Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges
Abstract
1. Introduction
2. Background
2.1. What Is Deep Learning?
2.2. Deep Learning in Oncology
2.3. Quantitative Imaging for Cancer Diagnosis, Characterization and Assessment of Treatment Response
2.4. Radiotherapy Treatment (RT) Planning and Optimization
2.5. Automatic Image Segmentation
Evaluating the Quality and Success of Segmentation
3. Literature Review
- non-DL segmentation techniques;
- segmentation applied to sites other than the pelvis;
- no training/validation of methods on real patient data;
- image modalities used other than CT and MRI;
- full articles published in languages other than English;
- no clinical application focus or published outcome
3.1. Bladder Cancer
3.2. Cervical Cancer
3.3. Prostate Cancer
3.4. Rectal Cancer
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Parekh, V.S.; Jacobs, M.A. Deep learning and radiomics in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 59–72. [Google Scholar] [CrossRef]
- Ashley, E.A. Towards precision medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef]
- Malayeri, A.A.; El Khouli, R.H.; Zaheer, A.; Jacobs, M.A.; Corona-Villalobos, C.P.; Kamel, I.R.; Macura, K.J. Principles and Applications of Diffusion-weighted Imaging in Cancer Detection, Staging, and Treatment Follow-up. Radiographics 2011, 31, 1773–1791. [Google Scholar] [CrossRef]
- Ma, D.; Gulani, V.; Seiberlich, N.; Liu, K.; Sunshine, J.L.; Duerk, J.L.; Griswold, M.A. Magnetic resonance fingerprinting. Nat. Cell Biol. 2013, 495, 187–192. [Google Scholar] [CrossRef]
- O’Connor, J.P.B.; Aboagye, E.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef]
- Nelms, B.E.; Tomé, W.; Robinson, G.; Wheeler, J. Variations in the Contouring of Organs at Risk: Test Case From a Patient With Oropharyngeal Cancer. Int. J. Radiat. Oncol. 2012, 82, 368–378. [Google Scholar] [CrossRef]
- Miles, E.A.; Clark, C.H.; Urbano, M.T.G.; Bidmead, M.; Dearnaley, D.P.; Harrington, K.J.; A’Hern, R.; Nutting, C.M. The impact of introducing intensity modulated radiotherapy into routine clinical practice. Radiother. Oncol. 2005, 77, 241–246. [Google Scholar] [CrossRef]
- Brouwer, C.L.; Steenbakkers, R.J.H.M.; Heuvel, E.V.D.; Duppen, J.C.; Navran, A.; Bijl, H.P.; Chouvalova, O.; Burlage, F.R.; Meertens, H.; Langendijk, J.A.; et al. 3D Variation in delineation of head and neck organs at risk. Radiat. Oncol. 2012, 7, 32. [Google Scholar] [CrossRef]
- Boldrini, L.; Cusumano, D.; Cellini, F.; Azario, L.; Mattiucci, G.C.; Valentini, V. Online adaptive magnetic resonance guided radiotherapy for pancreatic cancer: State of the art, pearls and pitfalls. Radiat. Oncol. 2019, 14, 71. [Google Scholar] [CrossRef]
- Mikeljevic, J.S.; Haward, R.; Johnston, C.; Crellin, A.; Dodwell, D.; Jones, A.; Pisani, P.; Forman, D. Trends in postoperative radiotherapy delay and the effect on survival in breast cancer patients treated with conservation surgery. Br. J. Cancer 2004, 90, 1343–1348. [Google Scholar] [CrossRef]
- Chen, Z.; King, W.; Pearcey, R.; Kerba, M.; Mackillop, W.J. The relationship between waiting time for radiotherapy and clinical outcomes: A systematic review of the literature. Radiother. Oncol. 2008, 87, 3–16. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Cardenas, C.E.; Yang, J.; Anderson, B.M.; Court, L.E.; Brock, K.B. Advances in Auto-Segmentation. Semin. Radiat. Oncol. 2019, 29, 185–197. [Google Scholar] [CrossRef]
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3–4, 100004. [Google Scholar] [CrossRef]
- Almeida, G.; Tavares, J.M.R. Deep Learning in Radiation Oncology Treatment Planning for Prostate Cancer: A Systematic Review. J. Med. Syst. 2020, 44, 179. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lin, C.-H.; Lu, H.-Y.; Chiang, H.-J.; Wang, H.-K.; Huang, Y.-T.; Ng, S.-H.; Hong, J.-H.; Yen, T.-C.; Lai, C.-H.; et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer. Eur. Radiol. 2020, 30, 1297–1305. [Google Scholar] [CrossRef]
- Ueda, D.; Shimazaki, A.; Miki, Y. Technical and clinical overview of deep learning in radiology. Jpn. J. Radiol. 2019, 37, 15–33. [Google Scholar] [CrossRef]
- Boldrini, L.; Bibault, J.-E.; Masciocchi, C.; Shen, Y.; Bittner, M.-I. Deep Learning: A Review for the Radiation Oncologist. Front. Oncol. 2019, 9, 977. [Google Scholar] [CrossRef]
- Meyer, P.; Noblet, V.; Mazzara, C.; Lallement, A. Survey on deep learning for radiotherapy. Comput. Biol. Med. 2018, 98, 126–146. [Google Scholar] [CrossRef]
- Kowalski, R. Computational Logic and Human Thinking: How to Be Artificially Intelligent; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Hebb, D.O. The Organization of Behavior: A Neuropsychological Theory; Wiley: New York, NY, USA, 1949. [Google Scholar]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Zhang, Q.-S.; Zhu, S.-C. Visual interpretability for deep learning: A survey. Front. Inf. Technol. Electron. Eng. 2018, 19, 27–39. [Google Scholar] [CrossRef]
- Jian, J.; Xiong, F.; Xia, W.; Zhang, R.; Gu, J.; Wu, X.; Meng, X.; Gao, X. Fully convolutional networks (FCNs)-based segmentation method for colorectal tumors on T2-weighted magnetic resonance images. Australas. Phys. Eng. Sci. Med. 2018, 41, 393–401. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, L.; Zhang, Z.; Fei, B. PSNet: Prostate segmentation on MRI based on a convolutional neural network. J. Med. Imaging 2018, 5, 021208. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, L.; Fei, B. Deep convolutional neural network for prostate MR segmentation. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1687–1696. [Google Scholar] [CrossRef]
- Ma, L.; Guo, R.; Zhang, G.; Tade, F.; Schuster, D.M.; Nieh, P.; Master, V.; Fei, B. Automatic segmentation of the prostate on CT images using deep learning and multi-atlas fusion. Proc. SPIE Int. Soc. Opt. Eng. 2017, 10133, 101332O. [Google Scholar]
- Song, Y.; Hu, J.; Wu, Q.; Xu, F.; Nie, S.; Zhao, Y.; Bai, S.; Yi, Z. Automatic delineation of the clinical target volume and organs at risk by deep learning for rectal cancer postoperative radiotherapy. Radiother. Oncol. 2020, 145, 186–192. [Google Scholar] [CrossRef]
- Chai, X.; van Herk, M.; Betgen, A.; Hulshof, M.C.; Bel, A. Automatic bladder segmentation on CBCT for multiple plan ART of bladder cancer using a patient-specific bladder model. Phys. Med. Biol. 2012, 57, 3945–3962. [Google Scholar] [CrossRef] [PubMed]
- Gulliford, S.L.; Webb, S.; Rowbottom, C.; Corne, D.W.; Dearnaley, D.P. Use of artificial neural networks to predict biological outcomes for patients receiving radical radiotherapy of the prostate. Radiother. Oncol. 2004, 71, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.-H.; Kim, H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 6994. [Google Scholar] [CrossRef] [PubMed]
- Han, X. MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Cao, X.; Gao, Y.; Wang, L.; Shen, D. Estimating CT Image from MRI Data Using 3D Fully Convolutional Networks. In Design, User Experience, and Usability: Design Thinking and Methods; Springer: Cham, Switzerland, 2016; pp. 170–178. [Google Scholar] [CrossRef]
- Zhen, X.; Chen, J.; Zhong, Z.; Hrycushko, B.; Zhou, L.; Jiang, S.; Albuquerque, K.; Gu, X. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: A feasibility study. Phys. Med. Biol. 2017, 62, 8246–8263. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Kovalchuk, N.; Buyyounouski, M.K.; Xing, L.; Yang, Y. Incorporating dosimetric features into the prediction of 3D VMAT dose distributions using deep convolutional neural network. Phys. Med. Biol. 2019, 64, 125017. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Maturen, K.; Prisciandaro, J.; Zhou, J.; Cao, Y.; Balter, J.; Jolly, S. Using MRI to Characterize Small Anatomic Structures Critical to Pelvic Floor Stability in Gynecologic Cancer Patients Undergoing Radiation Therapy. Int. J. Radiat. Oncol. 2015, 93, E608. [Google Scholar] [CrossRef]
- Colosio, A.; Soyer, P.; Rousset, P.; Barbe, C.; Nguyen, F.; Bouché, O.; Hoeffel, C. Value of diffusion-weighted and gadolinium-enhanced MRI for the diagnosis of pelvic recurrence from colorectal cancer. J. Magn. Reson. Imaging 2014, 40, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.J.; Yun, M.; Oh, Y.T.; Kim, J.W.; Kim, S.; Jung, Y.W.; Kim, S.W.; Kim, Y.T. Diagnosis and staging of primary ovarian cancer: Correlation between PET/CT, Doppler US, and CT or MRI. Gynecol. Oncol. 2010, 116, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Fütterer, J.J.; Briganti, A.; De Visschere, P.; Emberton, M.; Giannarini, G.; Kirkham, A.; Taneja, S.S.; Thoeny, H.; Villeirs, G.; Villers, A. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur. Urol. 2015, 68, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Valerio, M.; Donaldson, I.; Emberton, M.; Ehdaie, B.; Hadaschik, B.; Marks, L.S.; Mozer, P.; Rastinehad, A.R.; Ahmed, H.U. Detection of Clinically Significant Prostate Cancer Using Magnetic Resonance Imaging–Ultrasound Fusion Targeted Biopsy: A Systematic Review. Eur. Urol. 2015, 68, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.G.; Fütterer, J.J.; Gupta, R.T.; Katz, A.; Kirkham, A.; Kurhanewicz, J.; Moul, J.W.; Pinto, P.A.; Rastinehad, A.R.; Robertson, C.; et al. The role of magnetic resonance imaging (MRI) in focal therapy for prostate cancer: Recommendations from a consensus panel. BJU Int. 2014, 113, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Eldred-Evans, D.; Tam, H.; Smith, A.P.T.; Winkler, M.; Ahmed, H.U. Use of Imaging to Optimise Prostate Cancer Tumour Volume Assessment for Focal Therapy Planning. Curr. Urol. Rep. 2020, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, Y.; Hricak, H.; Fine, S.W.; Akin, O.; Shukla-Dave, A.; Ishill, N.M.; Moskowitz, C.S.; Grater, J.E.; Reuter, V.E.; Zakian, K.L.; et al. Prostate Tumor Volume Measurement with Combined T2-weighted Imaging and Diffusion-weighted MR: Correlation with Pathologic Tumor Volume. Radiology 2009, 252, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, C.C. Measures of Response: RECIST, WHO, and New Alternatives. J. Clin. Oncol. 2006, 24, 3245–3251. [Google Scholar] [CrossRef]
- Padhani, A.; Liu, G.; Mu-Koh, D.; Chenevert, T.L.; Thoeny, H.C.; Takahara, T.; Dzik-Jurasz, A.; Ross, B.D.; Van Cauteren, M.; Collins, D.; et al. Diffusion-Weighted Magnetic Resonance Imaging as a Cancer Biomarker: Consensus and Recommendations. Neoplasia 2009, 11, 102–125. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lin, G.; Hong, J.-H.; Lin, Y.-P.; Chen, F.-H.; Ng, S.-H.; Wang, C.-C.; Bsc, Y.-P.L. Diffusion radiomics analysis of intratumoral heterogeneity in a murine prostate cancer model following radiotherapy: Pixelwise correlation with histology. J. Magn. Reson. Imaging 2017, 46, 483–489. [Google Scholar] [CrossRef]
- Schob, S.; Meyer, H.J.; Pazaitis, N.; Schramm, D.; Bremicker, K.; Exner, M.; Höhn, A.K.; Garnov, N.; Surov, A. ADC Histogram Analysis of Cervical Cancer Aids Detecting Lymphatic Metastases—A Preliminary Study. Mol. Imaging Biol. 2017, 61, 69–962. [Google Scholar] [CrossRef]
- Lin, G.; Yang, L.-Y.; Lin, Y.-C.; Huang, Y.-T.; Liu, F.-Y.; Wang, C.-C.; Lu, H.-Y.; Chiang, H.-J.; Chen, Y.-R.; Wu, R.-C.; et al. Prognostic model based on magnetic resonance imaging, whole-tumour apparent diffusion coefficient values and HPV genotyping for stage IB-IV cervical cancer patients following chemoradiotherapy. Eur. Radiol. 2018, 29, 556–565. [Google Scholar] [CrossRef]
- Thiesse, P.; Ollivier, L.; Di Stefano-Louineau, D.; Négrier, S.; Savary, J.; Pignard, K.; Lasset, C.; Escudier, B. Response rate accuracy in oncology trials: Reasons for interobserver variability. Groupe Français d’Immunothérapie of the Fédération Nationale des Centres de Lutte Contre le Cancer. J. Clin. Oncol. 1997, 15, 3507–3514. [Google Scholar] [CrossRef]
- Pollard, J.M.; Wen, Z.; Sadagopan, R.; Wang, J.; Ibbott, G.S. The future of image-guided radiotherapy will be MR guided. Br. J. Radiol. 2017, 90, 20160667. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Erickson, B.; Chen, X.; Li, G.; Wu, G.; Paulson, E.; Knechtges, P.; Li, X.A. Appropriate magnetic resonance imaging techniques for gross tumor volume delineation in external beam radiation therapy of locally advanced cervical cancer. Oncotarget 2018, 9, 10100–10109. [Google Scholar] [CrossRef] [PubMed]
- Veera, J.; Lim, K.; Dowling, J.A.; O’Connor, C.; Holloway, L.C.; Vinod, S.K. DedicatedMRIsimulation for cervical cancer radiation treatment planning: Assessing the impact on clinical target volume delineation. J. Med. Imaging Radiat. Oncol. 2019, 63, 236–243. [Google Scholar] [CrossRef]
- Chavaudra, J.; Bridier, A. Definition of volumes in external radiotherapy: ICRU reports 50 and 62. Cancer Radiother. 2001, 5, 472. [Google Scholar] [CrossRef]
- The Royal College of Radiologists, Society of Radiographers, College, Institute of Physics in Medicine, and Engineering. On Target: Ensuring Geometric Accuracy in Radiotherapy; Technical Report; The Royal College of Radiologists RCR: London, UK, 2008. [Google Scholar]
- Chan, T.F.; Vese, L.A. Active Contour and Segmentation Models Using Geometric PDE’s for Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2002; pp. 63–75. [Google Scholar]
- Jiang, X.; Zhang, R.; Nie, S. Image Segmentation Based on Level Set Method. Phys. Procedia 2012, 33, 840–845. [Google Scholar] [CrossRef]
- Boykov, Y.; Jolly, M.-P. Interactive Organ Segmentation Using Graph Cuts. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Pittsburgh, PA, USA, 11–14 October 2000; Springer: Berlin/Heidelberg, Germany, 2000; pp. 276–286. [Google Scholar]
- Beucher, S. Use of watersheds in contour detection. In Proceedings of the International Workshop on Image Processing, Real-Time Edge and Motion Detection/Estimation, CCETT, Rennes, France, 17–21 September 1979. [Google Scholar]
- Naik, S.; Doyle, S.; Agner, S.; Madabhushi, A.; Feldman, M.; Tomaszewski, J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Paris, France, 14–17 May 2008; pp. 284–287. [Google Scholar]
- Zyout, I.; Abdel-Qader, I.; Jacobs, C. Bayesian Classifier with Simplified Learning Phase for Detecting Microcalcifications in Digital Mammograms. Int. J. Biomed. Imaging 2009, 2009, 767805. [Google Scholar] [CrossRef]
- Qiao, J.; Cai, X.; Xiao, Q.; Chen, Z.; Kulkarni, P.; Ferris, C.; Kamarthi, S.; Sridhar, S. Data on MRI brain lesion segmentation using K-means and Gaussian Mixture Model-Expectation Maximization. Data Brief 2019, 27, 104628. [Google Scholar] [CrossRef]
- Zhang, Y.; Brady, M.; Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 2001, 20, 45–57. [Google Scholar] [CrossRef]
- Iglesias, J.E.; Sabuncu, M.R. Multi-atlas segmentation of biomedical images: A survey. Med. Image Anal. 2015, 24, 205–219. [Google Scholar] [CrossRef]
- Blezek, D.J.; Miller, J.V. Atlas stratification. Med. Image Anal. 2007, 11, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Commowick, O.; Malandain, G. Efficient Selection of the Most Similar Image in a Database for Critical Structures Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Brisbane, Australia, 29 October–2 November 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 203–210. [Google Scholar]
- Commowick, O.; Warfield, S.K.; Malandain, G. Using Frankenstein’s Creature Paradigm to Build a Patient Specific Atlas. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, London, UK, 20–24 September 2009; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Yang, J.; Amini, A.; Williamson, R.; Zhang, L.; Zhang, Y.; Komaki, R.; Liao, Z.; Cox, J.; Welsh, J.; Court, L.; et al. Automatic contouring of brachial plexus using a multi-atlas approach for lung cancer radiation therapy. Pract. Radiat. Oncol. 2013, 3, e139–e147. [Google Scholar] [CrossRef] [PubMed]
- Sharp, G.; Fritscher, K.D.; Pekar, V.; Peroni, M.; Shusharina, N.; Veeraraghavan, H.; Yang, J. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med. Phys. 2014, 41, 050902. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.; Galvin, J.; Yu, Y.; Xiao, Y. SU-FF-J-172: Deformable Fusion and Atlas Based Autosegmentation: MimVista Vs. CMS Focal ABAS. Med. Phys. 2009, 36, 2517. [Google Scholar] [CrossRef]
- La Macchia, M.; Fellin, F.; Amichetti, M.; Cianchetti, M.; Gianolini, S.; Paola, V.; Lomax, A.J.; Widesott, L. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat. Oncol. 2012, 7, 160. [Google Scholar] [CrossRef]
- Menzel, H.-G. International Commission on Radiation Units and Measurements. J. Int. Comm. Radiat. Units Meas. 2014, 14, 1–2. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Çiçek, Ö. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar]
- Kamnitsas, K.; Ferrante, E.; Parisot, S.; Ledig, C.; Nori, A.V.; Criminisi, A.; Rueckert, D.; Glocker, B. DeepMedic for Brain Tumor Segmentation. In Proceedings of the International Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Athens, Greece, 17 October 2016; Springer: Cham, Switzerland, 2016; pp. 138–149. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv 2018, arXiv:1809.04430. [Google Scholar]
- Ge, F.; Wang, S.; Liu, T. New benchmark for image segmentation evaluation. J. Electron. Imaging 2007, 16, 033011. [Google Scholar] [CrossRef]
- Huttenlocher, D.; Klanderman, G.; Rucklidge, W. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 850–863. [Google Scholar] [CrossRef]
- Cha, K.H.; Hadjiiski, L.M.; Samala, R.; Chan, H.-P.; Cohan, R.H.; Caoili, E.M.; Paramagul, C.; Alva, A.; Weizer, A.Z. Bladder Cancer Segmentation in CT for Treatment Response Assessment: Application of Deep-Learning Convolution Neural Network—A Pilot Study. Tomography 2016, 2, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hadjiiski, L.M.; Wei, J.; Chan, H.; Cha, K.H.; Cohan, R.H.; Caoili, E.M.; Samala, R.; Zhou, C.; Lu, Y. U-Net based deep learning bladder segmentation in CT urography. Med. Phys. 2019, 46, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yuan, K.; Liu, F.; Xiao, P.; Lv, G.; Liang, Z. An Adaptive Window-Setting Scheme for Segmentation of Bladder Tumor Surface via MR Cystography. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 720–729. [Google Scholar] [CrossRef][Green Version]
- Duan, C.; Liang, Z.; Bao, S.; Zhu, H.; Wang, S.; Zhang, G.; Chen, J.J.; Lu, H. A Coupled Level Set Framework for Bladder Wall Segmentation With Application to MR Cystography. IEEE Trans. Med. Imaging 2010, 29, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Li, L.; Duan, C.; Zhang, H.; Zhao, Y.; Liang, Z. A unified EM approach to bladder wall segmentation with coupled level-set constraints. Med. Image Anal. 2013, 17, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Li, X.; Liu, Y.; Lu, H.; Yan, P. Adaptive Shape Prior Constrained Level Sets for Bladder MR Image Segmentation. IEEE J. Biomed. Health Inform. 2013, 18, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.; Hadjiiski, L.; Samala, R.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med. Phys. 2016, 43, 1882–1896. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, F.; Liu, B. Automatic bladder segmentation from CT images using deep CNN and 3D fully connected CRF-RNN. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 967–975. [Google Scholar] [CrossRef]
- Dolz, J.; Xu, X.; Rony, J.; Yuan, J.; Liu, Y.; Granger, E.; Desrosiers, C.; Zhang, X.; Ben Ayed, I.; Lu, H. Multiregion segmentation of bladder cancer structures in MRI with progressive dilated convolutional networks. Med. Phys. 2018, 45, 5482–5493. [Google Scholar] [CrossRef]
- Li, R.; Chen, H.; Gong, G.; Wang, L. Bladder Wall Segmentation in MRI Images via Deep Learning and Anatomical Constraints. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1629–1632. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Xiao, B.; Wang, S.; Miao, Z.; Sun, Y.; Zhang, F. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys. Med. 2020, 69, 184–191. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Guan, H.; Zhen, H.; Sun, Y.; Chen, Q.; Chen, Y.; Wang, S.; Qiu, J. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother. Oncol. 2020, 153, 172–179. [Google Scholar] [CrossRef]
- Wang, Z.; Chang, Y.; Peng, Z.; Lv, Y.; Shi, W.; Wang, F.; Pei, X.; Xu, X.G. Evaluation of deep learning-based auto-segmentation algorithms for delineating clinical target volume and organs at risk involving data for 125 cervical cancer patients. J. Appl. Clin. Med. Phys. 2020, 21, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yang, Z.; Jiang, S.; Zhou, Z.; Meng, M.; Wang, W. Automatic segmentation and applicator reconstruction for CT-based brachytherapy of cervical cancer using 3D convolutional neural networks. J. Appl. Clin. Med. Phys. 2020, 21, 158–169. [Google Scholar] [CrossRef]
- Rhee, D.J.; Jhingran, A.; Rigaud, B.; Netherton, T.; Cardenas, C.E.; Zhang, L.; Vedam, S.; Kry, S.; Brock, K.K.; Shaw, W.; et al. Automatic contouring system for cervical cancer using convolutional neural networks. Med. Phys. 2020, 47, 5648–5658. [Google Scholar] [CrossRef]
- Breto, A.; Zavala-Romero, O.; Asher, D.; Baikovitz, J.; Ford, J.; Stoyanova, R.; Portelance, L. A Deep Learning Pipeline for per-Fraction Automatic Segmentation of GTV and OAR in cervical cancer. Int. J. Radiat. Oncol. 2019, 105, S202. [Google Scholar] [CrossRef]
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother. Oncol. 2020, 144, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Kiljunen, T.; Akram, S.; Niemelä, J.; Löyttyniemi, E.; Seppälä, J.; Heikkilä, J.; Vuolukka, K.; Kääriäinen, O.-S.; Heikkilä, V.-P.; Lehtiö, K.; et al. A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study. Diagnostics 2020, 10, 959. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Nie, D.; Adeli, E.; Yin, J.; Lian, J.; Shen, D. High-Resolution Encoder–Decoder Networks for Low-Contrast Medical Image Segmentation. IEEE Trans. Image Process. 2020, 29, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Lei, Y.; Tian, S.; Wang, T.; Patel, P.; Curran, W.J.; Jani, A.B.; Liu, T.; Yang, X. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother. Oncol. 2019, 141, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; He, K.; Nie, D.; Zhou, S.; Gao, Y.; Shen, D. CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation. Med. Image Anal. 2019, 54, 168–178. [Google Scholar] [CrossRef]
- Liu, C.; Gardner, S.J.; Wen, N.; Elshaikh, M.A.; Siddiqui, F.; Movsas, B.; Chetty, I.J. Automatic Segmentation of the Prostate on CT Images Using Deep Neural Networks (DNN). Int. J. Radiat. Oncol. 2019, 104, 924–932. [Google Scholar] [CrossRef]
- Kearney, V.P.; Chan, J.W.; Wang, T.; Perry, A.; Yom, S.S.; Solberg, T.D. Attention-enabled 3D boosted convolutional neural networks for semantic CT segmentation using deep supervision. Phys. Med. Biol. 2019, 64, 135001. [Google Scholar] [CrossRef]
- He, K.; Cao, X.; Shi, Y.; Nie, D.; Gao, Y.; Shen, D. Pelvic Organ Segmentation Using Distinctive Curve Guided Fully Convolutional Networks. IEEE Trans. Med. Imaging 2019, 38, 585–595. [Google Scholar] [CrossRef]
- Kazemifar, S.; Balagopal, A.; Nguyen, D.; McGuire, S.; Hannan, R.; Jiang, S.B.; Owrangi, A.M. Segmentation of the prostate and organs at risk in male pelvic CT images using deep learning. Biomed. Phys. Eng. Express 2018, 4, 055003. [Google Scholar] [CrossRef]
- Balagopal, A.; Kazemifar, S.; Nguyen, D.; Lin, M.-H.; Hannan, R.; Owrangi, A.M.; Jiang, S.B. Fully automated organ segmentation in male pelvic CT images. Phys. Med. Biol. 2018, 63, 245015. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, W.; Gao, Y.; Shen, D. Does Manual Delineation only Provide the Side Information in CT Prostate Segmentation? In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; pp. 692–700. [Google Scholar] [CrossRef]
- Jia, H.; Xia, Y.; Song, Y.; Zhang, D.; Huang, H.; Zhang, Y.; Cai, W. 3D APA-Net: 3D Adversarial Pyramid Anisotropic Convolutional Network for Prostate Segmentation in MR Images. IEEE Trans. Med. Imaging 2019, 39, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Yahya, N.; Alsaih, K.; Meriaudeau, F. Segmentation of Prostate in MRI Images Using Depth Separable Convolution Operations. In Proceedings of the International Conference on Intelligent Human Computer Interaction, Daegu, Korea, 24–26 November 2020; pp. 132–141. [Google Scholar]
- Cem Birbiri, U.; Hamidinekoo, A.; Grall, A.; Malcolm, P.; Zwiggelaar, R. Investigating the Performance of Generative Adversarial Networks for Prostate Tissue Detection and Segmentation. J. Imaging 2020, 6, 83. [Google Scholar] [CrossRef]
- Dai, Z.; Carver, E.; Liu, C.; Lee, J.; Feldman, A.; Zong, W.; Pantelic, M.; Elshaikh, M.; Wen, N. Segmentation of the Prostatic Gland and the Intraprostatic Lesions on Multiparametic Magnetic Resonance Imaging Using Mask Region-Based Convolutional Neural Networks. Adv. Radiat. Oncol. 2020, 5, 473–481. [Google Scholar] [CrossRef]
- Zhu, Q.; Du, B.; Yan, P. Boundary-Weighted Domain Adaptive Neural Network for Prostate MR Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Li, X.; Zheng, Y.; Chen, Z.; Shi, Z.; Liu, L.; Fei, B. Graph-convolutional-network-based interactive prostate segmentation in MR images. Med. Phys. 2020, 47, 4164–4176. [Google Scholar] [CrossRef] [PubMed]
- Aldoj, N.; Biavati, F.; Michallek, F.; Stober, S.; Dewey, M. Automatic prostate and prostate zones segmentation of magnetic resonance images using DenseNet-like U-net. Sci. Rep. 2020, 10, 14315. [Google Scholar] [CrossRef] [PubMed]
- Rundo, L.; Han, C.; Zhang, J.; Hataya, R.; Nagano, Y.; Militello, C.; Ferretti, C.; Nobile, M.S.; Tangherloni, A.; Gilardi, M.C.; et al. CNN-Based Prostate Zonal Segmentation on T2-Weighted MR Images: A Cross-Dataset Study. In Neural Approaches to Dynamics of Signal Exchanges; Springer: Singapore, 2020. [Google Scholar]
- Savenije, M.H.F.; Maspero, M.; Sikkes, G.G.; van der Voort van Zyp, J.R.; Kotte, A.N.T.J.; Bol, G.H.; van den Berg, C.A. Clinical implementation of MRI-based organs-at-risk auto-segmentation with convolutional networks for prostate radiotherapy. Radiat. Oncol. 2020, 15, 104. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhao, M.; Pang, Y. CDA-Net for Automatic Prostate Segmentation in MR Images. Appl. Sci. 2020, 10, 6678. [Google Scholar] [CrossRef]
- Geng, L.; Wang, J.; Xiao, Z.; Tong, J.; Zhang, F.; Wu, J. Encoder-decoder with dense dilated spatial pyramid pooling for prostate MR images segmentation. Comput. Assist. Surg. 2019, 24, 13–19. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, W.; Lee, K.-H.; Lo, Y.-L.; Ng, Y.-L.; Dou, Q.; Vardhanabhuti, V.; Kwok, K.-W. A Two-Stage Approach for Automated Prostate Lesion Detection and Classification with Mask R-CNN and Weakly Supervised Deep Neural Network. In Proceedings of the Workshop on Artificial Intelligence in Radiation Therapy, Shenzhen, China, 17 October 2019; Springer: Cham, Switzerland, 2019; pp. 43–51. [Google Scholar]
- Zabihollahy, F.; Schieda, N.; Jeyaraj, S.K.; Ukwatta, E. Automated segmentation of prostate zonal anatomy on T2-weighted (T2W) and apparent diffusion coefficient (ADC) map MR images using U-Nets. Med. Phys. 2019, 46, 3078–3090. [Google Scholar] [CrossRef]
- Liu, Y.; Sung, K.; Yang, G.; Mirak, S.A.; Hosseiny, M.; Azadikhah, A.; Zhong, X.; Reiter, R.E.; Lee, Y.; Raman, S.S. Automatic Prostate Zonal Segmentation Using Fully Convolutional Network With Feature Pyramid Attention. IEEE Access 2019, 7, 163626–163632. [Google Scholar] [CrossRef]
- Nie, D.; Wang, L.; Gao, Y.; Lian, J.; Shen, D. STRAINet: Spatially Varying sTochastic Residual AdversarIal Networks for MRI Pelvic Organ Segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2018, 30, 1552–1564. [Google Scholar] [CrossRef]
- Taghanaki, S.A.; Zheng, Y.; Zhou, S.K.; Georgescu, B.; Sharma, P.; Xu, D.; Comaniciu, D.; Hamarneh, G. Combo loss: Handling input and output imbalance in multi-organ segmentation. Comput. Med. Imaging Graph. 2019, 75, 24–33. [Google Scholar] [CrossRef]
- Elguindi, S.; Zelefsky, M.J.; Jiang, J.; Veeraraghavan, H.; Deasy, J.; Hunt, M.A.; Tyagi, N. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys. Imaging Radiat. Oncol. 2019, 12, 80–86. [Google Scholar] [CrossRef]
- Tan, L.; Liang, A.; Li, L.; Liu, W.; Kang, H.; Chen, C. Automatic prostate segmentation based on fusion between deep network and variational methods. J. Xray Sci. Technol. 2019, 27, 821–837. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Wang, X.; Kim, J.; Khadra, M.; Fulham, M.; Feng, D. A propagation-DNN: Deep combination learning of multi-level features for MR prostate segmentation. Comput. Methods Programs Biomed. 2019, 170, 11–21. [Google Scholar] [CrossRef]
- Zhu, Y.; Wei, R.; Gao, G.; Ding, L.; Zhang, X.; Wang, X.; Zhang, J. Fully automatic segmentation on prostate MR images based on cascaded fully convolution network. J. Magn. Reson. Imaging 2019, 49, 1149–1156. [Google Scholar] [CrossRef]
- Alkadi, R.; Taher, F.; El-Baz, A.; Werghi, N. A Deep Learning-Based Approach for the Detection and Localization of Prostate Cancer in T2 Magnetic Resonance Images. J. Digit. Imaging 2019, 32, 793–807. [Google Scholar] [CrossRef]
- Ghavami, N.; Hu, Y.; Gibson, E.; Bonmati, E.; Emberton, M.; Moore, C.M.; Barratt, D.C. Automatic segmentation of prostate MRI using convolutional neural networks: Investigating the impact of network architecture on the accuracy of volume measurement and MRI-ultrasound registration. Med. Image Anal. 2019, 58, 101558. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Chen, W.; Chen, Y.; Tang, X. Prostate Segmentation Using Z-Net. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 11–14. [Google Scholar]
- Feng, Z.; Nie, D.; Wang, L.; Shen, D. Semi-supervised learning for pelvic MR image segmentation based on multi-task residual fully convolutional networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 885–888. [Google Scholar] [CrossRef]
- Han, C.; Zhang, J.; Hataya, R.; Nagano, Y.; Nakayama, H.; Rundo, L. Prostate zonal segmentation using deep learning. IEICE Tech. Rep. 2018, 117, 69–70. [Google Scholar]
- Brosch, T.; Peters, J.; Groth, A.; Stehle, T.; Weese, J. Deep Learning-Based Boundary Detection for Model-Based Segmentation with Application to MR Prostate Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018; pp. 515–522. [Google Scholar]
- Kang, J.; Samarasinghe, G.; Senanayake, U.; Conjeti, S.; Sowmya, A. Deep Learning for Volumetric Segmentation in Spatio-Temporal Data: Application to Segmentation of Prostate in DCE-MRI. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 61–65. [Google Scholar]
- Drozdzal, M.; Chartrand, G.; Vorontsov, E.; Shakeri, M.; Di Jorio, L.; Tang, A.; Romero, A.; Bengio, Y.; Pal, C.; Kadoury, S. Learning normalized inputs for iterative estimation in medical image segmentation. Med. Image Anal. 2018, 44, 1–13. [Google Scholar] [CrossRef]
- To, M.N.N.; Vu, D.Q.; Turkbey, B.; Choyke, P.L.; Kwak, J.T. Deep dense multi-path neural network for prostate segmentation in magnetic resonance imaging. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Karimi, D.; Samei, G.; Kesch, C.; Nir, G.; Salcudean, S. Prostate segmentation in MRI using a convolutional neural network architecture and training strategy based on statistical shape models. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1211–1219. [Google Scholar] [CrossRef]
- Zhu, Q.; Du, B.; Turkbey, B.; Choyke, P.L.; Yan, P. Deeply-supervised CNN for prostate segmentation. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 178–184. [Google Scholar]
- Zhu, Y.; Wang, L.; Liu, M.; Qian, C.; Yousuf, A.; Oto, A.; Shen, D. MRI-based prostate cancer detection with high-level representation and hierarchical classification. Med. Phys. 2017, 44, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Roth, H.R.; Lay, N.; Lu, L.; Turkbey, B.; Gandler, W.; McCreedy, E.S.; Pohida, T.; Pinto, P.A.; Choyke, P.; et al. Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J. Med. Imaging 2017, 4, 041302. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.; Zhang, J.; Baig, S.; Wong, A.; Haider, M.A.; Khalvati, F. Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks. J. Med. Imaging 2017, 4, 041307. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yang, X.; Chen, H.; Qin, J.; Heng, P. Volumetric ConvNets with Mixed Residual Connections for Automated Prostate Segmentation from 3D MR Images. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Guo, Y.; Gao, Y.; Shen, D. Deformable MR Prostate Segmentation via Deep Feature Learning and Sparse Patch Matching. IEEE Trans. Med. Imaging 2016, 35, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Gao, Y.; Oto, A.; Shen, D. Representation Learning: A Unified Deep Learning Framework for Automatic Prostate MR Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 254–261. [Google Scholar]
- Men, K.; Boimel, P.; Janopaul-Naylor, J.; Zhong, H.; Huang, M.; Geng, H.; Cheng, C.; Fan, Y.; Plastaras, J.P.; Ben-Josef, E.; et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys. Med. Biol. 2018, 63, 185016. [Google Scholar] [CrossRef]
- Men, K.; Dai, J.; Li, Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med. Phys. 2017, 44, 6377–6389. [Google Scholar] [CrossRef]
- Zhao, X.; Xie, P.; Wang, M.; Li, W.; Pickhardt, P.J.; Xia, W.; Xiong, F.; Zhang, R.; Xie, Y.; Jian, J.; et al. Deep learning–based fully automated detection and segmentation of lymph nodes on multiparametric-mri for rectal cancer: A multicentre study. EBioMedicine 2020, 56, 102780. [Google Scholar] [CrossRef]
- Wang, M.; Xie, P.; Ran, Z.; Jian, J.; Zhang, R.; Xia, W.; Yu, T.; Ni, C.; Gu, J.; Gao, X.; et al. Full convolutional network based multiple side-output fusion architecture for the segmentation of rectal tumors in magnetic resonance images: A multi-vendor study. Med. Phys. 2019, 46, 2659–2668. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, J.; Qin, G.; Shen, L.; Sun, Y.; Ying, H.; Zhang, Z.; Hu, W. Technical Note: A deep learning-based autosegmentation of rectal tumors in MR images. Med. Phys. 2018, 45, 2560–2564. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.; Dou, Q.; Wang, Z.-X.; Liu, L.-Z.; Wang, L.-S.; Chen, H.; Heng, P.-A.; Xu, R.-H. HL-FCN: Hybrid loss guided FCN for colorectal cancer segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 195–198. [Google Scholar]
- Trebeschi, S.; Van Griethuysen, J.J.M.; Lambregts, D.; Lahaye, M.J.; Parmar, C.; Bakers, F.C.H.; Peters, N.H.G.M.; Beets-Tan, R.G.H.; Aerts, H.J.W.L. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci. Rep. 2017, 7, 5301. [Google Scholar] [CrossRef]
- McVeigh, P.Z.; Syed, A.M.; Milosevic, M.; Fyles, A.; Haider, M.A. Diffusion-weighted MRI in cervical cancer. Eur. Radiol. 2008, 18, 1058–1064. [Google Scholar] [CrossRef]
- Niaf, E.; Rouviere, O.; Mège-Lechevallier, F.; Bratan, F.; Lartizien, C. Computer-aided diagnosis of prostate cancer in the peripheral zone using multiparametric MRI. Phys. Med. Biol. 2012, 57, 3833–3851. [Google Scholar] [CrossRef] [PubMed]
- Toth, R.; Ribault, J.; Gentile, J.; Sperling, D.; Madabhushi, A. Simultaneous segmentation of prostatic zones using Active Appearance Models with multiple coupled levelsets. Comput. Vis. Image Underst. 2013, 117, 1051–1060. [Google Scholar] [CrossRef]
- Qiu, W.; Yuan, J.; Ukwatta, E.; Sun, Y.; Rajchl, M.; Fenster, A. Dual optimization based prostate zonal segmentation in 3D MR images. Med. Image Anal. 2014, 18, 660–673. [Google Scholar] [CrossRef]
- Makni, N.; Iancu, A.; Colot, O.; Puech, P.; Mordon, S.; Betrouni, N. Zonal segmentation of prostate using multispectral magnetic resonance images. Med. Phys. 2011, 38, 6093–6105. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.; et al. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Bloch, N.; Madabhushi, A.; Huisman, H.; Freymann, J.; Kirby, J.; Grauer, M.; Enquobahrie, C.J.; Clarke, L.; Farahani, K. NCI-ISBI 2013 Challenge: Automated Segmentation of Prostate Structures. Cancer Imaging Arch. 2015, 370, 6. [Google Scholar]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. ProstateX Challenge Database. 2017. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=23691656 (accessed on 8 August 2021).
- Yu, L.; Cheng, J.-Z.; Dou, Q.; Yang, X.; Chen, H.; Qin, J.; Heng, P.-A. Automatic 3D Cardiovascular MR Segmentation with Densely-Connected Volumetric ConvNets. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland, 2017; pp. 287–295. [Google Scholar]
- Lemaître, G.; Martí, R.; Freixenet, J.; Vilanova, J.C.; Walker, P.M.; Meriaudeau, F. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: A review. Comput. Biol. Med. 2015, 60, 8–31. [Google Scholar] [CrossRef]
- Saidu, C.; Csato, L. Medical Image Analysis with Semantic Segmentation and Active Learning. Studia Univ. Babeș-Bolyai Inform. 2019, 64, 26–38. [Google Scholar] [CrossRef]
- The Brigham and Women’s Hospital, (BWH). BWH Prostate MR Image Database. 2008. Available online: https://central.xnat.org/data/projects/NCIGT_PROSTATE (accessed on 8 August 2021).
- Pekar, V.; McNutt, T.R.; Kaus, M.R. Automated model-based organ delineation for radiotherapy planning in prostatic region. Int. J. Radiat. Oncol. 2004, 60, 973–980. [Google Scholar] [CrossRef]
- Pasquier, D.; Lacornerie, T.; Vermandel, M.; Rousseau, J.; Lartigau, E.; Betrouni, N. Automatic Segmentation of Pelvic Structures From Magnetic Resonance Images for Prostate Cancer Radiotherapy. Int. J. Radiat. Oncol. 2007, 68, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Choi, H.; You, Y.N.; Rauch, G.M.; Jensen, C.T.; Hou, P.; Chang, G.J.; Skibber, J.M.; Ernst, R.D. MR Imaging for Preoperative Evaluation of Primary Rectal Cancer: Practical Considerations. Radiographics 2012, 32, 389–409. [Google Scholar] [CrossRef]
- Hernando-Requejo, O.; López, M.; Cubillo, A.; Rodriguez, A.; Ciervide, R.; Valero, J.; Sánchez, E.; Garcia-Aranda, M.; Rodriguez, J.; Potdevin, G.; et al. Complete pathological responses in locally advanced rectal cancer after preoperative IMRT and integrated-boost chemoradiation. Strahlenther. Onkol. 2014, 190, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Schipaanboord, B.; Boukerroui, D.; Peressutti, D.; van Soest, J.; Lustberg, T.; Dekker, A.; van Elmpt, W.; Gooding, M.J. An Evaluation of Atlas Selection Methods for Atlas-Based Automatic Segmentation in Radiotherapy Treatment Planning. IEEE Trans. Med. Imaging 2019, 38, 2654–2664. [Google Scholar] [CrossRef]
- Fritscher, K.D.; Peroni, M.; Zaffino, P.; Spadea, M.F.; Schubert, R.; Sharp, G. Automatic segmentation of head and neck CT images for radiotherapy treatment planning using multiple atlases, statistical appearance models, and geodesic active contours. Med. Phys. 2014, 41, 051910. [Google Scholar] [CrossRef] [PubMed]
- Losnegård, A.; Hysing, L.; Kvinnsland, Y.; Muren, L.; Munthe-Kaas, A.Z.; Hodneland, E.; Lundervold, A. Semi-Automatic Segmentaiton of the Large Intestine for Radiotherapy Planning Using the Fast-Marching Method. Radiother. Oncol. 2009, 92, S75. [Google Scholar] [CrossRef]
- Haas, B.; Coradi, T.; Scholz, M.; Kunz, P.; Huber, M.; Oppitz, U.; André, L.; Lengkeek, V.; Huyskens, D.; Van Esch, A.; et al. Automatic segmentation of thoracic and pelvic CT images for radiotherapy planning using implicit anatomic knowledge and organ-specific segmentation strategies. Phys. Med. Biol. 2008, 53, 1751–1771. [Google Scholar] [CrossRef]
- Gambacorta, M.; Valentini, C.; DiNapoli, N.; Mattiucci, G.; Pasini, D.; Barba, M.; Manfrida, S.; Boldrini, L.; Caria, N.; Valentini, V. Atlas-based Auto-segmentation Clinical Validation of Pelvic Volumes and Normal Tissue in Rectal Tumors. Int. J. Radiat. Oncol. 2012, 84, S347–S348. [Google Scholar] [CrossRef]
- Oakden-Rayner, L. Exploring Large-scale Public Medical Image Datasets. Acad. Radiol. 2020, 27, 106–112. [Google Scholar] [CrossRef]
- Cuocolo, R.; Stanzione, A.; Castaldo, A.; De Lucia, D.R.; Imbriaco, M. Quality control and whole-gland, zonal and lesion annotations for the PROSTATEx challenge public dataset. Eur. J. Radiol. 2021, 138, 109647. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Green, B.; Adam, H.; Yuille, A.; Chen, L.-C. Axial-DeepLab: Stand-Alone Axial-Attention for Panoptic Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Glasgow, UK, 23–28 August 2020; Springer: Cham, Switzerland, 2020; pp. 108–126. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 213–229. [Google Scholar]
- Ranjbarzadeh, R.; Kasgari, A.B.; Ghoushchi, S.J.; Anari, S.; Naseri, M.; Bendechache, M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci. Rep. 2021, 11, 10930. [Google Scholar] [CrossRef] [PubMed]
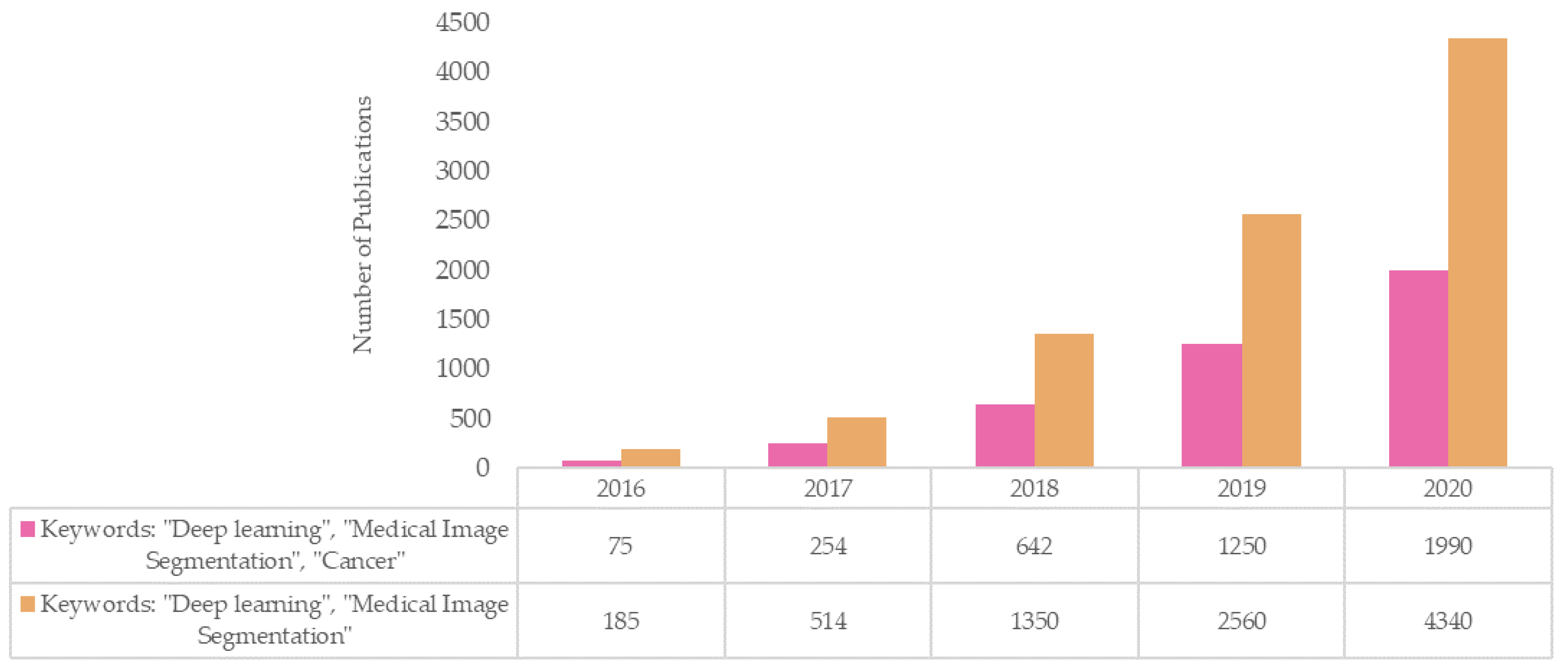
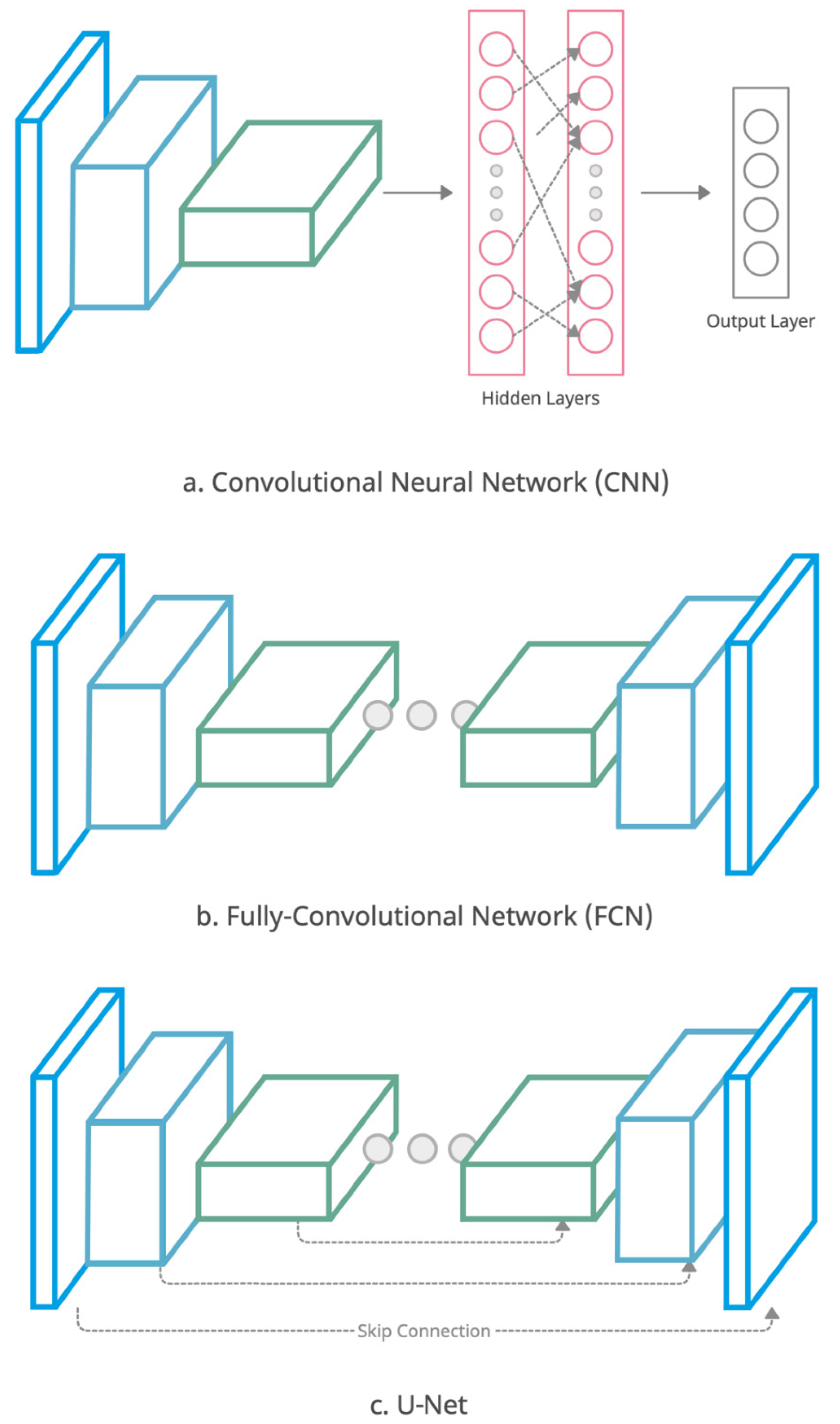

| Image Modality | Deep Learning Strategy | DL Network Dimension | Number of Patients (Train/Test) | Segmentation Evaluation Metrics | Year | Reference |
|---|---|---|---|---|---|---|
| (MR Acquisition Mode) | ||||||
| Bladder Cancer | ||||||
| CT | U-Net | 2D/3D | 81/92 | Bladder (IoU: 0.85/0.82) | 2019 | [88] |
| CT | CNN + FCN (CRF-RNN) | 3D | 100/24 | Bladder (DSC: 0.92) | 2018 | [94] |
| CT | CNN | 2D | 62 leave-one-out cross validation | Bladder Tumor (area under the ROC curve (AUC): 0.73) | 2016 | [87] |
| CT | CNN | 2D | 81/92 | Bladder (IoU: 0.76) | 2016 | [93] |
| T2W (2D), DW (2D) MRI | AE + modified residual network (BW-Net) | 2D | 144/25 | Bladder Wall (DSC: 0.85) | 2020 | [96] |
| T2W MRI (3D) | U-Net with progressive dilated convolutions (U-Net Progressive) | 2D | 40/15 | Bladder Tumor (DSC: 0.68), Outer Wall (DSC: 0.83), Inner Wall (DSC: 0.98) | 2018 | [95] |
| Cervical Cancer | ||||||
| CT | U-Net with context aggregation blocks (CabUNet) | 2D | 77/14 | Bladder (DSC: 0.90), Bone Marrow (DSC: 0.85), L Fem. Head (DSC: 0.90), R Fem. Head (DSC: 0.90), Rectum (DSC: 0.79), Small Intestine (DSC: 0.83), Spinal Cord (DSC: 0.82) | 2020 | [97] |
| CT | Dual path U-Net (DpnUNet) | 2.5D | 210 five-fold cross validation | CTV (DSC: 0.86), Bladder (DSC: 0.91), Bone Marrow (DSC: 0.85), L Fem. Head (DSC: 0.90), R Fem. Head (DSC: 0.90), Rectum (DSC: 0.82), Bowel Bag (DSC: 0.85), Spinal Cord (DSC: 0.82) | 2020 | [98] |
| CT | U-Net | 3D | 100/25 | CTV (DSC: 0.86), Bladder (DSC: 0.88), Rectum (DSC: 0.81), L Fem. Head (DSC: 0.88), R Fem. Head (DSC: 0.88), Small Intestine (DSC: 0.86) | 2020 | [99] |
| CT | U-Net with residual connection, dilated convolution and deep supervision (DSD-UNet) | 3D | 73/18 | High-risk CTV (DSC: 0.82, IOU: 0.72), Bladder (DSC: 0.86, IOU: 0.77), Rectum (DSC: 0.82, IOU: 0.71), Small Intestine (DSC: 0.80, IOU: 0.69), Sigmoid (DSC: 0.64, IOU: 0.52) | 2020 | [100] |
| CT | V-Net | 3D | 2464/140 (+30 external test patients) | Primary CTV (UteroCervix) (DSC: 0.85), Nodal CTV (DSC: 0.86), PAN CTV (DSC: 0.76), Bladder (DSC: 0.89), Rectum (DSC: 0.81), Spinal Cord (DSC: 0.90), L Femur (DSC: 0.94), R Femur (DSC: 0.93), L Kidney (DSC: 0.94), R Kidney (DSC: 0.95), Pelvic Bone (DSC: 0.93), Sacrum (DSC: 0.91), L4 Vertebral Body (DSC: 0.91), L5 Vertebral Body (DSC: 0.90) | 2020 | [101] |
| MRI (unspecified) | Mask R-CNN | 2D | 5 (646 images split 9:1 for training and testing) | GTV + Cervix (DSC: 0.84), Uterus (DSC: 0.92), Sigmoid (DSC: 0.89), Bladder (DSC: 0.90), Rectum (DSC: 0.89), Parametrium (DSC: 0.66), Vagina (DSC: 0.71), Mesorectum (DSC: 0.68), Femur (DSC: 0.81) | 2019 | [102] |
| DW MRI (2D) | U-Net | 2D | 144/25 | Cervical Tumor (DSC: 0.82) | 2019 | [17] |
| Prostate Cancer | ||||||
| CT | U-Net (External commercial software) | 2D | 328/20 | Prostate (DSC: 0.79), Bladder (DSC: 0.97), Rectum (DSC: 0.78), Fem. Head (DSC: 0.91), Seminal Vesicles (DSC: 0.64) | 2020 | [103] |
| CT | U-Net | 3D | 900/30 | Prostate (DSC: 0.82), Bladder (DSC: 0.93), Rectum (DSC: 0.84), L Fem. Head (DSC: 0.68), R Fem. Head (DSC: 0.69), Lymph Nodes (DSC: 0.80), Seminal Vesicles (DSC: 0.72) | 2020 | [104] |
| CT | High-resolution multi-scale encoder-decoder network (HMEDN) | 2D | 180/100 | Prostate (DSC: 0.88), Bladder (DSC: 0.94), Rectum (DSC: 0.87) | 2019 | [105] |
| CT/ Synthetic T2W MRI | CT-to-MR synthesis + Deep Attention U-Net (DAUNet) | 3D | 112/28 five-fold cross validation | Prostate (DSC: 0.87), Bladder (DSC: 0.95), Rectum (DSC: 0.89) | 2019 | [106] |
| CT | Modified U-Net | 3D | 313 five-fold cross validation | Prostate: (DSC: 0.89), Bladder: (DSC: 0.94), Rectum: (DSC: 0.89) | 2019 | [107] |
| CT | Deep Neural Network (DNN) | 3D | 771/140 | Prostate (DSC: 0.88) | 2019 | [108] |
| CT | Deeply-supervised attention-enabled boosted convolutional neural network (DAB-CNN) | 3D | 80/20 | Prostate (DSC: 0.90), Bladder (DSC: 0.93), Rectum (DSC: 0.83), Penile bulb (DSC: 0.72) | 2019 | [109] |
| CT | Distinctive curve guided fully convolutional network (FCN) | 2D | 313 five-fold cross validation | Prostate (DSC: 0.89), Bladder (DSC: 0.94), Rectum (DSC: 0.89) | 2019 | [110] |
| CT | U-Net | 2D | 60/25 | Prostate: (DSC: 0.88), Bladder: DSC: 0.95), Rectum: (DSC: 0.92) | 2018 | [111] |
| CT | 2D U-Net + 3D U-Net with aggregated residual networks (ResNeXt) | 2D/3D | 108/28 four-fold cross validation | Prostate (DSC: 0.90), Bladder (DSC: 0.95), Rectum (DSC: 0.84), L Fem. Head (DSC: 0.96), R Fem. Head (DSC: 0.95) | 2018 | [112] |
| CT | CNN + multi-atlas fusion | 2D | 92 five-fold cross validation | Prostate (DSC: 0.86) | 2017 | [31] |
| CT | FCN (based on LeNet) | 2D | 22 two-fold cross validation | Prostate (DSC: 0.89) | 2017 | [113] |
| T2W MRI (2D) | Adversarial pyramid anisotropic convolutional deep neural network (APA-Net) | 3D | 110 three-fold cross validation | Whole Prostate Gland (DSC: 0.90) | 2020 | [114] |
| T2W MRI (2D/3D) | DeeplabV3+ | 2D | 40 | Prostate Central Gland (DSC: 0.81), Peripheral Zone (DSC: 0.70) | 2020 | [115] |
| T2W (2D), DW (2D) MRI | Conditional GAN (cGAN)/Cycle-consistent GAN (Cycle-GAN) | 2D | 40/50 | Whole Prostate Gland (DSC: 0.75) | 2020 | [116] |
| T2W (2D), DW (2D) MRI | Mask R-CNN | 2D | 54/16 (+12 external test patients) | Whole Prostate Gland (DSC: 0.86), Prostate Tumor (DSC: 0.56) | 2020 | [117] |
| T2W MRI (2D) | Boundary-weighted domain adaptive neural network (BOWDA-Net) | 3D | 40/146 | Whole Prostate Gland (DSC: 0.91) Prostate Base (DSC: 0.89) Prostate Apex (DSC: 0.89) | 2020 | [118] |
| T2W MRI (2D) | Graph convolutional network (GCN) | 2D | 140 five-fold cross validation | Whole Prostate Gland (DSC: 0.93) | 2020 | [119] |
| T2W MRI (2D) | Dense U-Net | 2D | 141/47 four-fold cross validation | Whole Prostate Gland (DSC: 0.92), Central Gland (DSC: 0.89), Peripheral Zone (DSC: 0.78) | 2020 | [120] |
| T2W MRI (2D) | U-Net/Pix2pix | 2D | 40 four-fold cross validation | Prostate Central Gland (DSC: 0.86–0.88), Peripheral Zone (DSC: 0.90–0.83) | 2020 | [121] |
| T1W (3D), T2W (unspecified) MRI | Multi-scale DeepMedic | 3D | 97/53 three-fold cross validation | Bladder (DSC: 0.96), Rectum (DSC: 0.88), L femur (DSC: 0.97), R femur (DSC: 0.97) | 2020 | [122] |
| T2W MRI (2D) | Cascaded dual attention network (CDA-Net) | 3D | 40/109 | Whole Prostate Gland (DSC: 0.92) | 2020 | [123] |
| T2W MRI (2D) | Encoder-Decoder structure with dense dilated spatial pyramid pooling (DDSPP) | 2D | 150 | Whole Prostate Gland (DSC: 0.95) | 2019 | [124] |
| T2W (2D), DW (2D) MRI | Mask R-CNN | 2D | 36 (split 7:2:1 for training, validation and testing) | Whole Prostate Gland (IoU: 0.84), Prostate Tumor (IoU: 0.40), Central Gland (IoU: 0.78), Peripheral Zone (IoU: 0.51) | 2019 | [125] |
| T2W (2D), DW (2D) MRI | U-Net | 2D | 100/125 | Whole Prostate Gland (DSC: 0.84), Central Gland (DSC: 0.78), Peripheral Zone (DSC: 0.69) | 2019 | [126] |
| T2W MRI (2D) | FCN with feature pyramid attention | 2D | 250/63 (+46 external test patients) | Prostate Transition Zone (DSC: 0.79), Peripheral zone (DSC: 0.74) | 2019 | [127] |
| T2W MRI (3D) | Spatially-varying stochastic residual adversarial network (STRAINet) | 3D | 50 five-fold cross validation | Whole Prostate Gland (DSC: 0.91), Bladder (DSC: 0.97), Rectum (DSC: 0.91) | 2019 | [128] |
| T2W MRI (2D) | U-Net with “combo loss” | 3D | 700/258 | Whole Prostate Gland (DSC: 0.91) | 2019 | [129] |
| T2W MRI (unspecified) | DeepLabV3+ | 2D | 40/50 | CTV (DSC: 0.83), Bladder (DSC: 0.93), Rectum (DSC: 0.82), Penile Bulb (DSC: 0.74), Urethra (DSC: 0.69), Rectal Spacer (DSC: 0.81) | 2019 | [130] |
| T2W MRI (2D) | V-Net + variational methods | 3D | 85 | Whole Prostate Gland (DSC: 0.64) | 2019 | [131] |
| T2W MRI (2D) | Propagation Deep Neural Network (P-DNN) | 2D | 50/30 | Whole Prostate Gland: (DSC: 0.84) | 2019 | [132] |
| T2W (2D), DW (2D) MRI | Cascaded U-Net | 2D | 76/51 | Whole Prostate Gland (DSC: 0.92), Peripheral zone (DSC: 0.79) | 2019 | [133] |
| T2W MRI (3D) | Multi-view CNN | 2D | 19 leave-one-out cross validation | Prostate Tumor (DSC: 0.92, IoU: 0.67), Prostate Central Gland (IoU: 0.65), Peripheral Zone (IoU: 0.59) | 2019 | [134] |
| T2W MRI (2D) | Investigative CNN study (U-Net, V-Net, HighRes3dNet, HolisticNet, Dense V-Net, Adapted U-Net) | 3D | 173/59 | Whole Prostate Gland (DSC: 0.87) | 2019 | [135] |
| T2W MRI (2D) | Z-Net | 2D | 45/30 | Whole Prostate Gland (DSC: 0.90) | 2019 | [136] |
| T2W MRI (3D) | FCN | 3D | 60/10 | Whole Prostate Gland (DSC: 0.89), Bladder (DSC: 0.95), Rectum (DSC: 0.88) | 2018 | [137] |
| T2W MRI (2D) | SegNet | 2D | 16/5 (+19 external test patients) | Whole Prostate Gland (DSC: 0.75) | 2018 | [138] |
| T2W MRI (2D) | CNN + Boundary Detection | 3D | 50 five-fold cross validation | Whole Prostate Gland (DSC: 0.90) | 2018 | [139] |
| Dynamic Contrast-Enhanced (DCE) MRI (3D) | U-Net + Long-Short-Term Memory (LSTM) | 3D | (15/2) three-fold cross validation | Whole Prostate Gland (DSC: 0.86) | 2018 | [140] |
| T2W MRI (2D) | FCN | 2D | 50/30 | Whole Prostate Gland (DSC: 0.87) | 2018 | [141] |
| T2W MRI (2D) | CNN | 2D | 20 | Whole Prostate Gland (DSC: 0.85) | 2018 | [30] |
| T2W MRI (2D) | CNN (PSNet) | 3D | 112/28 five-fold cross validation | Whole Prostate Gland (DSC: 0.85) | 2018 | [29] |
| T2W (2D), DW (2D) MRI | Deep dense multi-path CNN | 3D | 100/50 (+30 external test patients) | Whole Prostate Gland (DSC: 0.95) | 2018 | [142] |
| T2W MRI (2D) | U-Net | 3D | 26 | Whole Prostate Gland (DSC: 0.88) | 2018 | [143] |
| T2W MRI (2D) | Deeply-supervised CNN | 2D | 77/4 | Whole Prostate Gland (DSC: 0.89) | 2017 | [144] |
| T2W (2D), DW (2D) MRI | Auto-Encoder | 2D | 21 leave-one-out cross validation | Prostate Tumor (section-based evaluation (SBE): 0.89, sensitivity: 91%, specificity: 88%) | 2017 | [145] |
| T2W MRI (2D) | Holistically-nested FCN | 2D | 250 five-fold cross validation | Whole Prostate Gland (DSC: 0.89, IoU: 0.81) | 2017 | [146] |
| DW MRI (2D) | Modified U-Net with inception blocks | 2D | 141 four-fold cross validation | Whole Prostate Gland (DSC: 0.93), Transition Zone (DSC: 0.88) | 2017 | [147] |
| T2W MRI (2D) | ConvNet with mixed residual connections | 3D | 50/30 | Whole Prostate Gland (DSC: 0.87) | 2017 | [148] |
| T2W MRI (2D) | Stacked Sparse AE (SSAE) + Sparse patch matching | 2D | 66 two-fold cross validation | Whole Prostate Gland (DSC: 0.87) | 2016 | [149] |
| T2W MRI (2D) | V-Net | 3D | 50/30 | Whole Prostate Gland (DSC: 0.87) | 2016 | [79] |
| T2W MRI (unspecified) | Stacked independent subspace analysis (ISA) | 2D | 30 leave-one-out cross validation | Whole Prostate Gland (DSC: 0.86) | 2013 | [150] |
| Rectal Cancer | ||||||
| CT | DeepLabV3+ | 2D | 98/63 | CTV (DSC: 0.88), Bladder (DSC: 0.90), Small Intestine (DSC: 0.76), L Fem. Head (DSC: 0.93), R Fem. Head (DSC: 0.93) | 2020 | [32] |
| CT/ T2W MRI (2D) | CNN with cascaded atrous convolution (CAC) and spatial pyramid pooling module (SPP) | 2D | 100/70 five-fold cross validation | Rectal Tumor (DSC: 0.78) CTV (DSC: 0.85) | 2018 | [151] |
| CT | Dilated CNN (transfer learning from VGG-16) | 2D | 218/60 | CTV (DSC: 0.87), Bladder (DSC: 0.93), L Fem. Head (DSC: 0.92), R Fem. Head (DSC: 0.92), Intestine (DSC: 0.65), Colon (DSC: 0.62) | 2017 | [152] |
| T2W (2D), DW (2D) MRI | Mask R-CNN | 2D | 293/31 (+50 external test patients) | Lymph Nodes (DSC: 0.81) | 2020 | [153] |
| T2W MRI (2D) | CNN (transfer learning from ResNet50) | 2D | 461/107 | Rectal Tumor (DSC: 0.82) | 2019 | [154] |
| T2W MRI (3D) | U-Net | 2D | 93 ten-foldcross validation | Rectal GTV (DSC: 0.74, IoU: 0.60) | 2018 | [155] |
| T2W MRI (2D) | FCN (transfer learning from VGG-16) | 2D | 410/102 | Rectal Tumor (DSC: 0.84) | 2018 | [28] |
| T2W MRI (2D) | Hybrid loss FCN (HL-FCN) | 3D | 64 four-fold cross validation | Rectal Tumor (DSC: 0.72) | 2018 | [156] |
| T2W (unspecified), DW (2D) MRI | CNN | 2D | 70/70 | Rectal Tumor (DSC: 0.69) | 2017 | [157] |
| Dataset | Image Modality (MRI Acquisition Mode) | Number of Patients | Ground-Truth Contours | URL | Studies |
|---|---|---|---|---|---|
| PROMISE12 [163] | T2W MRI (2D) | 80 | Whole Prostate Gland | https://promise12.grand-challenge.org/ [Accessed 21 October 2021] | [29,79,114,116,118,119,123,124,128,130,131,132,133,136,141,142,143,147,148] |
| I2CVB [167] | T2W (2D/3D), | 40 | Whole Prostate Gland, Peripheral Zone, Central Gland, Prostate Tumor | https://i2cvb.github.io/ [Accessed 21 October 2021] | [115,125,134,138,140,168] |
| DW (2D), | |||||
| DCE (3D), | |||||
| MRSI (3D) MRI | |||||
| BWH [169] | T1W (2D/3D), | 230 | Whole Prostate Gland | https://prostatemrimagedatabase.com/ [Accessed 21 October 2021] | [118,131] |
| T2W (2D) MRI | |||||
| ASPS13 [164] | T1W (2D), | 156 | Whole Prostate Gland, Peripheral Zone | https://wiki.cancerimagingarchive.net/display/Public/NCI-ISBI+2013+Challenge+-+Automated+Segmentation+of+Prostate+Structures [Accessed 21 October 2021] | [29,114,123,124] |
| T2W (2D), | |||||
| DCE (3D) MRI | |||||
| PROSTATEx [165] | T2W (2D), | 330 (malignant lesions: 76, benign lesions: 245) | Prostate Tumor | https://prostatex.grand-challenge.org/ [Accessed 21 October 2021] | [120,125,127,129] |
| DW (2D), | |||||
| PDW (3D), | |||||
| DCE (3D) MRI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalantar, R.; Lin, G.; Winfield, J.M.; Messiou, C.; Lalondrelle, S.; Blackledge, M.D.; Koh, D.-M. Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges. Diagnostics 2021, 11, 1964. https://doi.org/10.3390/diagnostics11111964
Kalantar R, Lin G, Winfield JM, Messiou C, Lalondrelle S, Blackledge MD, Koh D-M. Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges. Diagnostics. 2021; 11(11):1964. https://doi.org/10.3390/diagnostics11111964
Chicago/Turabian StyleKalantar, Reza, Gigin Lin, Jessica M. Winfield, Christina Messiou, Susan Lalondrelle, Matthew D. Blackledge, and Dow-Mu Koh. 2021. "Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges" Diagnostics 11, no. 11: 1964. https://doi.org/10.3390/diagnostics11111964
APA StyleKalantar, R., Lin, G., Winfield, J. M., Messiou, C., Lalondrelle, S., Blackledge, M. D., & Koh, D.-M. (2021). Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges. Diagnostics, 11(11), 1964. https://doi.org/10.3390/diagnostics11111964







