | |
| Specificity | - | 100% | - | - | 100% | - | - | - | - | - | |
| P. malariae (n = 2) | P/N | 2/0 | 0/2 | 0/2 | 2/0 | - | - | - | 2/0 | 0/2 | 2/0 |
| Sensitivity | 100% | - | - | 100% | - | - | - | 100% | - | 100% | |
| Specificity | - | 100% | 100% | - | - | - | - | - | 100% | - | |
| P. ovale (n = 1) | P/N | 1/0 | 0/1 | 0/1 | 1/0 | - | - | - | 1/0 | 0/1 | 1/0 |
| Sensitivity | 100% | - | - | 100% | - | - | - | 100% | - | 100% | |
| Specificity | - | 100% | 100% | - | - | - | - | - | 100% | - | |
| Non-infection (n = 90) | P/N | 0/90 | 0/90 | 0/90 | 90/0 | 0/90 | 0/90 | 90/0 | - | - | - |
| Sensitivity | - | - | - | 100% | - | - | 100% | - | - | - | |
| Specificity | 100% | 100% | 100% | - | 100% | 100% | - | - | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, W.S.; Lim, D.H.; Choe, Y.; Jee, H.; Moon, K.C.; Kim, C.; Choi, M.; Park, I.S.; Lim, C.S. Development of a Multiplex Loop-Mediated Isothermal Amplification Assay for Diagnosis of Plasmodium spp., Plasmodium falciparum and Plasmodium vivax. Diagnostics 2021, 11, 1950. https://doi.org/10.3390/diagnostics11111950
Jang WS, Lim DH, Choe Y, Jee H, Moon KC, Kim C, Choi M, Park IS, Lim CS. Development of a Multiplex Loop-Mediated Isothermal Amplification Assay for Diagnosis of Plasmodium spp., Plasmodium falciparum and Plasmodium vivax. Diagnostics. 2021; 11(11):1950. https://doi.org/10.3390/diagnostics11111950
Chicago/Turabian StyleJang, Woong Sik, Da Hye Lim, YoungLan Choe, Hyunseul Jee, Kyung Chul Moon, Chaewon Kim, Minkyeong Choi, In Su Park, and Chae Seung Lim. 2021. "Development of a Multiplex Loop-Mediated Isothermal Amplification Assay for Diagnosis of Plasmodium spp., Plasmodium falciparum and Plasmodium vivax" Diagnostics 11, no. 11: 1950. https://doi.org/10.3390/diagnostics11111950
APA StyleJang, W. S., Lim, D. H., Choe, Y., Jee, H., Moon, K. C., Kim, C., Choi, M., Park, I. S., & Lim, C. S. (2021). Development of a Multiplex Loop-Mediated Isothermal Amplification Assay for Diagnosis of Plasmodium spp., Plasmodium falciparum and Plasmodium vivax. Diagnostics, 11(11), 1950. https://doi.org/10.3390/diagnostics11111950







