Point of Care Diagnostics in the Age of COVID-19
Abstract
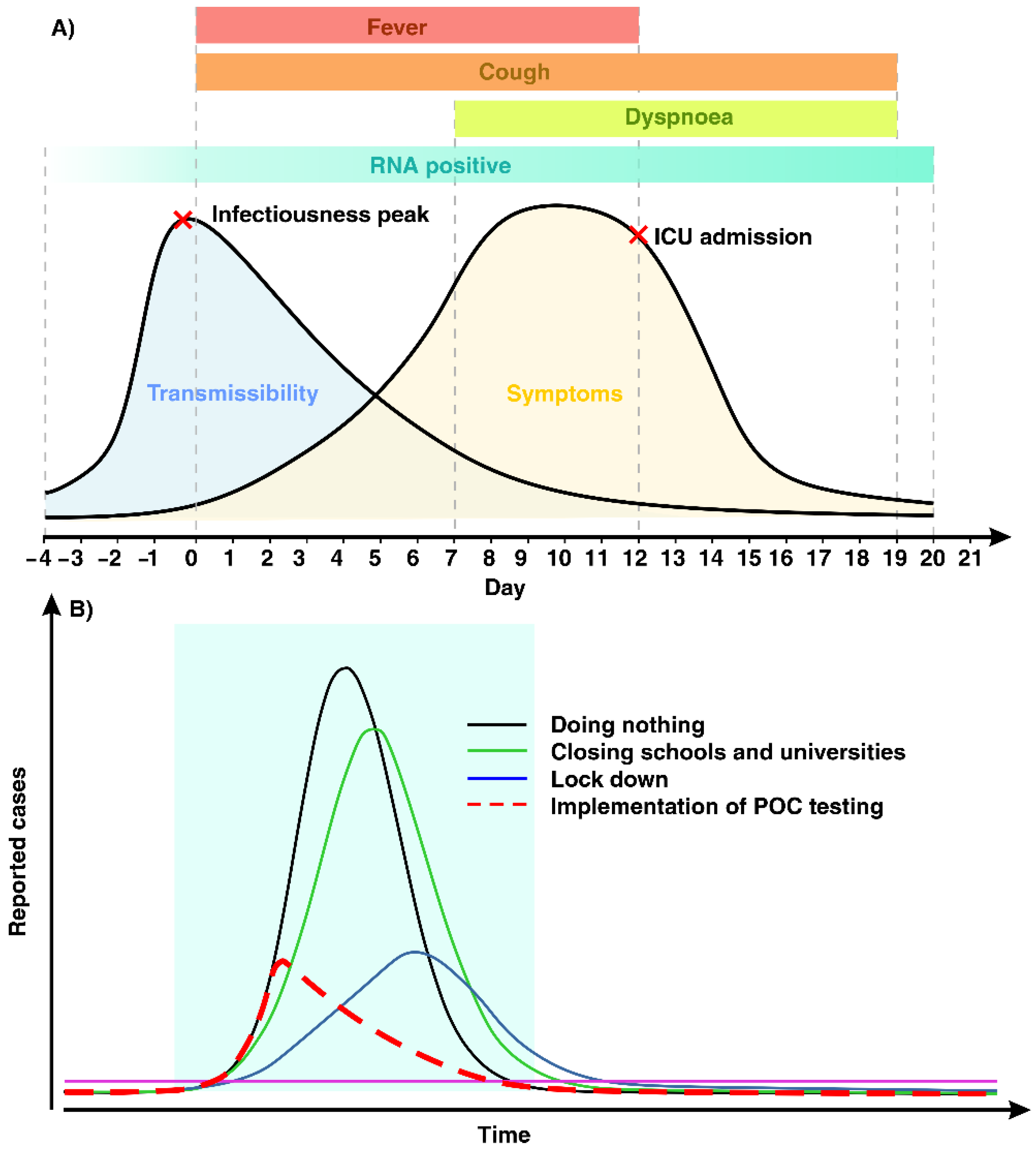
1. Introduction
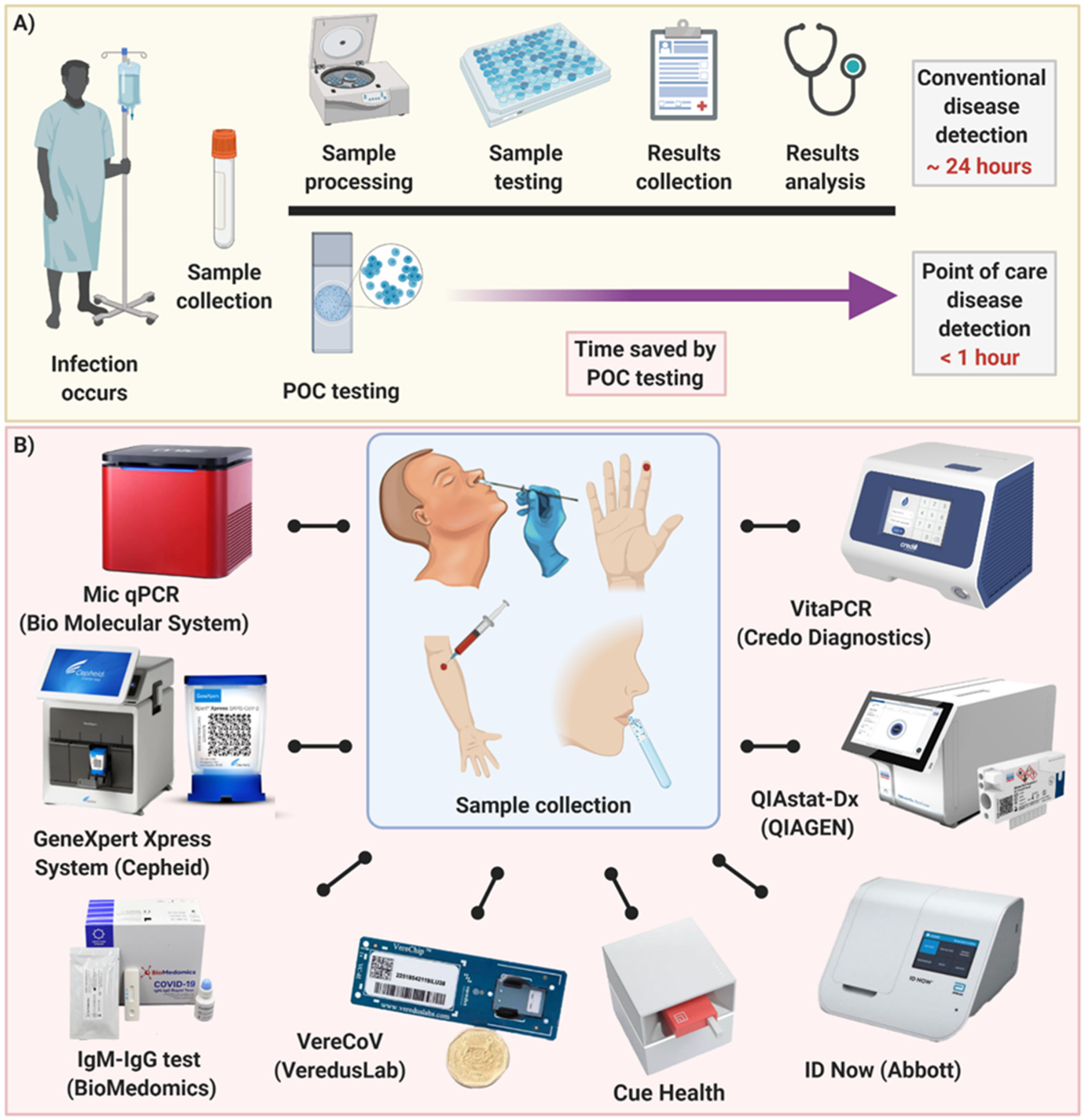
2. COVID-19 Detection at POC Level
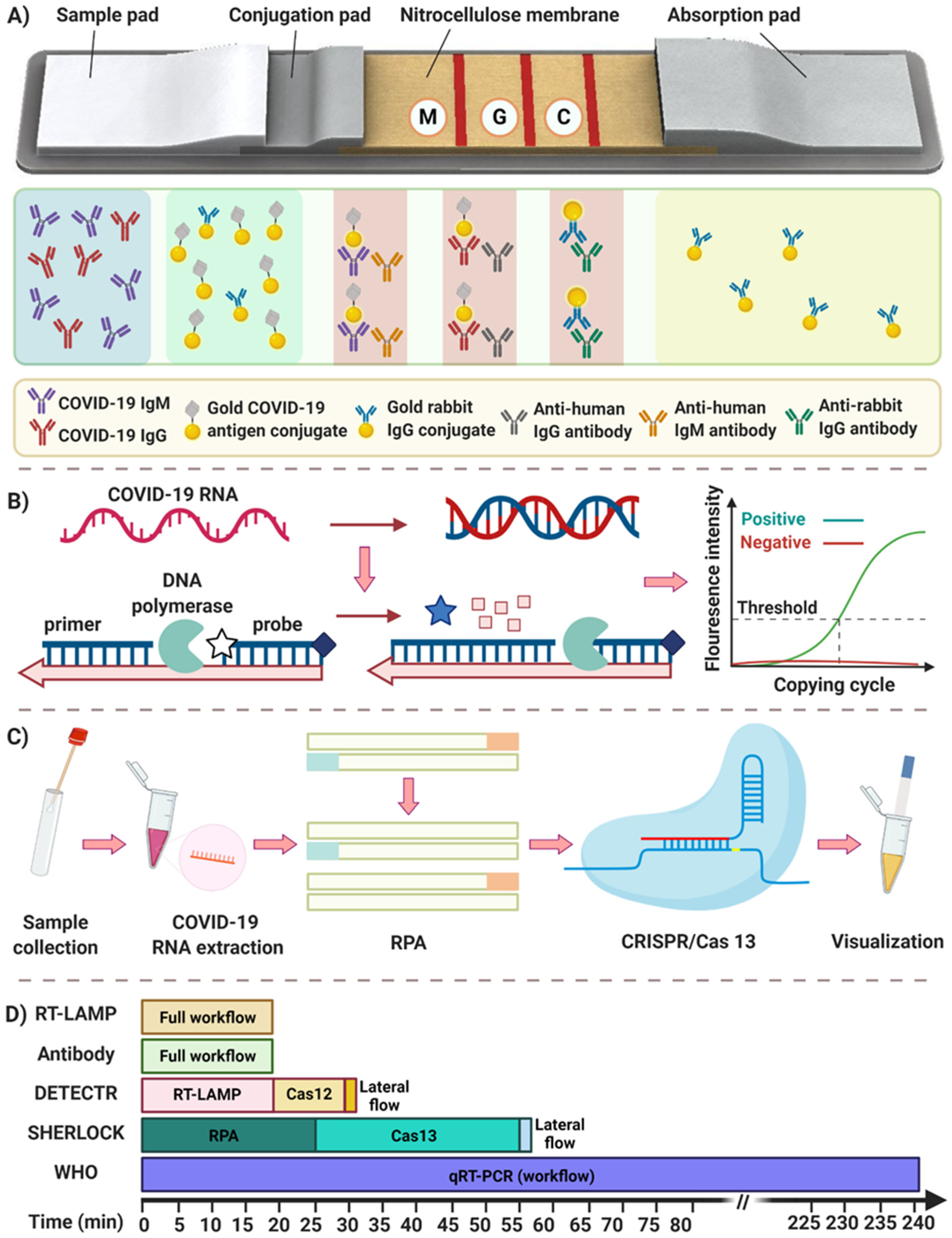
2.1. Immunoassays
2.1.1. Antibody Detection
2.1.2. Antigen Detection
2.2. Nucleic Acid Assays
2.2.1. Rapid PCR-Based Methods
2.2.2. Isothermal Amplification
2.2.3. CRISPR-Cas (12/13) Based Detection Methods
3. Future Direction and Outlook
| Immunoassays (Antibody) | Test/Author | Time (min) | Sensitivity | Specificity | LOD | Sample |
| Li et al. [21] | 15 | 88.66% | 90.63% | - | Whole blood, serum, plasma | |
| Pan et al. [22] | 15 | 92.9% intermediate stage, 96.8% late stage | - | - | Whole blood, serum, plasma | |
| BioMedomics [23] | 10–15 | 100% | ~99% | - | Whole blood, serum, plasma | |
| Pharmact company [24] | 20 | 98.2% | 99.7% | - | Whole blood, serum | |
| Chembio diagnostics [25] | 15–20 | 96% | 98.7 | - | Whole blood, serum, plasma | |
| Immunoassays (Antigen) | CareStart [38] | 10 | 88.4% | 100% | 8 × 102–6.4 × 103 TCID50/mL | Nasopharyngeal |
| Panbio [39] | 10 | 91% | 100% | 2.5 × 101.8 TCID50/mL | Nasopharyngeal | |
| Rapid Response [40] | 15 | 94% | 100% | - | Nasopharyngeal, oropharyngeal | |
| Sofia [43] | 15 | 96% | 100% | - | Nasopharyngeal | |
| Standard Q [41] | 20 | 96% | 99% | - | Nasopharyngeal | |
| Wantai kit [42] | 94% | 98% | 20 pg/mL | Nasopharyngeal, oropharyngeal | ||
| BD Veritor [44] | 15 | 84% | 100% | 1.4 × 102 TCID50/mL | Nasopharyngeal, oropharyngeal | |
| Rapid PCR | Xpert Xpress [61] | 25 | 99.4% | 96.8% | - | Nasopharyngeal swab, nasal swab, and nasal wash/aspirates |
| QIAstat-Dx [63] | 60 | 95% | 100% | 500 copies/mL | nasopharyngeal swabs | |
| NxTAG COV [64,108] | 60 | 97.8% | 100% | - | Nasopharyngeal | |
| VereCoV OneMix [66] | 120 | - | - | 20 copies/mL | Nasopharyngeal | |
| VERI-Q Kit [67] | 55 | - | - | 8.9–9 copies/reaction | nasopharyngeal, oropharyngeal, sputum specimens | |
| Isothermal amplification | Yang et al. [83] | 30 | - | 99% | 1000 copies/mL | Nasopharyngeal |
| El-Tholoth et al. [84] | 50 | 100% | - | 7 copies/reaction | Nasal swab | |
| ID NOW [90] | 13 | 95% | 97.9% | 125 copies/mL | Nasal, Throat, Nasopharyngeal | |
| Cue™ COVID-19 [93] | 25 | 99% | 98% | 20 copies/sample | nasal swab | |
| CRISPR-Cas | DETECTR [104] | 40 | - | - | 10 copies/µL | Nasopharyngeal, oropharyngeal, mid-turbinate nasal swabs, anterior nasal swabs, nasopharyngeal wash/aspirate and nasal aspirate |
| Sherlock [101,109] | 60 | 100% | 100% | 6.75 copies/µL | nasopharyngeal, oropharyngeal, bronchoalveolar lavage | |
| iSCAN [106] | 60 | - | - | 10 copies/reaction | Oropharyngeal, nasopharyngeal |
Funding
Acknowledgments
Conflicts of Interest
References
- Sigrist, C.; Bridge, A.; Le Mercier, P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020, 177, 104759. [Google Scholar] [CrossRef]
- World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Zhand, S.; Saghaeian Jazi, M.; Mohammadi, S.; Tarighati Rasekhi, R.; Rostamian, G.; Kalani, M.R.; Rostamian, A.; George, J.; Douglas, M.W. COVID-19: The immune responses and clinical therapy candidates. Int. J. Mol. Sci. 2020, 21, 5559. [Google Scholar] [CrossRef]
- World Health Organization. Advice on the Use of Point-of-Care Immunodiagnostic Tests for COVID-19; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Shuaib, W.; Stanazai, H.; Abazid, A.G.; Mattar, A.A. Re-Emergence of Zika Virus: A Review on Pathogenesis, Clinical Manifestations, Diagnosis, Treatment, and Prevention. Am. J. Med. 2016, 129, 879.e7–879.e12. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Gallina, L.; Palladini, A.; Prosperi, S.; Battilani, M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. Sci. World J. 2012, 2012, 989514. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology 2020, 296, 200343. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Zoëga Andreasen, S.; Wolff, A.; Duong Bang, D. From lab on a chip to point of care devices: The role of open source microcontrollers. Micromachines 2018, 9, 403. [Google Scholar] [CrossRef]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody tests in detecting SARS-CoV-2 infection: A meta-analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Grossi, V.; Lari, B.; Bambi, R.; Perri, A.; Manneschi, M.; Terenzi, G.; Liotti, I.; Ciotta, G.; Taddei, C. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: An Italian experience. J. Med. Virol. 2020, 92, 1671–1675. [Google Scholar] [CrossRef] [PubMed]
- Padoan, A.; Cosma, C.; Sciacovelli, L.; Faggian, D.; Plebani, M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. (Cclm) 2020, 58, 1081–1088. [Google Scholar] [CrossRef]
- Zhong, L.; Chuan, J.; Gong, B.; Shuai, P.; Zhou, Y.; Zhang, Y.; Jiang, Z.; Zhang, D.; Liu, X.; Ma, S. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci. China Life Sci. 2020, 63, 777–780. [Google Scholar] [CrossRef]
- Wen, T.; Huang, C.; Shi, F.J.; Zeng, X.Y.; Lu, T.; Ding, S.N.; Jiao, Y.J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef]
- Sajid, M.; Kawde, A.-N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Liu, R.; Liu, X.; Han, H.; Shereen, M.A.; Niu, Z.; Li, D.; Liu, F.; Wu, K.; Luo, Z.; Zhu, C. The comparative superiority of IgM-IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Pan, Y.; Li, X.; Yang, G.; Fan, J.; Tang, Y.; Zhao, J.; Long, X.; Guo, S.; Zhao, Z.; Liu, Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- BioMedomics. COVID-19 IgM/IgG Rapid Test. 2020. Available online: https://www.biomedomics.com/products/infectious-disease/covid-19-rt/ (accessed on 5 December 2020).
- PharmACT. Rapid Test: Corona Virus. 2020. Available online: https://www.pharmact.eu/ (accessed on 5 December 2020).
- Chembio. DPP COVID-19 IgM/IgG System. 2020. Available online: https://chembio.com/products/dpp-covid-19-igm-igg-system-ous-europe/ (accessed on 5 December 2020).
- FIND. SARS-COV-2 DIAGNOSTIC PIPELINE. 2020. Available online: https://www.finddx.org/covid-19/pipeline/ (accessed on 5 December 2020).
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- McArdle, M. China Supplied Faulty Coronavirus Test Kits to Spain, Czech Republic. Available online: https://www.nationalreview.com/news/china-supplied-faulty-coronavirus-test-kits-to-spain-czech-republic/ (accessed on 5 December 2020).
- Sinéad Baker and Ruqayyah Moynihan. Spain, Europe’s Worst-Hit Country after Italy, Says Coronavirus Tests It Bought from China Are Failing to Detect Positive Cases. Available online: https://www.businessinsider.com/coronavirus-spain-says-rapid-tests-sent-from-china-missing-cases-2020-3?r=DE&IR=T (accessed on 5 December 2020).
- Prague Morning. 80% of Rapid COVID-19 Tests the Czech Republic Bought from China Are Wrong. Available online: https://www.praguemorning.cz/80-of-rapid-covid-19-tests-the-czech-republic-bought-from-china-are-wrong/ (accessed on 5 December 2020).
- Chakraborty, B. Netherlands Becomes Latest Country to Reject China-Made Coronavirus Test Kits, Gear. Available online: https://www.foxnews.com/world/netherlands-becomes-latest-country-to-reject-china-made-coronavirus-test-kits-gear (accessed on 5 December 2020).
- Bruning, A.H.L.; Leeflang, M.M.G.; Vos, J.; Spijker, R.; de Jong, M.D.; Wolthers, K.C.; Pajkrt, D. Rapid Tests for Influenza, Respiratory Syncytial Virus, and Other Respiratory Viruses: A Systematic Review and Meta-analysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017, 65, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Wen, K.; Chen, J.; Liu, Y.; Yuan, Z.; Han, C.; Chen, J.; Pan, Y.; Chen, L.; Dan, Y. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv 2020. [Google Scholar] [CrossRef]
- Cheng, M.P.; Papenburg, J.; Desjardins, M.; Kanjilal, S.; Quach, C.; Libman, M.; Dittrich, S.; Yansouni, C.P. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus 2: A Narrative Review. Ann. Intern. Med. 2020, 172, 726–734. [Google Scholar] [CrossRef]
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 10. [Google Scholar] [CrossRef]
- Li, M.; Jin, R.; Peng, Y.; Wang, C.; Ren, W.; Lv, F.; Gong, S.; Fang, F.; Wang, Q.; Li, J. Generation of antibodies against COVID-19 virus for development of diagnostic tools. medRxiv 2020. [Google Scholar] [CrossRef]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Access Bio. ACCESS BIO’S CARESTART™ COVID-19. Antigen. Available online: https://accessbiodiagnostics.net/carestart-covid-19-antigen/ (accessed on 5 December 2020).
- Abbott Laboratories. COVID-19 Ag Rapid Test Device. Available online: https://www.who.int/diagnostics_laboratory/eual/eul_0564_032_00_panbi_covid19_ag_rapid_test_device.pdf (accessed on 5 December 2020).
- BTNX. COVID-19 Antigen Rapid Test Device. Available online: https://www.btnx.com/Product?id=2010 (accessed on 5 December 2020).
- SD Biosensor. STANDARD Q COVID-19 Ag. Available online: http://sdbiosensor.com/xe/product/7672 (accessed on 5 December 2020).
- Wantai BioPharm Ltd. Wantai SARS-CoV-2 Ag Rapid Test Kit. Available online: http://www.ystwt.cn/covid-19 (accessed on 5 December 2020).
- Quidel Corporation. Sofia® SARS Antigen. Available online: https://www.quidel.com/immunoassays/rapid-sars-tests/sofia-sars-antigen-fia (accessed on 5 December 2020).
- Dickinson, B. BD Veritor™ System for Rapid Detection of SARS-CoV-2. Available online: https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/point-of-care-testing/bd-veritor-plus-system-for-rapid-covid-19-sars-cov-2-testing (accessed on 5 December 2020).
- World Health Organization. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance, 11 September 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Wan, Z.; Zhang, Y.N.; He, Z.; Liu, J.; Lan, K.; Hu, Y.; Zhang, C. A Melting Curve-Based Multiplex RT-qPCR Assay for Simultaneous Detection of Four Human Coronaviruses. Int. J. Mol. Sci. 2016, 17, 1880. [Google Scholar] [CrossRef]
- ECDC. An Overview of the Rapid Test Situation for COVID-19 Diagnosis in the EU/EEA; ECDC: Stockholm, Sweden, 2020. [Google Scholar]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef]
- Vashist, S.K. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics 2020, 10, 202. [Google Scholar] [CrossRef]
- Guo, W.-L.; Jiang, Q.; Ye, F.; Li, S.-Q.; Hong, C.; Chen, L.-Y.; Li, S.-Y. Effect of throat washings on detection of 2019 novel coronavirus. Clin. Infect. Dis. 2020, 71, 1980–1981. [Google Scholar] [CrossRef] [PubMed]
- Adachi, D.; Johnson, G.; Draker, R.; Ayers, M.; Mazzulli, T.; Talbot, P.; Tellier, R. Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J. Virol. Methods 2004, 122, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ahrberg, C.D.; Manz, A.; Chung, B.G. Polymerase chain reaction in microfluidic devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef]
- Stilla. A 3-Color Crystal Digital PCR™ Kit for Detection of COVID-19. Available online: https://www.stillatechnologies.com/a-3-color-crystal-digital-pcr-kit-for-detection-of-covid-19/ (accessed on 5 December 2020).
- Bio-Rad. SARS-CoV-2 Droplet Digital PCR (ddPCR) Kit. Available online: https://www.bio-rad.com/en-al/product/sars-cov-2-droplet-digital-pcr-ddpcr-kit?ID=Q9KNF4RT8IG9 (accessed on 5 December 2020).
- Kilic, T.; Weissleder, R.; Lee, H.J.I. Molecular and immunological diagnostic tests of COVID-19—Current status and challenges. iScience 2020, 23, 101406. [Google Scholar] [CrossRef] [PubMed]
- Marzinotto, S.; Mio, C.; Verardo, R.; Pipan, C.; Schneider, C.; Curcio, F. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. medRxiv 2020. [Google Scholar] [CrossRef]
- Grant, P.R.; Turner, M.A.; Shin, G.Y.; Nastouli, E.; Levett, L.J. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bruce, E.A.; Huang, M.-L.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA ex-traction step. bioRxiv 2020. [Google Scholar] [CrossRef]
- Deiana, M.; Mori, A.; Piubelli, C.; Scarso, S.; Favarato, M.; Pomari, E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Xu, G.; Hsieh, T.-M.; Lee, D.Y.; Ali, E.M.; Xie, H.; Looi, X.L.; Koay, E.S.-C.; Li, M.-H.; Ying, J.Y. A self-contained all-in-one cartridge for sample preparation and real-time PCR in rapid influenza diagnosis. Lab Chip 2010, 10, 3103–3111. [Google Scholar] [CrossRef]
- Xpert® Xpress SARS-CoV-2. Xpert® Xpress SARS-CoV-2 Has Received FDA Emergency Use Authorization. Available online: https://www.cepheid.com/coronavirus (accessed on 5 December 2020).
- Etherington, D. Mesa Biotech Gains Emergency FDA Approval for Rapid, Point-of-Care COVID-19 Test. 2020. Available online: https://techcrunch.com/2020/03/24/mesa-biotech-gains-emergency-fda-approval-for-rapid-point-of-care-covid-19-test/ (accessed on 5 December 2020).
- QIAGEN. QIAGEN Launches QIAstat-Dx Test Kit for Detection of SARS-CoV-2 Coronavirus in Europe Following CE Marking. 2020. Available online: https://corporate.qiagen.com/newsroom/press-releases/2020/20200318_qiastat_covid19_ce-ivd?cmpid=CCOM_IR_IR_CEIVD_0320_SM_FacebookOrg_PR (accessed on 5 December 2020).
- Luminex. SARS-CoV-2 Detection. 2020. Available online: http://info.luminexcorp.com/covid19 (accessed on 5 December 2020).
- Mollajan, M.; Razavi Bazaz, S.; Abouei Mehrizi, A. A Thoroughgoing Design of a Rapid-cycle Microfluidic Droplet-based PCR Device to Amplify Rare DNA Strands. J. Appl. Fluid Mech. 2018, 11, 21–29. [Google Scholar] [CrossRef]
- VereCoV. VereCoV™ OneMix Detection Kit for VerePLEX™ Biosystem. Available online: https://vereduslabs.com/covid-19/ (accessed on 5 December 2020).
- MiCo BioMed. VERI-Q COVID-19 Multiplex Detection Kit. Available online: http://www.micobiomed.com/en/covid/main.php (accessed on 5 December 2020).
- Star Array. COVID-19 Detection. Available online: https://www.star-array.com/copy-of-antimicrobial-resistance (accessed on 5 December 2020).
- Kalorama Information. In Breaking Transmission Chain of Covid-19 and Other Diseases, Point-of-Care Is Best Hope. Available online: https://kaloramainformation.com/point-of-care-testing-for-covid-19/ (accessed on 5 December 2020).
- ElveFlow. Fastgene versus Pathogens Aka RT-PCR on Chip: A Valorization Success Story. Available online: https://www.elveflow.com/microfluidics-research-horizon-europe/european-projects/fastgene-versus-pathogens-an-epidemical-tracking-system/ (accessed on 5 December 2020).
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Gao, Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta 2015, 853, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef]
- Yehia, N.; Arafa, A.-S.; Abd El Wahed, A.; El-Sanousi, A.A.; Weidmann, M.; Shalaby, M.A. Development of reverse transcription recombinase polymerase amplification assay for avian influenza H5N1 HA gene detection. J. Virol. Methods 2015, 223, 45–49. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Soropogui, B.; Patel, P.; Abd El Wahed, A.; Loucoubar, C.; Fall, G.; Kiory, D.; Magassouba, N.F.; Keita, S. Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Eurosurveillance 2015, 20, 30053. [Google Scholar] [CrossRef]
- Magro, L.; Jacquelin, B.; Escadafal, C.; Garneret, P.; Kwasiborski, A.; Manuguerra, J.-C.; Monti, F.; Sakuntabhai, A.; Vanhomwegen, J.; Lafaye, P. based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Xia, S.; Chen, X. Ultrasensitive and whole-course encapsulated field detection of 2019-nCoV gene applying exponential amplification from RNA combined with chemical probes. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Xia, S.; Chen, X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT–RPA. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Goto, M.; Shimada, K.; Sato, A.; Takahashi, E.; Fukasawa, T.; Takahashi, T.; Ohka, S.; Taniguchi, T.; Honda, E.; Nomoto, A. Rapid detection of Pseudomonas aeruginosa in mouse feces by colorimetric loop-mediated isothermal amplification. J. Microbiol. Methods 2010, 81, 247–252. [Google Scholar] [CrossRef]
- Park, G.-S.; Ku, K.; Beak, S.-H.; Kim, S.J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) Assays Targeting SARS-CoV-2. J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Dang, X.; Wang, Q.; Xu, M.; Zhao, Q.; Zhou, Y.; Zhao, H.; Wang, L.; Xu, Y.; Wang, J. Rapid Detection of SARS-CoV-2 Using Reverse transcription RT-LAMP method. medRxiv 2020. [Google Scholar] [CrossRef]
- El-Tholoth, M.; Bau, H.H.; Song, J. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Woo, C.H.; Jang, S.; Shin, G.; Jung, G.Y.; Lee, J.W. Sensitive one-step isothermal detection of pathogen-derived RNAs. medRxiv 2020. [Google Scholar] [CrossRef]
- Qian, J.; Ferguson, T.M.; Shinde, D.N.; Ramírez-Borrero, A.J.; Hintze, A.; Adami, C.; Niemz, A. Sequence dependence of isothermal DNA amplification via EXPAR. Nucleic Acids Res. 2012, 40, e87. [Google Scholar] [CrossRef]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Böhmer, A.; Schildgen, V.; Lüsebrink, J.; Ziegler, S.; Tillmann, R.L.; Kleines, M.; Schildgen, O. Novel application for isothermal nucleic acid sequence-based amplification (NASBA). J. Virol. Methods 2009, 158, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Abbott Laboratories. ID NOW™ COVID-19 Molecular. In Minutes. On the Front Line. Available online: https://www.abbott.com/intended-for-us-residents.html (accessed on 5 December 2020).
- Basu, A.; Zinger, T.; Inglima, K.; Woo, K.-m.; Atie, O.; Yurasits, L.; See, B.; Aguero-Rosenfeld, M.E. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef]
- Rendu Biotechnology. The High-Throughput, Whole-Process Integration of the New Crown Nucleic Acid Detection Platform. 2020. Available online: http://www.rdbio.com/gongsidongtai/122.html (accessed on 5 December 2020).
- CueHealth. Cue COVID-19 Test. 2020. Available online: https://www.cuehealth.com/what-is-cue (accessed on 5 December 2020).
- TwistDx. Twista. 2020. Available online: https://www.twistdx.co.uk/docs/default-source/twista/twista_manual_revb.pdf?sfvrsn=4 (accessed on 5 December 2020).
- Optigene. Genie® II. 2020. Available online: http://www.optigene.co.uk/instruments/instrument-genie-ii/ (accessed on 5 December 2020).
- Dara, M.; Talebzadeh, M. CRISPR/Cas as a Potential Diagnosis Technique for COVID-19. Avicenna J. Med. Biotechnol. 2020, 12, 201–202. [Google Scholar] [PubMed]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and Evaluation of A CRISPR-based Diagnostic For 2019-novel Coronavirus. medRxiv 2020. [Google Scholar] [CrossRef]
- Bai, H.; Cai, X.; Zhang, X. Landscape Coronavirus Disease 2019 Test (COVID-19 Test) in Vitro—A Comparison of PCR Vs Immunoassay Vs Crispr-based Test. OSF Prepr. 2020. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444. [Google Scholar] [CrossRef]
- Broad Communications. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics. Available online: https://www.broadinstitute.org/news/enabling-coronavirus-detection-using-crispr-cas13-open-access-sherlock-research-protocols-and (accessed on 31 March 2020).
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection using Cas13. Nature 2020. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, W.; Fasching, C.L.; Singh, J.; Chiu, C.Y.; Chen, J.S. A Protocol for Rapid Detection of the 2019 Novel Coronavirus SARS-CoV-2 Using CRISPR Diagnostics: SARS-CoV-2 DETECTR. 2020. Available online: https://www.protocols.io/view/a-protocol-for-rapid-detection-of-the-2019-novel-c-bfhcjj2w (accessed on 5 December 2020).
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Liu, C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV virus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chen, J.H.-K.; Yip, C.C.-Y.; Chan, J.F.-W.; Poon, R.W.-S.; To, K.K.-W.; Chan, K.-H.; Cheng, V.C.-C.; Yuen, K.-Y. Clinical Performance of the Luminex NxTAG CoV Extended Panel for SARS-CoV-2 Detection in Nasopharyngeal Specimens from COVID-19 Patients in Hong Kong. J. Clin. Microbiol. 2020, 58, e00936-20. [Google Scholar] [CrossRef] [PubMed]
- Sherlock. Sherlock™ CRISPR SARS-CoV-2. 2020. Available online: https://sherlock.bio/crispr-sars-cov-2/ (accessed on 5 December 2020).
- Rostami, A.; Sepidarkish, M.; Leeflang, M.M.G.; Riahi, S.M.; Nourollahpour Shiadeh, M.; Esfandyari, S.; Mokdad, A.H.; Hotez, P.J.; Gasser, R.B. SARS-CoV-2 seroprevalence worldwide: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251. [Google Scholar] [CrossRef] [PubMed]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 104500. [Google Scholar] [CrossRef]
- World Health Organization. Report of the Who-China Joint Mission on Coronavirus Disease 2019 (COVID-19); World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv 2020. [Google Scholar] [CrossRef]
- Moazzam, P.; Tavassoli, H.; Razmjou, A.; Warkiani, M.E.; Asadnia, M. Mist harvesting using bioinspired polydopamine coating and microfabrication technology. Desalination 2018, 429, 111–118. [Google Scholar] [CrossRef]
- Kwon, T.; Prentice, H.; De Oliveira, J.; Madziva, N.; Warkiani, M.E.; Hamel, J.-F.P.; Han, J. Microfluidic cell retention device for perfusion of mammalian suspension culture. Sci. Rep. 2017, 7, 6703. [Google Scholar] [CrossRef]
- Vasilescu, S.A.; Bazaz, S.R.; Jin, D.; Shimoni, O.; Warkiani, M.E. 3D printing enables the rapid prototyping of modular microfluidic devices for particle conjugation. Appl. Mater. Today 2020, 20, 100726. [Google Scholar] [CrossRef]
- Svoboda, E. A sticking point for rapid flu tests? Nature 2019, 573, S56–S57. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezaei, M.; Razavi Bazaz, S.; Zhand, S.; Sayyadi, N.; Jin, D.; Stewart, M.P.; Ebrahimi Warkiani, M. Point of Care Diagnostics in the Age of COVID-19. Diagnostics 2021, 11, 9. https://doi.org/10.3390/diagnostics11010009
Rezaei M, Razavi Bazaz S, Zhand S, Sayyadi N, Jin D, Stewart MP, Ebrahimi Warkiani M. Point of Care Diagnostics in the Age of COVID-19. Diagnostics. 2021; 11(1):9. https://doi.org/10.3390/diagnostics11010009
Chicago/Turabian StyleRezaei, Meysam, Sajad Razavi Bazaz, Sareh Zhand, Nima Sayyadi, Dayong Jin, Martin P. Stewart, and Majid Ebrahimi Warkiani. 2021. "Point of Care Diagnostics in the Age of COVID-19" Diagnostics 11, no. 1: 9. https://doi.org/10.3390/diagnostics11010009
APA StyleRezaei, M., Razavi Bazaz, S., Zhand, S., Sayyadi, N., Jin, D., Stewart, M. P., & Ebrahimi Warkiani, M. (2021). Point of Care Diagnostics in the Age of COVID-19. Diagnostics, 11(1), 9. https://doi.org/10.3390/diagnostics11010009







