Analysis of Brain MRI Images Using Improved CornerNet Approach
Abstract
:1. Introduction
- I.
- Proposed an improved CornerNet approach with DenseNet-41 for keypoints extraction, which enhanced the brain tumor classification accuracy while reducing both the training and testing time complexity.
- II.
- Precise detection of the cancerous region of the human brain because of the robustness of the CornerNet framework.
- III.
- Proposed low-cost solution to brain tumor classification as CornerNet uses a one-stage object identification framework.
- IV.
- Rigorous evaluation has been conducted in comparison to other state-of-the-art brain tumor detection approaches over standard databases comprising diverse images with several distortions, i.e., noise, blurriness, color, light changes, angle, size, and location variations, to demonstrate the efficacy of the presented approach.
2. Literature Review
3. Proposed Methodology
3.1. Annotations
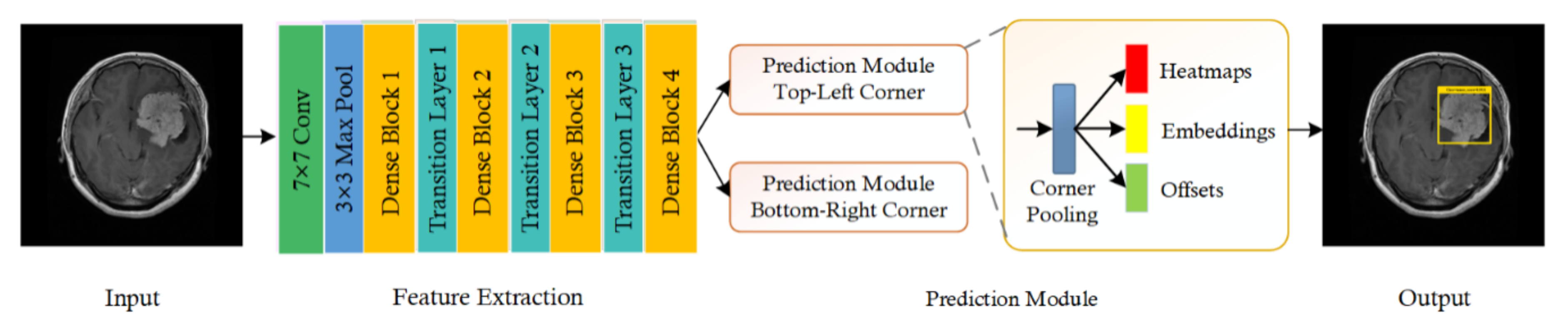
3.2. CornerNet Model
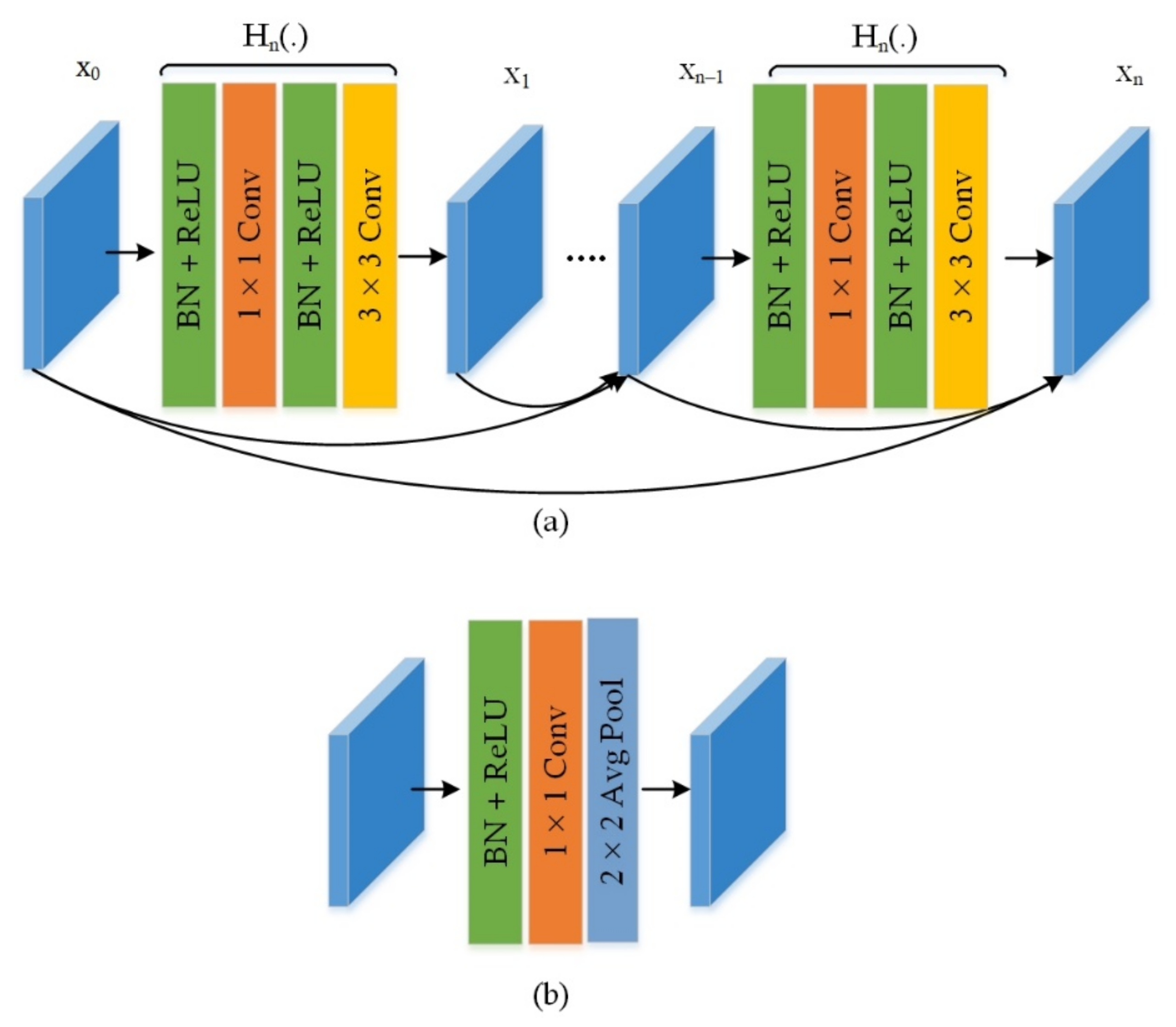
3.3. Feature Extraction Using Customized Backbone Network
4. Results
4.1. Dataset
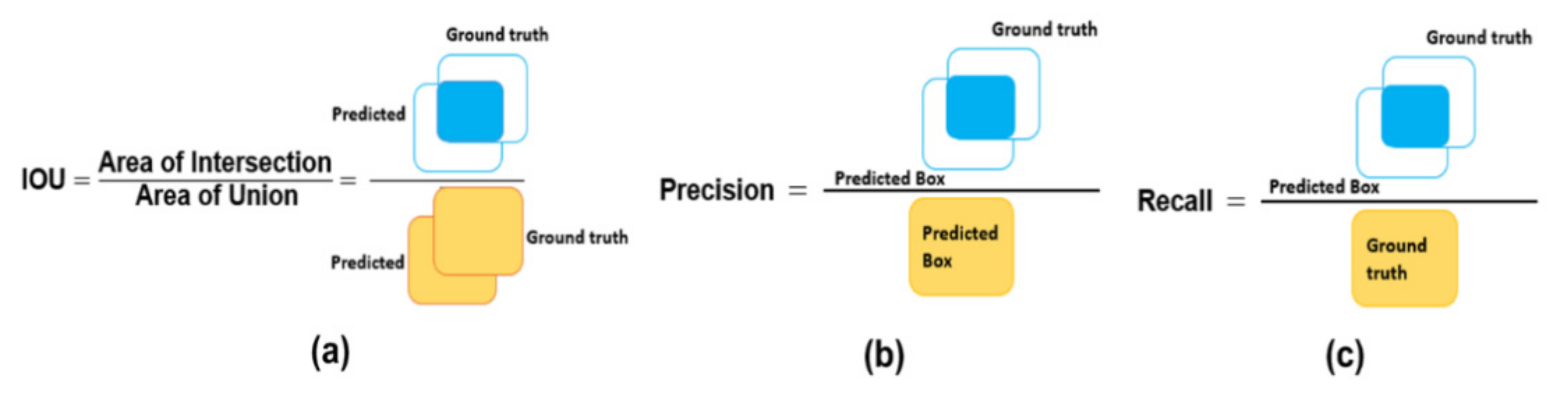
4.2. Evaluation Metrics
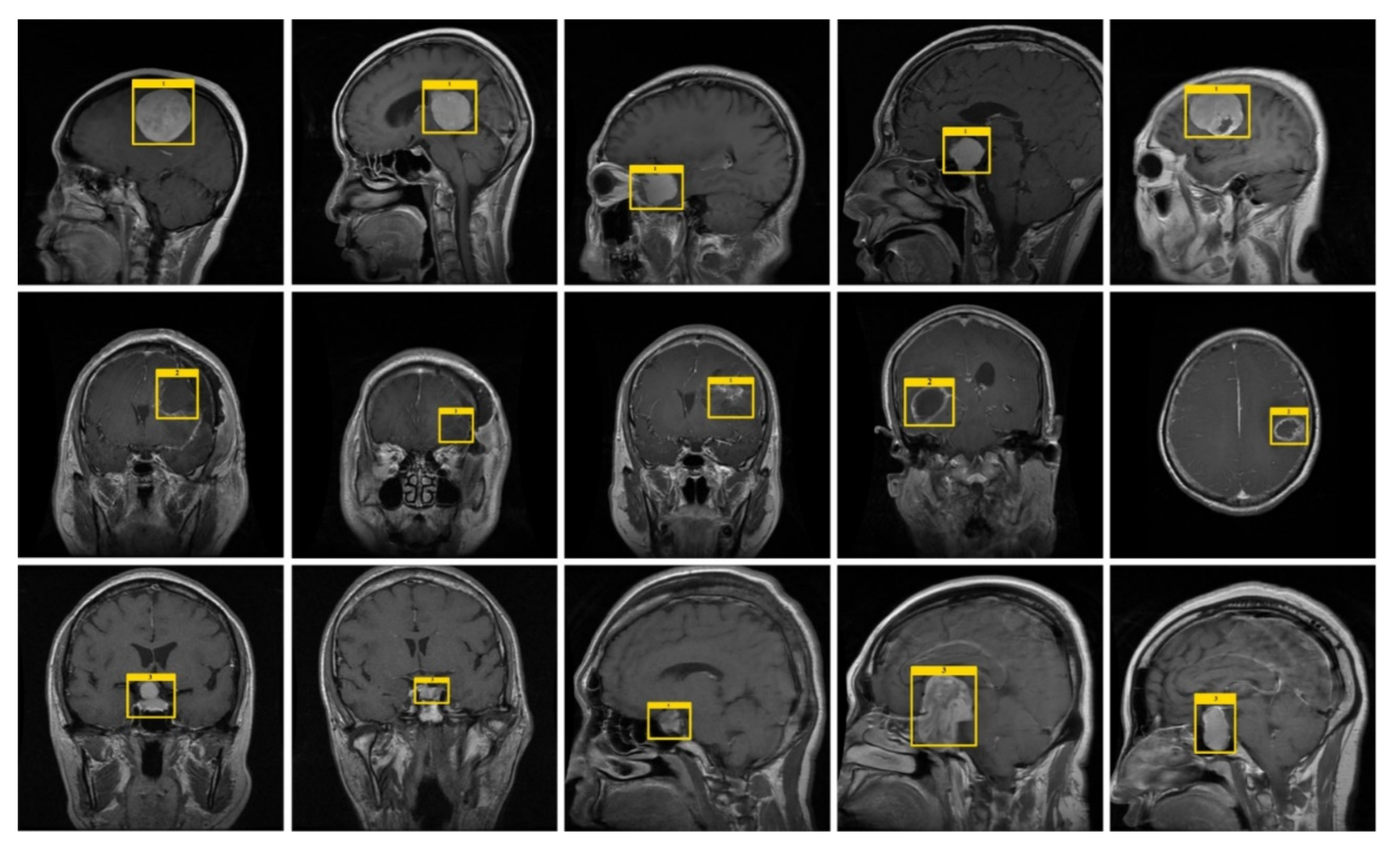
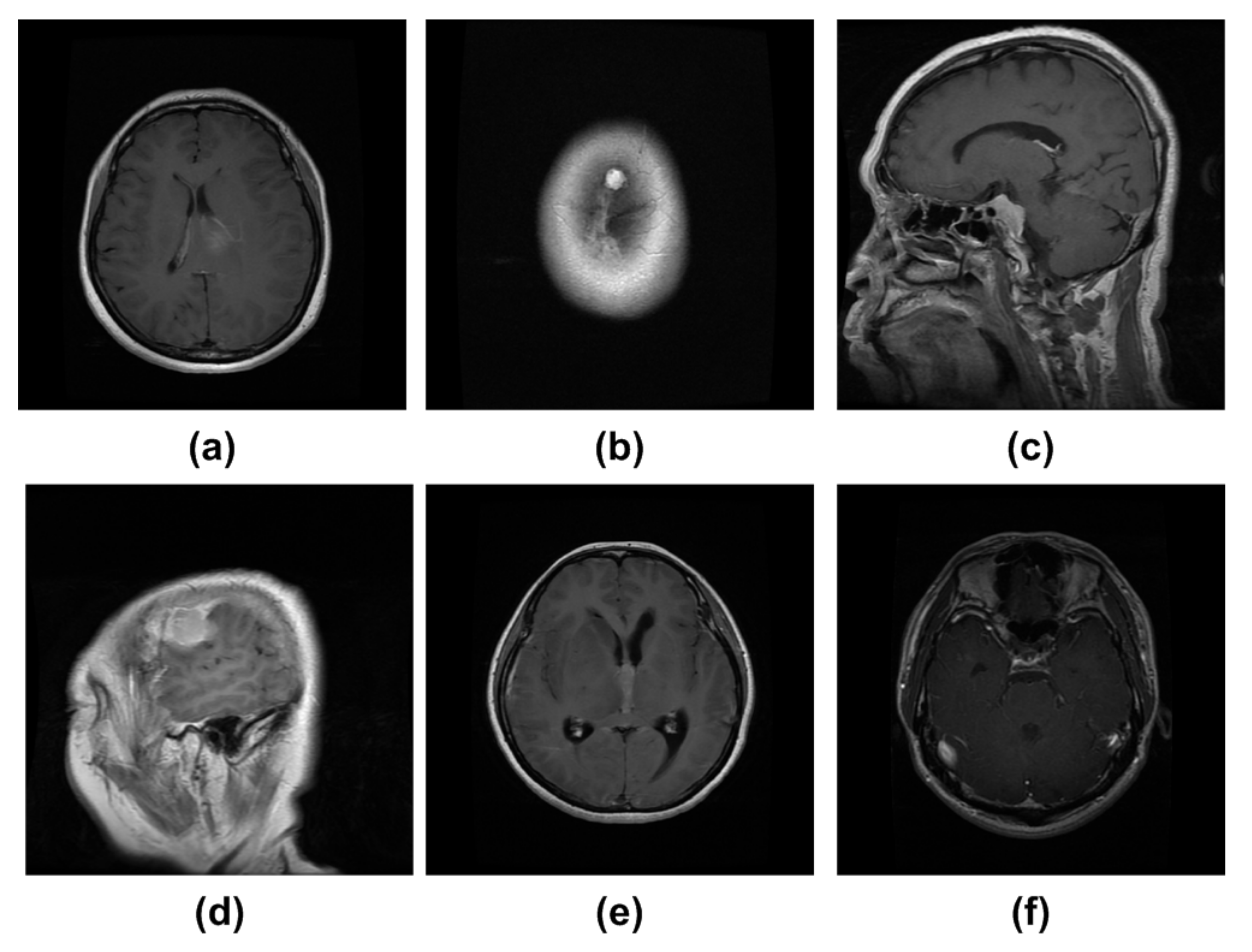
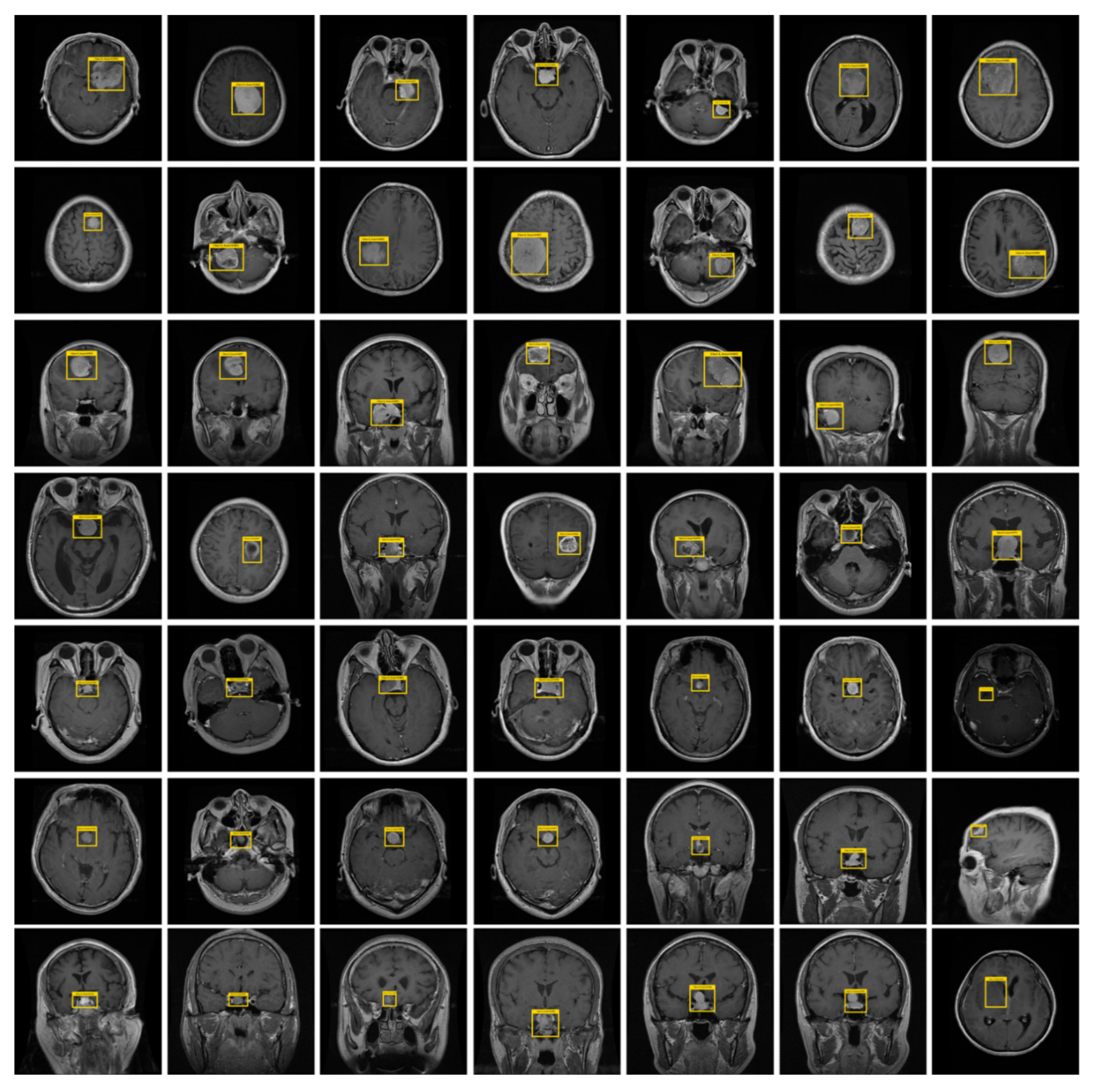
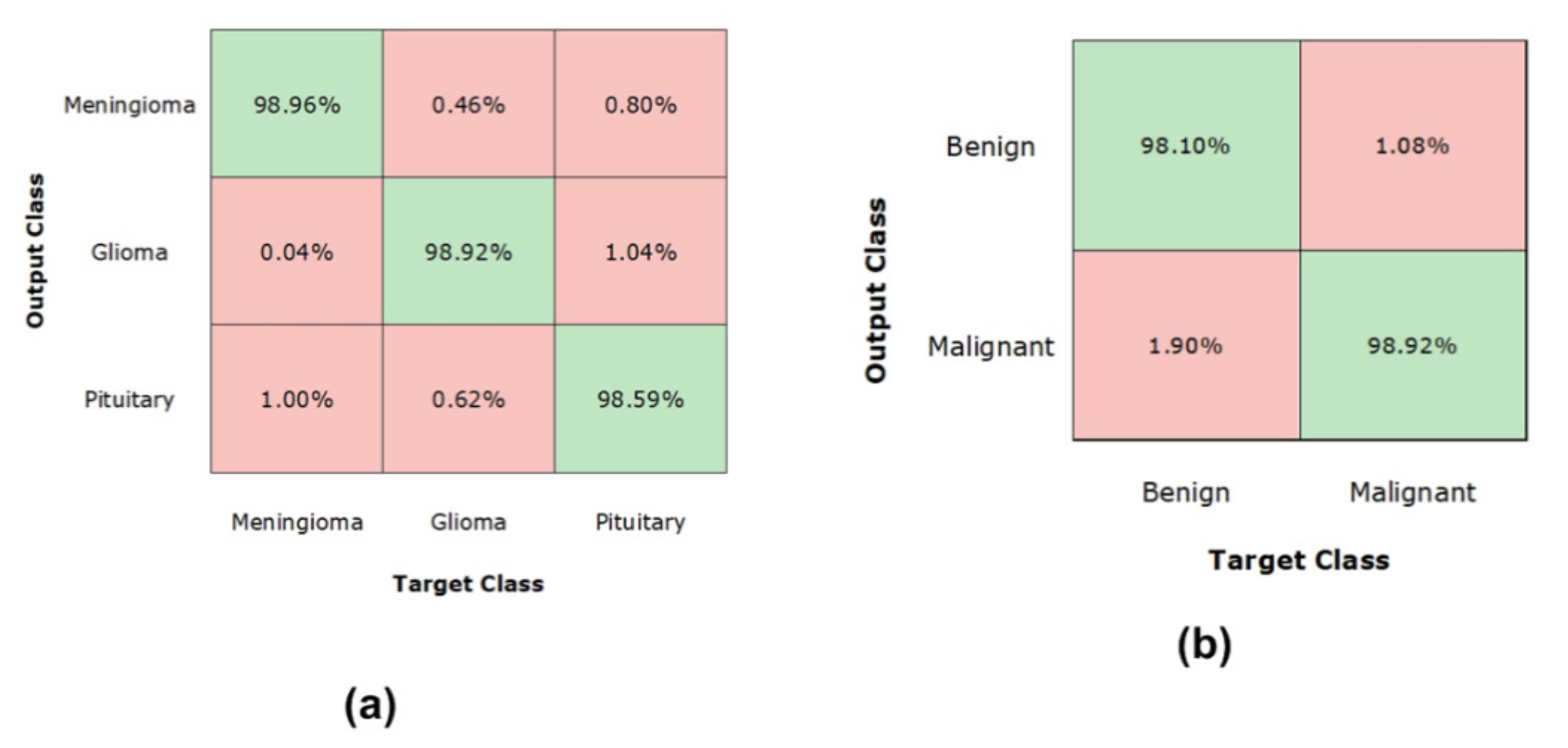
4.3. Experimental Results and Discussion
4.4. Evaluation of DenseNet
4.5. Comparison with Other Object Detection Methods
4.6. Comparison with Other ML-Based Classifiers
4.7. Comparison with the State-of-the-Art Techniques
4.8. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, M.A.; Lali, I.U.; Rehman, A.; Ishaq, M.; Sharif, M.; Saba, T.; Zahoor, S.; Akram, T. Brain tumor detection and classification: A framework of marker-based watershed algorithm and multilevel priority features selection. Microsc. Res. Tech. 2019, 82, 909–922. [Google Scholar] [CrossRef]
- Nazar, U.; Khan, M.A.; Lali, I.U.; Lin, H.; Ali, H.; Ashraf, I.; Tariq, J. Review of automated computerized methods for brain tumor segmentation and classification. Curr. Med. Imaging 2020, 16, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recognit. Lett. 2020, 129, 181–189. [Google Scholar] [CrossRef]
- Akil, M.; Saouli, R.; Kachouri, R. Fully automatic brain tumor segmentation with deep learning-based selective attention using overlapping patches and multi-class weighted cross-entropy. Med Image Anal. 2020, 63, 101692. [Google Scholar]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Hussain, U.N.; Khan, M.A.; Lali, I.U.; Javed, K.; Ashraf, I.; Tariq, J.; Ali, H.; Din, A. A unified design of ACO and skewness based brain tumor segmentation and classification from MRI scans. J. Control. Eng. Appl. Inform. 2020, 22, 43–55. [Google Scholar]
- Sharif, M.I.; Khan, M.A.; Alhussein, M.; Aurangzeb, K.; Raza, M.A. A decision support system for multimodal brain tumor classification using deep learning. Complex Intell. Syst. 2021, 1–14. [Google Scholar] [CrossRef]
- Khan, M.A.; Arshad, H.; Nisar, W.; Javed, M.Y.; Sharif, M. An Integrated Design of Fuzzy C-Means and NCA-Based Multi-properties Feature Reduction for Brain Tumor Recognition. In Signal and Image Processing Techniques for the Development of Intelligent Healthcare Systems; Springer: Berlin, Germany, 2021; pp. 1–28. [Google Scholar]
- Coburger, J.; Merkel, A.; Scherer, M.; Schwartz, F.; Gessler, F.; Roder, C.; Pala, A.; König, R.; Bullinger, L.; Nagel, G.; et al. Low-grade glioma surgery in intraoperative magnetic resonance imaging: Results of a multicenter retrospective assessment of the German Study Group for Intraoperative Magnetic Resonance Imaging. Neurosurgery 2016, 78, 775–786. [Google Scholar] [CrossRef] [Green Version]
- Aziz, A.; Attique, M.; Tariq, U.; Nam, Y.; Nazir, M.; Jeong, C.W.; Sakr, R.H. An Ensemble of Optimal Deep Learning Features for brain tumor classification. Comput. Mater. Contin. 2021, 69, 1–15. [Google Scholar]
- Miner, R.C. Image-guided neurosurgery. J. Med Imaging Radiat. Sci. 2017, 48, 328–335. [Google Scholar] [CrossRef] [Green Version]
- Tahir, A.B.; Khan, M.A.; Alhaisoni, M.; Khan, J.A.; Nam, Y.; Wang, S.H.; Javed, K. Deep Learning and Improved Particle Swarm Optimization Based Multimodal Brain Tumor Classification. CMC Comput. Mater. Contin. 2021, 68, 1099–1116. [Google Scholar]
- Nadeem, M.W.; Ghamdi, M.A.; Hussain, M.; Khan, M.A.; Khan, K.M.; Almotiri, S.H.; Butt, S.A. Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges. Brain Sci. 2020, 10, 118. [Google Scholar] [CrossRef] [Green Version]
- Manic, K.S.; Biju, R.; Patel, W.; Khan, M.A.; Raja, N.; Uma, S. Extraction and Evaluation of Corpus Callosum from 2D Brain MRI Slice: A Study with Cuckoo Search Algorithm. Comput. Math. Methods Med. 2021, 2021, 5524637. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Wiest, R.; Nolte, L.P.; Reyes, M. A survey of MRI-based medical image analysis for brain tumor studies. Phys. Med. Biol. 2013, 58, R97–R129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Muhammad, K.; Sharif, M.; Akram, T.; de Albuquerque, V.H. Multi-Class Skin Lesion Detection and Classification via Teledermatology. IEEE J. Biomed. Health Inform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Myronenko, A. 3D MRI brain tumor segmentation using autoencoder regularization. Int. MICCAI Brainlesion Workshop 2018, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Goetz, M.; Weber, C.; Bloecher, J.; Stieltjes, B.; Meinzer, H.P.; Maier-Hein, K. Extremely randomized trees based brain tumor segmentation. Proc. BRATS Chall. MICCAI 2014, 4, 6–11. [Google Scholar]
- Reza, S.; Iftekharuddin, K. Improved brain tumor tissue segmentation using texture features. Proc. MICCAI BraTS 2014, 9035, 27–30. [Google Scholar]
- Khan, M.A.; Sharif, M.; Akram, T.; Raza, M.; Saba, T.; Rehman, A. Hand-crafted and deep convolutional neural network features fusion and selection strategy: An application to intelligent human action recognition. Appl. Soft Comput. 2020, 87, 105986. [Google Scholar] [CrossRef]
- Kleesiek, J.; Biller, A.; Urban, G.; Kothe, U.; Bendszus, M.; Hamprecht, F. Ilastik for multi-modal brain tumor segmentation. Proc. MICCAI 2014, 4, 12–17. [Google Scholar]
- Bauer, S.; Nolte, L.-P.; Reyes, M. Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Heisenberg, Germany, 18–22 September 2011; pp. 354–361. [Google Scholar]
- Zikic, D.; Glocker, B.; Konukoglu, E.; Criminisi, A.; Demiralp, C.; Shotton, J.; Thomas, O.M.; Das, T.; Jena, R.; Price, S.J. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Berlin, Germany, 1–5 October 2012; pp. 369–376. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Tustison, N.J.; Shrinidhi, K.L.; Wintermark, M.; Durst, C.R.; Kandel, B.M.; Gee, J.C.; Grossman, M.C.; Avants, B.B. Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics 2015, 13, 209–225. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef]
- Hu, K.; Gan, Q.; Zhang, Y.; Deng, S.; Xiao, F.; Huang, W.; Cao, C.; Gao, X. Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 2019, 7, 92615–92629. [Google Scholar] [CrossRef]
- Pinto, A.; Pereira, S.; Correia, H.; Oliveira, J.; Rasteiro, D.M.; Silva, C.A. Brain tumour segmentation based on extremely randomized forest with high-level features. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; Volume 2015, pp. 3037–3040. [Google Scholar]
- Doyle, S.; Vasseur, F.; Dojat, M.; Forbes, F. Fully automatic brain tumor segmentation from multiple MR sequences using hidden Markov fields and variational EM. Procs. NCI MICCAI BraTS 2013, 1, 18–22. [Google Scholar]
- Prastawa, M.; Bullitt, E.; Ho, S.; Gerig, G. A brain tumor segmentation framework based on outlier detection. Med Image Anal. 2004, 8, 275–283. [Google Scholar] [CrossRef]
- Prastawa, M.; Bullitt, E.; Ho, S.; Gerig, G. Robust estimation for brain tumor segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Montreal, QC, Canada, 15–18 November 2003. [Google Scholar]
- Khotanlou, H.; Colliot, O.; Atif, J.; Bloch, I. 3D brain tumor segmentation in MRI using fuzzy classification, symmetry analysis and spatially constrained deformable models. Fuzzy Sets Syst. 2009, 160, 1457–1473. [Google Scholar] [CrossRef] [Green Version]
- Popuri, K.; Cobzas, D.; Murtha, A.; Jägersand, M. 3D variational brain tumor segmentation using Dirichlet priors on a clustered feature set. Int. J. Comput. Assist. Radiol. Surg. 2012, 7, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Hamamci, A.; Kucuk, N.; Karaman, K.; Engin, K.; Unal, G. Tumor-cut: Segmentation of brain tumors on contrast enhanced MR images for radiosurgery applications. IEEE Trans. Med Imaging 2011, 31, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Subbanna, N.; Precup, D.; Arbel, T. Iterative multilevel MRF leveraging context and voxel information for brain tumour segmentation in MRI. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–38 June 2014; pp. 400–405. [Google Scholar]
- Chandra, S.K.; Bajpai, M.K. Brain tumor detection and segmentation using mesh-free super-diffusive model. Multimed. Tools Appl. 2020, 79, 2653–2670. [Google Scholar] [CrossRef]
- Chandra, S.K.; Bajpai, M.K. Efficient three-dimensional super-diffusive model for benign brain tumor segmentation. Eur. Phys. J. Plus 2020, 135, 419. [Google Scholar] [CrossRef]
- Hussain, C.A.; Gopi, C.; Kishore, D.S.; Reddy, G.G.; Sai, G.C. Brain tumor detection and segmentation using anisotropic filtering for MRI images. J. Eng. Sci. 2020, 11, 1–10. [Google Scholar]
- Nandi, A. Detection of human brain tumour using MRI image segmentation and morphological operators. In Proceedings of the 2015 IEEE International Conference on Computer Graphics, Vision and Information Security (CGVIS), Bhubaneswar, India, 2–3 November 2015; pp. 55–60. [Google Scholar]
- Rajan, P.G.; Sundar, C. Brain tumor detection and segmentation by intensity adjustment. J. Med Syst. 2019, 43, 282. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.; Tanvir, U.; Munir, E.U.; Khan, M.A.; Yasmin, M. Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J. Ambient. Intell. Humaniz. Comput. 2018, 1–20. [Google Scholar] [CrossRef]
- Kaya, I.E.; Pehlivanlı, A.Ç.; Sekizkardeş, E.G.; Ibrikci, T. PCA based clustering for brain tumor segmentation of T1w MRI images. Comput. Methods Programs Biomed. 2017, 140, 19–28. [Google Scholar] [CrossRef]
- Iqbal, S.; Ghani Khan, M.U.; Saba, T.; Mehmood, Z.; Javaid, N.; Rehman, A.; Abbasi, R. Deep learning model integrating features and novel classifiers fusion for brain tumor segmentation. Microsc. Res. Tech. 2019, 82, 1302–1315. [Google Scholar] [CrossRef]
- Iqbal, S.; Ghani, M.U.; Saba, T.; Rehman, A. Brain tumor segmentation in multi-spectral MRI using convolutional neural networks (CNN). Microsc. Res. Tech. 2018, 81, 419–427. [Google Scholar] [CrossRef]
- Qasem, S.N.; Nazar, A.; Qamar, S.A. A Learning Based Brain Tumor Detection System. Comput. Mater. Contin. 2019, 59, 713–727. [Google Scholar] [CrossRef]
- Naz, A.R.; Naseem, U.; Razzak, I.; Hameed, I.A. Deep autoencoder-decoder framework for semantic segmentation of brain tumor. Aust. J. Intell. Inf. Process. Syst. 2019, 15, 53–60. [Google Scholar]
- Sobhaninia, Z.; Rezaei, S.; Noroozi, A.; Ahmadi, M.; Zarrabi, H.; Karimi, N.; Emami, A.; Samavi, S. Brain tumor segmentation using deep learning by type specific sorting of images. arXiv 2018, arXiv:1809.077862017. [Google Scholar]
- Rayhan, F. Fr-mrinet: A deep convolutional encoder-decoder for brain tumor segmentation with relu-RGB and sliding-window. Int. J. Comput. Appl. 2018, 975, 8887. [Google Scholar]
- Saba, T.; Mohamed, A.S.; El-Affendi, M.; Amin, J.; Sharif, M. Brain tumor detection using fusion of hand crafted and deep learning features. Cogn. Syst. Res. 2020, 59, 221–230. [Google Scholar] [CrossRef]
- Alaraimi, S.; Okedu, K.E.; Tianfield, H.; Holden, R.; Uthmani, O. Transfer learning networks with skip connections for classification of brain tumors. Int. J. Imaging Syst. Technol. 2021, 31, 1564–1582. [Google Scholar] [CrossRef]
- Law, H.; Deng, J. CornerNet: Detecting Objects as Paired Keypoints. Int. J. Comput. Vis. 2019, 128, 642–656. [Google Scholar] [CrossRef] [Green Version]
- Lin, T. Labelimg. 2020. Available online: https://github.com/tzutalin/labelImg/blob/master/README (accessed on 8 April 2021).
- Girshick, R. Fast r-cnn. In Proceedings of the 2015 IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv 2015, arXiv:1506.01497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.E.; Fu, C.; Berg, A.C. SSD: Single Shot MultiBox Detector. Comput. Vis. ECCV 2016, 9905, 21–37. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2016, arXiv:1804.02767. [Google Scholar]
- Girshick, R.B.; Donahue, J.; Darrell, T.; Malik, J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 142–158. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2017, arXiv:1608.06993. [Google Scholar]
- Albahli, S.; Nawaz, M.; Javed, A.; Irtaza, A. An improved faster-RCNN model for handwritten character recognition. Arab. J. Sci. Eng. 2021, 46, 8509–8523. [Google Scholar] [CrossRef]
- Albahli, S.; Nazir, T.; Irtaza, A.; Javed, A. Recognition and Detection of Diabetic Retinopathy Using Densenet-65 Based Faster-RCNN. Comput. Mater. Contin. 2021, 67, 1333–1351. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Javed, M.Y.; Muhammad, N.; Yasmin, M. An implementation of optimized framework for action classification using multilayers neural network on selected fused features. Pattern Anal. Appl. 2019, 22, 1377–1397. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, M.A.; Ahmed, F.; Mittal, M.; Goyal, L.M.; Hemanth, D.J.; Satapathy, S.C. Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recognit. Lett. 2020, 131, 193–204. [Google Scholar] [CrossRef]
- Pathak, D.; Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional multi-class multiple instance learning. arXiv 2014, arXiv:1412.7144. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wu, L.; Wang, Y.; Shao, L.; Wang, M. 3-D Person VLAD: Learning Deep. Computing 2014, 32, 270–286. [Google Scholar]
- Islam, J.; Zhang, Y. An ensemble of deep convolutional neural networks for Alzheimer’s disease detection and classification. arXiv 2017, arXiv:1712.01675. [Google Scholar]
- Chelghoum, R.; Ikhlef, A.; Hameurlaine, A.; Jacquir, S. Transfer learning using convolutional neural network architectures for brain tumor classification from MRI images. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, Neos Marmaras, Greece, 5–7 June 2020. [Google Scholar]
- Polat, Ö.; Güngen, C. Classification of brain tumors from MR images using deep transfer learning. J. Supercomput. 2021, 77, 7236–7252. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Masood, M.; Nazir, T.; Nawaz, M.; Mehmood, A.; Rashid, J.; Kwon, H.Y.; Mahmood, T.; Hussain, A. A Novel Deep Learning Method for Recognition and Classification of Brain Tumors from MRI Images. Diagnostics 2021, 11, 744. [Google Scholar] [CrossRef]
- Shafiee, M.J.; Chywl, B.; Li, F.; Wong, A. Fast YOLO: A fast you only look once system for real-time embedded object detection in video. arXiv 2017, arXiv:1709.05943. [Google Scholar]
- Pashaei, A.; Sajedi, H.; Jazayeri, N. Brain tumor classification via convolutional neural network and extreme learning machines. In Proceedings of the 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 25–26 October 2018; IEEE: Piscataway Township, NJ, USA, 2018. [Google Scholar]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Anaraki, A.K.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Shaik, N.S.; Naralasetti, V.; Mundukur, N.B. Joint training of two-channel deep neural network for brain tumor classification. Signal Image Video Process. 2021, 15, 753–760. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. Tumor type detection in brain MR images of the deep model developed using hypercolumn technique, attention modules, and residual blocks. Med Biol. Eng. Comput. 2021, 59, 57–70. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumour classification using siamese neural network and neighbourhood analysis in embedded feature space. Int. J. Imaging Syst. Technol. 2021, 31, 1655–1669. [Google Scholar] [CrossRef]

| References | Method | Limitation |
|---|---|---|
| GM-based | ||
| [32] | A posterior probability-based technique to identify the matching cases from history. | The work is not robust to the detection of varying shapes of brain tumors. |
| [34] | An atlas-based method was used to locate the presence of the brain tumor. | Needs the expertise of trained human experts. |
| [35] | An atlas-based approach together with brain symmetry was employed for detecting cancerous cells. | Performance degrades for samples having less texture information. |
| ML-based | ||
| [39] | A gradient computation-based approach to localize the tumorous cells. | It may not perform well over large intensity changes within MRI images. |
| [41] | The AF method along with the adjustment-based segmentation technique was employed to identify the brain tumor. | Unable to detect a tumor of small size. |
| [43] | The K-Mean and Fuzzy C-Means clustering along with the co-occurrence matrix was employed for feature computation, while the SVM classifier was used for brain tumor classification. | The technique may not perform well over the samples with huge light changes. |
| [44] | The GA algorithm along with the SVM classifier was employed to detect the brain tumor from the MRI images. | This method may not accurately detect the tumors along the boundaries of images. |
| [45] | The PCA technique along with FCM and K-means clustering was used for locating the cancerous tissues of the human brain. | The approach requires huge training data. |
| DL-based | ||
| [29] | The MCCNN framework along with the CRFs was used for brain tumor detection. | The method is economically inefficient. |
| [46] | An approach merging both combining both the CoveNet and the LSTM framework was introduced to identify the brain regions containing the tumor. | The approach is suffering from high computational complexity. |
| [47] | Three DL frameworks named Interpolated Network, SkipNet, and SE-Net were applied for brain tumor segmentation. | The work may not be generalized well to real-world scenarios. |
| [19] | A watershed segmentation technique together with the KNN for brain tumor detection. | This method is not robust to identify the brain tumor from the MRI samples having organizational complexities. |
| [50] | An encoder–decoder-based method was used for identifying the tumorous cells. | The technique is not robust to brain tumors of small sizes. |
| [51] | A 33-layer deep network was used to locate the cancerous brain cells. | The technique is computationally expensive. |
| [52] | The LBP, HOG descriptors along the VGG-19 framework were used for feature computation. While the SVM, KNN, LDA, LD, and DT were used for classification. | The approach may not perform well over the samples with extensive color changes. |
| Layer | Operator | Stride | |
|---|---|---|---|
| Convolutional Layer | 2 | ||
| Pooling | 2 | ||
| DB1 | 1 | ||
| TL1 | Convolutional Layer | ||
| Pooling Layer | |||
| DB2 | 1 | ||
| TL2 | Convolutional Layer | ||
| Pooling Layer | |||
| DB3 | 1 | ||
| TL3 | Convolutional Layer | ||
| Pooling Layer | |||
| DB4 | 1 | ||
| Classification Layer | FC layer | ||
| Model | No of Parameters (Million) | Accuracy (%) | Execution Time (s) |
|---|---|---|---|
| VGG16 | 119.6 | 98.06 | 1051 |
| VGG19 | 143.6 | 97.97 | 1312 |
| ResNet50 | 23.6 | 96.67 | 1583 |
| DenseNet121 | 7.1 | 98.15 | 2165 |
| Proposed | 6.1 | 98.7 | 1022 |
| Method | Evaluation Parameters | |||
|---|---|---|---|---|
| Accuracy | mAP | Sensitivity | Time (s) | |
| Two-Stage Frameworks | ||||
| RCNN | 0.920 | 0.910 | 0.950 | 0.47 |
| Faster RCNN | 0.940 | 0.940 | 0.940 | 0.25 |
| Mask-RCNN | 0.983 | 0.949 | 0.953 | 0.20 |
| One-Stage Frameworks | ||||
| YOLO | 0.873 | 0.830 | 0.808 | 0.25 |
| SSD | 0.893 | 0.851 | 0.824 | 0.23 |
| Proposed | 0.987 | 0.952 | 0.969 | 0.19 |
| Classifier | Accuracy (%) |
|---|---|
| Deep features + KELM [76] | 93.68 |
| Deep features + SVM [77] | 98.00 |
| Deep features + GA [78] | 94.20 |
| Proposed | 98.70 |
| References | Method | Accuracy (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| [74] | Custom Mask-RCNN | 98.34 | - | 95.3 |
| [79] | Two-Channel DNN | 98.04 | - | - |
| [80] | Attention module, Hyper-column technique, and Residual block | 97.69 | 96.24 | 96.22 |
| [81] | SNN + KNN | 92.6 | 95.3 | - |
| Proposed | CornerNet with DenseNet-41 | 98.7 | 97.4 | 96.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, M.; Nazir, T.; Masood, M.; Mehmood, A.; Mahum, R.; Khan, M.A.; Kadry, S.; Thinnukool, O. Analysis of Brain MRI Images Using Improved CornerNet Approach. Diagnostics 2021, 11, 1856. https://doi.org/10.3390/diagnostics11101856
Nawaz M, Nazir T, Masood M, Mehmood A, Mahum R, Khan MA, Kadry S, Thinnukool O. Analysis of Brain MRI Images Using Improved CornerNet Approach. Diagnostics. 2021; 11(10):1856. https://doi.org/10.3390/diagnostics11101856
Chicago/Turabian StyleNawaz, Marriam, Tahira Nazir, Momina Masood, Awais Mehmood, Rabbia Mahum, Muhammad Attique Khan, Seifedine Kadry, and Orawit Thinnukool. 2021. "Analysis of Brain MRI Images Using Improved CornerNet Approach" Diagnostics 11, no. 10: 1856. https://doi.org/10.3390/diagnostics11101856
APA StyleNawaz, M., Nazir, T., Masood, M., Mehmood, A., Mahum, R., Khan, M. A., Kadry, S., & Thinnukool, O. (2021). Analysis of Brain MRI Images Using Improved CornerNet Approach. Diagnostics, 11(10), 1856. https://doi.org/10.3390/diagnostics11101856








