Abstract
The reactivation of hepatitis B virus (HBVr) in patients undergoing pharmacological immunosuppression is a potentially fatal clinical event that may occur in patients with overt or occult HBV infection. The risk of HBVr is mainly determined by the type of immunosuppressive therapy and the HBV serologic profile, with a higher risk in patients positive for the hepatitis B surface antigen (HBsAg), and a lower risk in HBsAg-negative/antibodies to core antigen-positive subjects. Notably, a considerable proportion of patients experiencing HBVr showed a high degree of variability of the HBV S gene, possibly leading to immune escape mutants. These mutations, usually in the “a-determinant” of the HBsAg, can cause diagnostic problems and consequently hamper the appropriate management strategy of patients at risk of HBVr. Here, we describe a case of HBVr in a patient with a diagnosis of chronic myeloid leukemia and a previous history of kidney transplant, providing evidence of the potential usefulness of hepatitis B core-related antigen measurement in patients with HBV immune-escape mutants at risk of viral reactivation.
1. Introduction
Hepatitis B virus reactivation (HBVr) encompasses a wide spectrum of clinical conditions ranging from an asymptomatic increase in circulating HBV DNA to hepatitis flares and even acute liver failure [1]. HBVr can occur both in hepatitis B surface antigen (HBsAg)-carriers or in subjects with a resolved HBV infection; the risk is primarily determined by the duration and potency of pharmacological immunosuppression [2].
HBV covalently closed-circular (ccc)DNA is the replicative intermediate of HBV responsible for the persistence of the virus within the hepatocytes [3]. HBV cccDNA acts as a template for all viral transcripts, including sub-genomic RNAs, pre-core RNA and pre-genomic RNA, that encode for the structural and soluble proteins of HBV [4]. Following the resolution of HBV infection, the replicative activity of HBV cccDNA is inhibited by the surveillance pressure of a competent innate and adaptative immune response; however, it can silently persist for decades in the liver of HBsAg-negative subjects with serum markers of past HBV exposure, a condition referred to as occult HBV infection [5,6].
Patients with occult HBV infection and hematological malignancies receiving cytotoxic or immunosuppressive therapies are at risk of HBVr [7]. Furthermore, HBVr is usually associated with the emergence of immune-escape mutations within the HBV S gene (i.e., the HBV gene that encodes for the HBsAg) that confers the virus a significant survival advantage, while hampering circulating HBsAg detection using commercial serological assays [8]. A recent study pointed out a high prevalence of HBV S gene mutations in patients with malignant blood diseases, reporting up to 15 different mutations in a single patients’ isolate [9].
Recently, Seto and colleagues investigated risk factors associated to HBVr in oncohematological patients with resolved HBV infection undergoing rituximab-based chemotherapy or allogeneic hematopoietic stem cell transplantation [10]. Interestingly, the authors observed that higher baseline hepatitis B core-related antigen (HBcrAg) levels were independently associated to HBVr, suggesting a potential clinical value of the biomarker for the identification of patients that might benefit from prophylactic nucleos(t)ide analogue (Nuc) treatment [10].
Here, we describe a case of reactivation of Hepatitis B virus with multiple gene S mutations, providing evidence for the possible clinical value of the measurement of the HBcrAg in immunosuppressed patients at risk of viral reactivation.
2. Case Report
In July 2019, a 74-year-old male patient was admitted to the Infectious Disease Section of the Verona University Hospital for investigation; he had HBV in his blood with a titer of 26,100,000 IU/mL (cobas® HBV, Roche Molecular Diagnostics, Branchburg, NJ, USA) but exhibited normal liver function. Although he displayed the hepatitis B e antigen (HBeAg), he was negative for the HBsAg, but positive for the homologous anti-HBs (ADVIA Centaur HBV assays, Siemens Healthcare GmbH, Erlangen, Germany).
In August 2013, the patient received a kidney transplant for nephroangiosclerosis. At this time, the serologic screening for HBV had shown that he was HBsAg-negative, anti-HBs-positive (12 mIU/mL), HBeAg-negative and positive for antibodies to the HBeAg and to the hepatitis B core antigen (anti-HBc). No antibody markers of a hepatitis C and hepatitis D virus infection were detected; serum HBV DNA had not been determined. The indices of hepatic cytolysis were normal. The kidney donor was negative for HBV. Post-transplant, the patient received immunosuppressive induction with basiliximab, tacrolimus, mycophenolate and steroids, and was then included in the follow-up program as per protocol; the HBsAg remained negative throughout, accompanied by normal liver biochemistry.
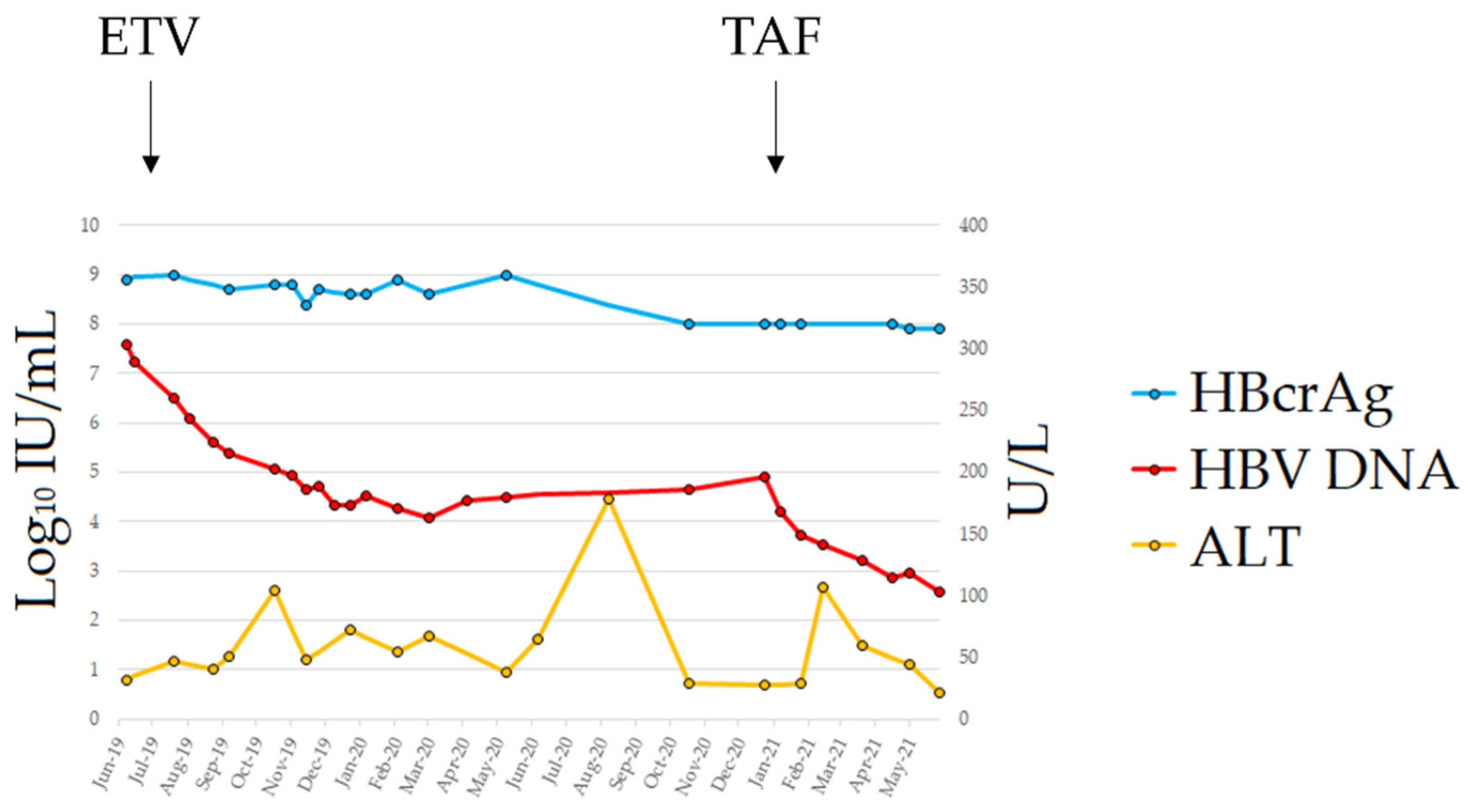
In May 2019, the patient developed chronic myeloid leukemia (CML). Before treatment with imatinib mesylate, the patient repeated the HBV serology and the HBeAg was again detected in his blood in the absence of circulating HBsAg; further testing using a real-time PCR showed that he had HBV DNA in serum at a titer of 26,100,000 IU/mL. The patient was still anti-HBs-positive (15 mIU/mL) with normal liver enzymes. A diagnosis of HBVr was made and, in July 2019, the patient started entecavir (ETV) treatment at 0.5 mg/day, which was subsequently reduced in April 2020 to 0.5 mg/48 h because of renal function deterioration. The patient underwent a close follow-up (every 2 weeks) for the first 6 months of treatment and, after, with monthly scheduling. The therapy led to a consistent decrease in viremia, reaching a plateau of 3 Log10 HBV DNA reduction after 6 months of treatment (Figure 1); however, viremia did not decrease further in the following months. No classic ETV-resistant mutations were observed in the HBV reverse transcriptase (rt) gene. However, we detected the mutation rtL269I, previously reported to confer a reduced susceptibility to ETV [11]. On January 2021, the patient switched to tenofovir alafenamide (TAF) treatment; HBV DNA rapidly declined and reached a value of 378 IU/mL at the last FU. The single ALT elevation (107 U/L) in the 2nd month of TAF treatment was documented in the course of an infection of the residual left limb requiring surgical toileting. Unfortunately, the patient died from coronary heart disease in September 2021.
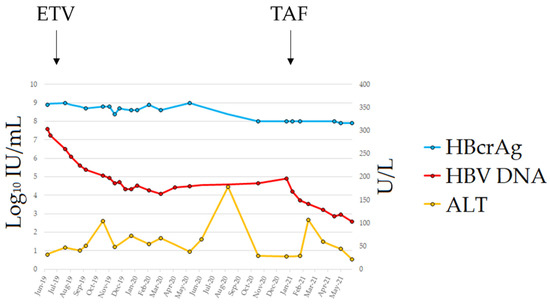
Figure 1.
Clinical course from HBVr diagnosis to last follow-up. Abbreviations: alanine aminotransferase (ALT); entecavir (ETV); hepatitis B core-related antigen (HBcrAg); hepatitis B surface antigen (HBsAg); tenofovir alafenamide (TAF).
The serum samples collected from July 2019 were tested for the HBsAg with the highly sensitive Lumipulse® G HBsAg-Quant assay (Fujirebio Inc., Tokyo, Japan) as well as for the HBcrAg (Lumipulse® G HBcrAg, Fujirebio Inc., Tokyo, Japan) [12]. The nucleotide sequences of the HBV S gene and the rt domain were determined using the Sanger sequencing method at BMR Genomics service (BMR Genomics, Padua, Italy) and analyzed using ClustalW2 software (https://www.ebi.ac.uk/Tools/msa/clustalw2/; accessed on 23 December 2019 and on 6 December 2020) [13]. The HBV genotype was determined based on a reverse hybridization line probe assay (INNO-LiPA HBV Genotyping, Fujirebio Europe, Gent, Belgium) [14].
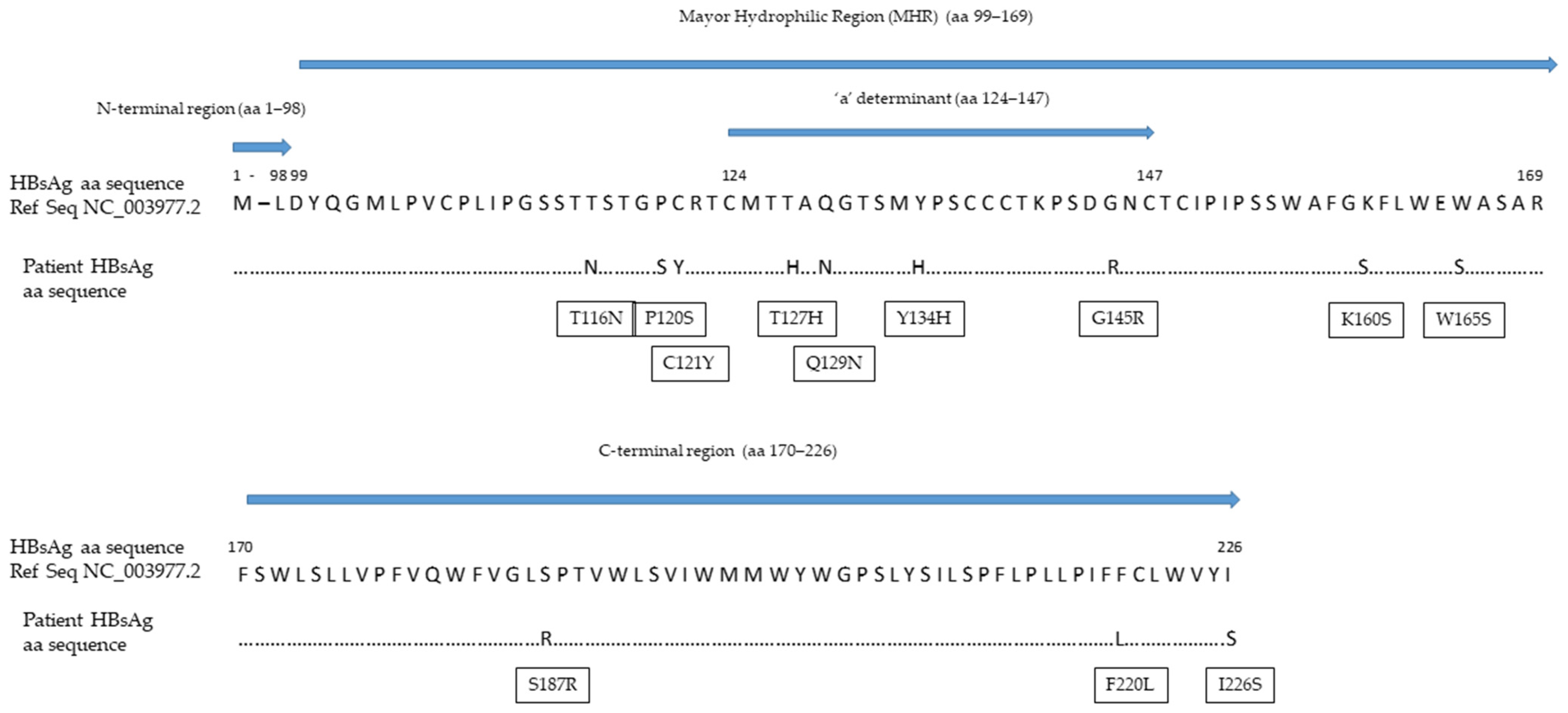
Despite its high analytical sensitivity for HBsAg mutants [15], the HBsAg assay failed to detect a circulating HBs antigen. The HBV S gene sequence analysis identified four mutations within the a-determinant (aa 124–147) (P/T127H, Q129N, F/Y134H and G145R) and five additional mutations within the mayor hydrophilic region (MHR, aa 99–169) (T116N, P120S, C121Y, K/R160S and W165S) (Figure 2). The values of the HBcrAg from baseline to the end of the follow-up were persistently above the upper limit threshold of the assay (>7.0 Log10 U/mL). Viral genotyping showed that the patient was infected with HBV genotype D.
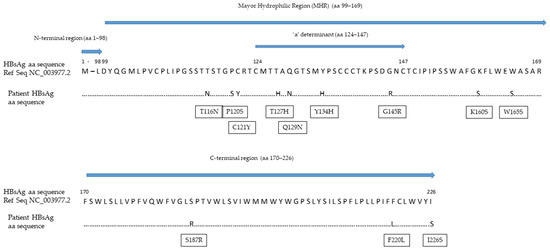
Figure 2.
Mutations within the HBV S gene detected in our patient. Abbreviations: amino acid (aa); hepatitis B surface antigen (HBsAg).
3. Discussion
HBsAg-negative/anti-HBc-positive patients undergoing immunosuppressive therapy following a solid organ transplant or tyrosine kinase inhibitors are at moderate risk of HBVr (from 1% to 10%) [16]; in these patients, pre-emptive therapy, rather than HBV prophylaxis, is recommended [17]. In case of HBsAg seroreversion, treatment with Nucs should be started as early as possible, independently from ALT levels [17]. However, in our patient, as well as in several other cases reported in the literature [18,19,20,21,22,23], HBVr occurred in default of the detectable HBsAg in serum. In this regard, the search for escape mutants should be recommended in patients with CML and markers of previous HBV exposure when undergoing pharmacological immunosuppression.
As far as the viral sequencing is concerned, we observed a high degree of HBV S gene variability accompanied by several mutations responsible for the changes in the antigenic pattern of the HBsAg. These mutations can prevent both neutralizing and diagnostic anti-HBs from interacting with the HBsAg; as a result, it is not uncommon to find anti-HBs antibodies in patients experiencing HBsAg-negative HBVr [24].
In our patient, four mutations were observed within the a-determinant. These included the most common HBV immune escape mutation at position 145, present as a glycine to arginine change, and another three mutations at positions 127, 129 and 134, which are frequently observed in patients with HBVr and known to be responsible for changes in the loop structure of the HBsAg [25]. Among the mutations observed outside the a-determinant but within the MHR, P120S can alter the antigenicity of the HBsAg by interfering with the loop structure of the a-determinant, while T116M is responsible for the introduction of additional N-linked glycosylation sites that can mask B-cells epitopes and, thus, increase HBV evasion from the humoral immune response [18,25]. Remarkably, the rate of mutations observed in our case was so high that the detection of circulating HBsAg by the most sensitive commercial assay was inhibited [15]. Nonetheless, we recommend the use of highly sensitive HBsAg quantification methods with high reactivity against HBV S gene mutants. A previous study showed an excellent analytical sensitivity for the Lumipulse® G HBsAg-Quant assay; among 1000 seronegative subjects, the assay detected 29 serum samples positive for the HBsAg, with concentrations between 0.005 and 0.05 IU/mL [26]. Furthermore, the Lumipulse® G HBsAg-Quant assay proved higher reactivity against the HBsAg mutants as compared to other commercial assays; the analytical method involves the HBsAg denaturation in linear form, allowing for the full exposure of the inner and outer portion of the a-determinant, thus increasing the number of epitopes that can be recognized by capture antibodies [15]. In our case, we suspected HBVr from the seroreversion of the HBeAg; this prompted the molecular testing, which revealed a highly replicative virus in the blood with an HBV DNA concentration of 26,100,000 IU/mL. HBVr was not recognized before, because national and international guidelines [17,27] do not recommend the assessment of HBV DNA in HBsAg-negative/anti-HBc-positive patients at moderate risk of HBVr.
To avoid overlooking HBV replication, a simple and cheaper alternative is the measurement of the HBcrAg. This is a composite biomarker that simultaneously detects the hepatitis B core antigen (HBcAg), the HBeAg and the core-related protein, p22cr [28]. These three core-related proteins share an identical 149 amino acid sequence and are expressed by the pre-core/core HBV gene [29]. The HBcAg is the major structural protein of the HBV capsid. The HBeAg is a soluble non-structural pre-core protein, while p22cr is an aberrantly processed pre-core protein that lacks the C-terminal arginine-rich domain responsible for HBV genome packaging [30]. The HBcrAg is measured in serum samples through a fully automated procedure; currently, it is considered the most reliable surrogate for intrahepatic HBV cccDNA quantity and transcriptional activity [12,31].
In conclusion, considering the economic burden of serial HBV DNA assessment and given the high correlation between HBV DNA and the HBcrAg [32], the combined use of the highly sensitive HBsAg and HBcrAg may represent a cost-effective monitoring option for HBsAg-negative/anti-HBc-positive patients with low-to-moderate risk of HBVr.
Author Contributions
Conceptualization, G.P.C., A.Z., A.O. and G.C.; methodology, G.P.C., A.Z., A.O. and G.C.; software, G.P.C.; formal analysis, G.P.C. and A.O.; investigation, G.P.C., A.Z., A.O. and G.C.; resources, A.Z. and A.O.; data curation, G.P.C., A.Z., A.O. and G.C.; writing—original draft preparation, G.P.C.; writing—review and editing, A.Z., M.R., M.M., A.O. and G.C.; visualization, G.P.C.; supervision, M.R. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted following the rules of the Declaration of Helsinki of 1975, revised in 2013. Ethical review and approval were waived for this study, since serum samples were obtained during standard diagnostic procedures and additional analysis were performed as part of the diagnostic workup. However, informed consent was obtained as stated below.
Informed Consent Statement
Written informed consent was obtained from the patient involved in the study. Written informed consent has been obtained from the patient to publish this paper.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors thank Corinna Orsini (Fujirebio Italia s.r.l.) for providing the Lumipulse® G HBsAg-Quant assay (Fujirebio Inc., Tokyo, Japan), and James Michael Hughes (Department of Medical Sciences, University of Turin) for English revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Viganò, M.; Serra, G.; Casella, G.; Grossi, G.; Lampertico, P. Reactivation of hepatitis B virus during targeted therapies for cancer and immune-mediated disorders. Expert Opin. Biol. Ther. 2016, 16, 917–926. [Google Scholar] [CrossRef]
- Svicher, V.; Salpini, R.; Malagnino, V.; Piermatteo, L.; Alkhatib, M.; Cerva, C.; Sarmati, L. New Markers in Monitoring the Reactivation of Hepatitis B Virus Infection in Immunocompromised Hosts. Viruses 2019, 11, 783. [Google Scholar] [CrossRef] [Green Version]
- Nassal, M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015, 64, 1972–1984. [Google Scholar] [CrossRef] [Green Version]
- Armandi, A.; Rosso, C.; Ribaldone, D.G.; Caviglia, G.P. Moving towards core antigen for the management of patients with overt and occult HBV infection. Panminerva Med. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, G.P.; Abate, M.L.; Tandoi, F.; Ciancio, A.; Amoroso, A.; Salizzoni, M.; Saracco, G.M.; Rizzetto, M.; Romagnoli, R.; Smedile, A. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: A new tool to detect occult infection. J. Hepatol. 2018, 69, 301–307. [Google Scholar] [CrossRef]
- Raimondo, G.; Locarnini, S.; Pollicino, T.; Levrero, M.; Zoulim, F.; Lok, A.S.; Taormina Workshop on Occult HBV Infection Faculty Members. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 2019, 71, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Zannella, A.; Marignani, M.; Begini, P. Hematological Malignancies and HBV Reactivation Risk: Suggestions for Clinical Management. Viruses 2019, 11, 858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, J.; Nakamura, T.; Masamune, A. Roles of Hepatitis B Virus Mutations in the Viral Reactivation after Immunosuppression Therapies. Viruses 2019, 11, 457. [Google Scholar] [CrossRef] [Green Version]
- Konopleva, M.V.; Belenikin, M.S.; Shanko, A.V.; Bazhenov, A.I.; Kiryanov, S.A.; Tupoleva, T.A.; Sokolova, M.V.; Pronin, A.V.; Semenenko, T.A.; Suslov, A.P. Detection of S-HBsAg Mutations in Patients with Hematologic Malignancies. Diagnostics 2021, 11, 969. [Google Scholar] [CrossRef] [PubMed]
- Seto, W.K.; Wong, D.K.; Chan, T.S.; Hwang, Y.Y.; Fung, J.; Liu, K.S.; Gill, H.; Lam, Y.F.; Cheung, K.S.; Lie, A.K.; et al. Association of Hepatitis B Core-Related Antigen with Hepatitis B Virus Reactivation in Occult Viral Carriers Undergoing High-Risk Immunosuppressive Therapy. Am. J. Gastroenterol. 2016, 111, 1788–1795. [Google Scholar] [CrossRef]
- Yamada, N.; Sugiyama, R.; Nitta, S.; Murayama, A.; Kobayashi, M.; Okuse, C.; Suzuki, M.; Yasuda, K.; Yotsuyanagi, H.; Moriya, K.; et al. Resistance mutations of hepatitis B virus in entecavir-refractory patients. Hepatol. Commun. 2017, 1, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Caviglia, G.P.; Olivero, A.; Ciancio, A.; Tandoi, F.; Troshina, G.; Rosso, C.; Abate, M.L.; Younes, R.; Ribaldone, D.G.; Smedile, A.; et al. Analytical and clinical evaluation of a novel assay for anti-HBc IgG measurement in serum of subjects with overt and occult HBV infection. Diagn. Microbiol. Infect. Dis. 2020, 96, 114985. [Google Scholar] [CrossRef] [PubMed]
- Tandoi, F.; Caviglia, G.P.; Pittaluga, F.; Abate, M.L.; Smedile, A.; Romagnoli, R.; Salizzoni, M. Prediction of occult hepatitis B virus infection in liver transplant donors through hepatitis B virus blood markers. Dig. Liver Dis. 2014, 46, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, G.P.; Abate, M.L.; Noviello, D.; Olivero, A.; Rosso, C.; Troshina, G.; Ciancio, A.; Rizzetto, M.; Saracco, G.M.; Smedile, A. Hepatitis B core-related antigen kinetics in chronic hepatitis B virus genotype D-infected patients treated with nucleos(t)ide analogues or pegylated-interferon-α. Hepatol. Res. 2017, 47, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, M.; Kagita, M.; Yoshioka, N.; Tsukamoto, H.; Takao, M.; Tahara, K.; Maeda, I.; Hidaka, Y.; Yamauchi, S.; Kaneko, A.; et al. Evaluation of the highly sensitive chemiluminescent enzyme immunoassay “Lumipulse HBsAg-HQ” for hepatitis B virus screening. J. Clin. Lab. Anal. 2018, 32, e22334. [Google Scholar] [CrossRef]
- Loomba, R.; Liang, T.J. Hepatitis B Reactivation Associated with Immune Suppressive and Biological Modifier Therapies: Current Concepts, Management Strategies, and Future Directions. Gastroenterology 2017, 152, 1297–1309. [Google Scholar] [CrossRef] [Green Version]
- European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Shiga, Y.; Takahashi, A.; Kai, T.; Kimura, H.; Takeyama, K.; Noji, H.; Ogawa, K.; Nakamura, A.; Ohira, H.; et al. Fatal hepatitis B virus reactivation in a chronic myeloid leukemia patient during imatinib mesylate treatment. Leuk. Lymphoma 2006, 47, 155–157. [Google Scholar] [CrossRef]
- Kim, S.G.; Chun, J.M.; Jin, R.; Kim, J.Y.; Won, D.I.; Hwang, Y.J. Living donor liver transplantation for acute hepatic failure caused by reactivation of hepatitis B virus infection after chemotherapy for hematologic malignancy: Case reports. Transplant. Proc. 2010, 42, 843–845. [Google Scholar] [CrossRef]
- Kang, B.W.; Lee, S.J.; Moon, J.H.; Kim, S.N.; Chae, Y.S.; Kim, J.G.; Hwang, Y.J.; Sohn, S.K. Chronic myeloid leukemia patient manifesting fatal hepatitis B virus reactivation during treatment with imatinib rescued by liver transplantation: Case report and literature review. Int. J. Hematol. 2009, 90, 383–387. [Google Scholar] [CrossRef]
- Wang, Y.D.; Cui, G.H.; Li, M.; Gowrea, B.; Xia, J.; Hu, Y. Hepatitis B virus reactivation in a chronic myeloid leukemia patient treated with imatinib mesylate. Chin. Med. J. 2012, 125, 2636–2637. [Google Scholar] [PubMed]
- Lai, G.M.; Yan, S.L.; Chang, C.S.; Tsai, C.Y. Hepatitis B reactivation in chronic myeloid leukemia patients receiving tyrosine kinase inhibitor. World J. Gastroenterol. 2013, 19, 1318–1321. [Google Scholar] [CrossRef]
- Lazarevic, I.; Banko, A.; Miljanovic, D.; Cupic, M. Immune-Escape Hepatitis B Virus Mutations Associated with Viral Reactivation upon Immunosuppression. Viruses 2019, 11, 778. [Google Scholar] [CrossRef] [Green Version]
- Salpini, R.; Colagrossi, L.; Bellocchi, M.C.; Surdo, M.; Becker, C.; Alteri, C.; Aragri, M.; Ricciardi, A.; Armenia, D.; Pollicita, M.; et al. Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology 2015, 61, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Kreutz, C. Molecular, immunological and clinical properties of mutated hepatitis B viruses. J. Cell. Mol. Med. 2002, 6, 113–143. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, N.; Matsuura, K.; Sugauchi, F.; Watanabe, T.; Murakami, S.; Iio, E.; Ogawa, S.; Nojiri, S.; Joh, T.; Tanaka, Y. Application of a newly developed high-sensitivity HBsAg chemiluminescent enzyme immunoassay for hepatitis B patients with HBsAg seroclearance. J. Clin. Microbiol. 2013, 51, 3484–3491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzano, A.; Bruno, R.; Fagiuoli, S.; Marignani, M.; Madonia, S.; Toniutto, P.; Viganò, M. Gestione Clinica Della Epatite B Negli Immunocompromessi: Aggiornamento Italiano 2017. Available online: https://www.webaisf.org/wpcontent/uploads/2019/02/gestione_clinica_della_epatite_b_negli_immunocompromessi.2017.pdf (accessed on 29 October 2020).
- Caviglia, G.P.; Noviello, D.; Pellicano, R.; Olivero, A. Role of serum hepatitis B core-related antigen in chronic hepatitis B infection. Minerva Biotecnol. 2018, 30, 29–35. [Google Scholar] [CrossRef]
- Hadziyannis, E.; Laras, A. Viral Biomarkers in Chronic HBeAg Negative HBV Infection. Genes 2018, 9, 469. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T.; Ohno, N.; Terada, N.; Rokuhara, A.; Matsumoto, A.; Yagi, S.; Tanaka, E.; Kiyosawa, K.; Ohno, S.; Maki, N. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 2005, 280, 21713–21719. [Google Scholar] [CrossRef] [Green Version]
- Testoni, B.; Lebossé, F.; Scholtes, C.; Berby, F.; Miaglia, C.; Subic, M.; Loglio, A.; Facchetti, F.; Lampertico, P.; Levrero, M.; et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J. Hepatol. 2019, 70, 615–625. [Google Scholar] [CrossRef]
- Yoshida, K.; Desbiolles, A.; Feldman, S.F.; Ahn, S.H.; Alidjinou, E.K.; Atsukawa, M.; Bocket, L.; Brunetto, M.R.; Buti, M.; Carey, I.; et al. Assay for Hepatitis B Core-related Antigen Identify Patients with High Viral Load: Systematic Review and Meta-analysis of Individual Participant Data. Clin. Gastroenterol. Hepatol. 2021, 19, 46–60.e8. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).