Clinical Perspective and Translational Oncology of Liquid Biopsy
Abstract
1. Liquid Biopsy
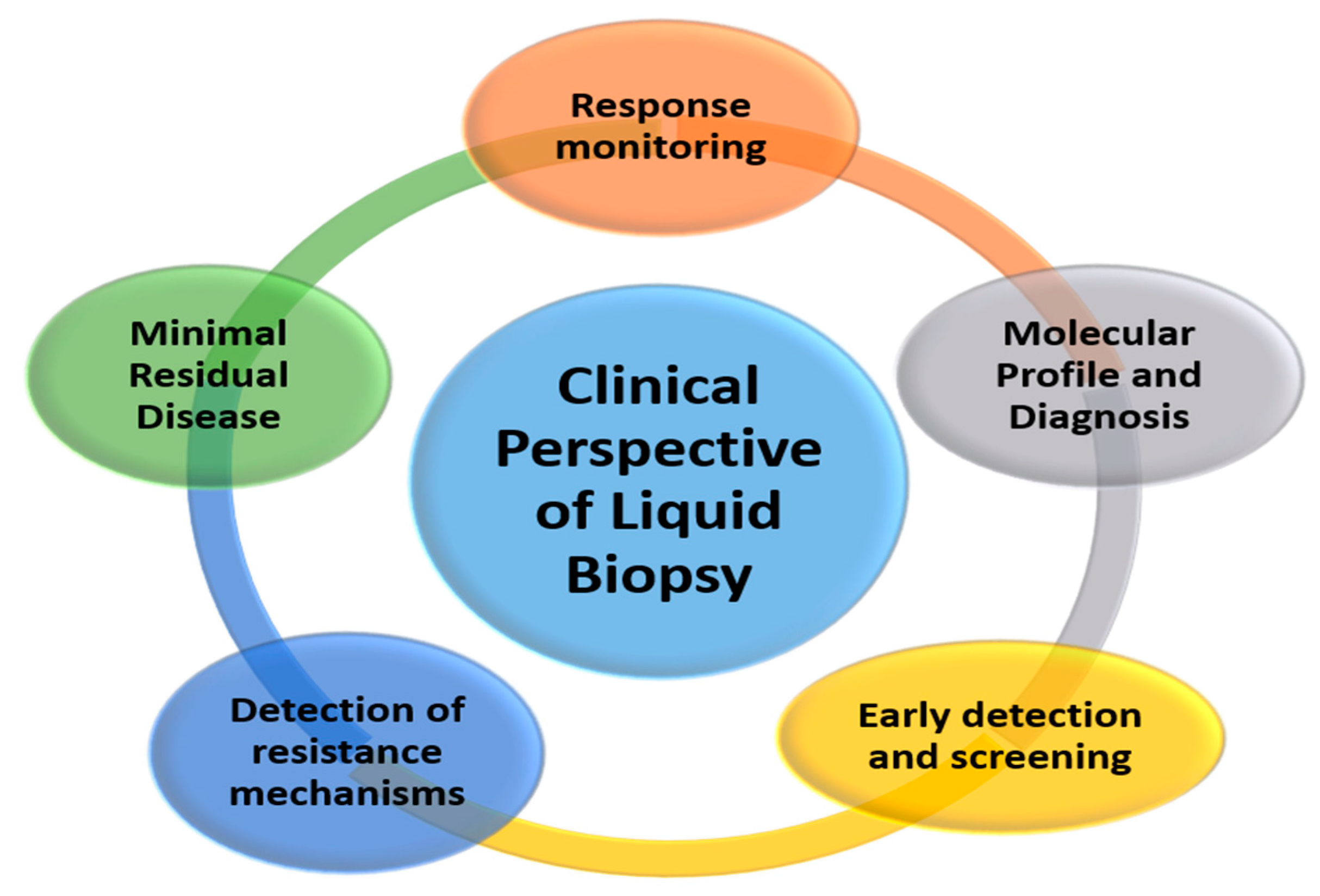
2. Clinical Perspective of Liquid Biopsy
2.1. Molecular Profile and Diagnosis
2.2. Response Monitoring and Early Detection of Resistance Mechanisms
2.3. Minimal Residual Disease
2.4. Early Detection and Screening
3. Translational Oncology Application of Liquid Biopsies in Cancer Therapy
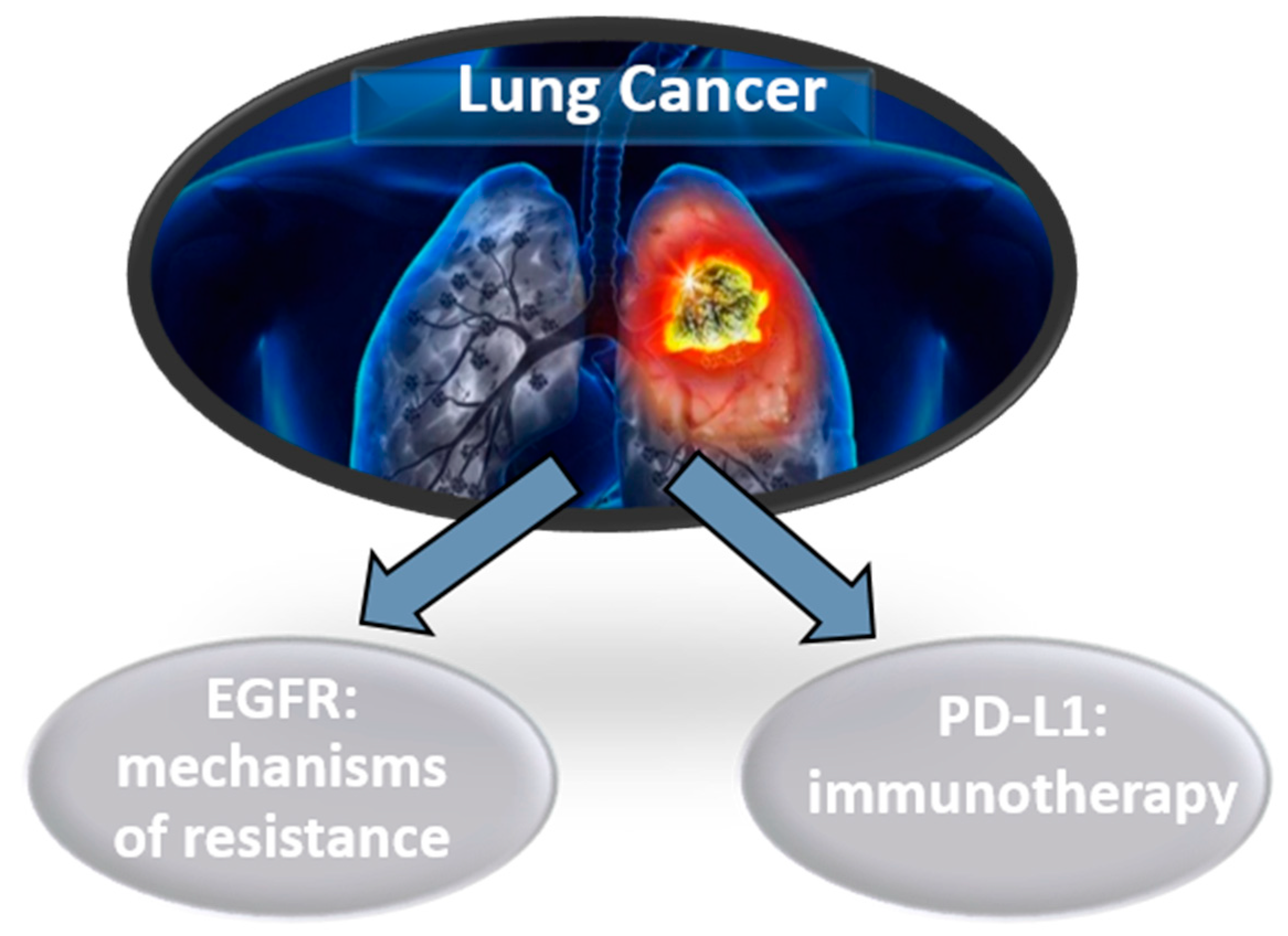
3.1. Lung Cancer
3.2. Colorectal Cancer
3.3. Pancreatic Cancer
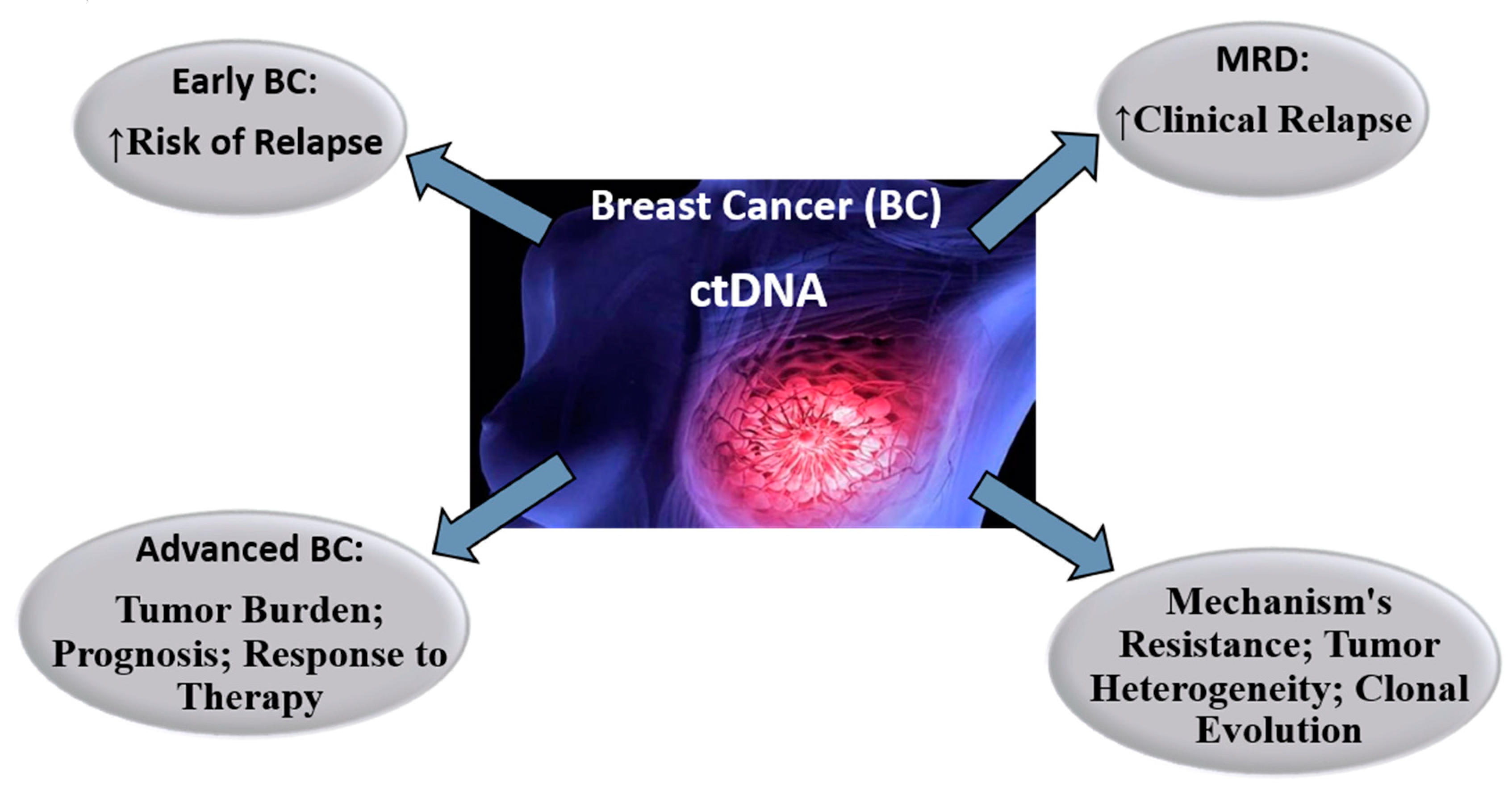
3.4. Breast Cancer
3.5. Prostate Cancer
3.6. Melanoma
4. Pitfalls of Using Liquid Biopsy for Precision Medicine
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Venesio, T.; Siravegna, G.; Bardelli, A.; Sapino, A. Liquid biopsies for monitoring temporal genomic heterogeneity in breast and colon cancers. Pathobiology 2018, 85, 146–154. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fernández-Lázaro, D.; García, J.L.; Caballero García, A.; Córdova, A.; Mielgo-Ayuso, J.; Cruz, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Buyse, M.; Sargent, D.J.; Grothey, A.; Matheson, A.; De Gramont, A. Biomarkers and surrogate end points—The challenge of statistical validation. Nat. Rev. Clin. Oncol. 2010, 7, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2018, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Kim, C.-J.; Sunkara, V.; Kim, M.-H.; Cho, Y.-K. Liquid biopsy in lung cancer: Clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Valda, P.; Cidado, J.; Blair, B.G.; et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef]
- Czeiger, D.; Shaked, G.; Eini, H.; Vered, I.; Belochitski, O.; Avriel, A.; Douvdevani, A. Measurement of circulating cell-free DNA levels by a new simple fluorescent test in patients with primary colorectal cancer. Am. J. Clin. Pathol. 2011, 135, 264–270. [Google Scholar] [CrossRef]
- Bardelli, A.; Pantel, K. Liquid biopsies, what we do not know (yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef]
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Arita, T.; Konishi, H.; Shiozaki, A.; et al. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J. Gastroenterol. 2016, 22, 5627–5641. [Google Scholar] [CrossRef] [PubMed]
- Maltoni, R.; Casadio, V.; Ravaioli, S.; Foca, F.; Tumedei, M.M.; Salvi, S.; Martignano, F.; Calistri, D.; Rocca, A.; Schirone, A.; et al. Cell-free DNA detected by “liquid biopsy” as a potential prognostic biomarker in early breast cancer. Oncotarget 2017, 8, 16642–16649. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Ignatiadis, M. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 2019, 79, 2798–2804. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Carneiro, B.A.; Pamarthy, S.; Shah, A.N.; Sagar, V.; Unno, K.; Han, H.; Yang, X.J.; Costa, R.B.; Nagy, R.J.; Lanman, R.B.; et al. Anaplastic lymphoma kinase mutation (ALK F1174C) in small cell carcinoma of the prostate and molecular response to alectinib. Clin. Cancer Res. 2018, 24, 2732–2739. [Google Scholar] [CrossRef]
- El Sayed, R.; El Jamal, L.; El Iskandarani, S.; Kort, J.; Salam, M.A.; Assi, H. Endocrine and Targeted Therapy for Hormone-Receptor-Positive, HER2-Negative Advanced Breast Cancer: Insights to Sequencing Treatment and Overcoming Resistance Based on Clinical Trials. Front. Oncol. 2019, 21, 510. [Google Scholar] [CrossRef]
- Schochter, F.; Friedl, T.W.; deGregorio, A.; Krause, S.; Huober, J.; Rack, B.; Janni, W. Are Circulating Tumor Cells (CTCs) Ready for Clinical Use in Breast Cancer? An Overview of Completed and Ongoing Trials Using CTCs for Clinical Treatment Decisions. Cells 2019, 8, 1412. [Google Scholar] [CrossRef]
- García, J.L.; Lozano, R.; Misiewicz-Krzeminska, I.; Fernández-Mateos, J.; Krzeminski, P.; Alfonso, S.; Marcos, R.A.; García, R.; Gómez-Veiga, F.; Virseda, Á.; et al. A novel capillary nano-immunoassay for assessing androgen receptor splice variant 7 in plasma. Correlation with CD133 antigen expression in circulating tumor cells. A pilot study in prostate cancer patients. Clin. Transl. Oncol. 2017, 19, 1350–1357. [Google Scholar] [CrossRef][Green Version]
- Goodman, C.R.; Seagle, B.-L.L.; Friedl, T.W.; Rack, B.; Lato, K.; Fink, V.; Cristofanilli, M.; Donnelly, E.D.; Janni, W.; Shahabi, S.; et al. Association of circulating tumor cell status with benefit of radiotherapy and survival in early-stage breast cancer. JAMA Oncol. 2018, 4, e180163. [Google Scholar] [CrossRef]
- Fribbens, C.; Garcia Murillas, I.; Beaney, M.; Hrebien, S.; O’Leary, B.; Kilburn, L.; Howarth, K.; Epstein, M.; Green, E.; Rosenfeld, N.; et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann. Oncol. 2018, 29, 145–153. [Google Scholar] [CrossRef]
- Condorelli, R.; Spring, L.; O’shaughnessy, J.; Lacroix, L.; Bailleux, C.; Scott, V.; Dubois, J.; Nagy, R.J.; Lanman, R.B.; Iafrate, A.J.; et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann. Oncol. 2018, 29, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Hu, Y.; Mileham, K.F.; Husain, H.; Costa, D.B.; Tracy, P.; Feeney, N.; Sholl, L.M.; Dahlberg, S.E.; Redig, A.J.; et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M–positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018, 4, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Schreuer, M.; Meersseman, G.; Van Den Herrewegen, S.; Jansen, Y.; Chevolet, I.; Bott, A.; Wilgenhof, S.; Seremet, T.; Jacobs, B.; Buyl, R.; et al. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J. Transl. Med. 2016, 14, 95. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Litière, S.; Rothé, F.; Riethdorf, S.; Proudhon, C.; Fehm, T.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091–10093, BIG 1–12, Treat CTC): A randomized phase II trial. Ann. Oncol. 2018, 29, 1777–1783. [Google Scholar] [CrossRef]
- van Dalum, G.; Stam, G.-J.; Scholten, L.F.; Mastboom, W.J.; Vermes, I.; Tibbe, A.G.; De Groot, M.R.; Terstappen, L.W. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 2015, 46, 1361–1368. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.-M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Andrew Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non–small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Rossi, G.; Mu, Z.; Rademaker, A.W.; Austin, L.K.; Strickland, K.S.; Costa, R.L.B.; Nagy, R.J.; Zagonel, V.; Taxter, T.J.; Behdad, A.; et al. Cell-free DNA and circulating tumor cells: Comprehensive liquid biopsy analysis in advanced breast cancer. Clin. Cancer Res. 2018, 24, 560–568. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Ammar, A.; Javed, A.A.; Wong, F.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Chan, K.A.; Woo, J.K.; King, A.; Zee, B.C.; Lam, W.J.; Chan, S.L.; Chan, S.L.; Chu, S.W.I.; Mak, C.; Tse, I.O.L.; et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Update on the types and usage of liquid biopsies in the clinical setting: A systematic review. BMC Cancer 2018, 18, 527. [Google Scholar] [CrossRef]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Boutin, A.T.; Liao, W.-T.; Wang, M.; Hwang, S.S.; Karpinets, T.V.; Cheung, H.; Chu, G.C.; Jiang, S.; Hu, J.; Chang, K.; et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017, 31, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef]
- Montagut, C.; Argilés, G.; Ciardiello, F.; Poulsen, T.T.; Dienstmann, R.; Kragh, M.; Kopetz, S.; Lindsted, T.; Ding, C.; Vidal, J.; et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses: A phase 2 randomized clinical trial. JAMA Oncol. 2018, 4, e175245. [Google Scholar] [CrossRef]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019, 110, 1148–1155. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–102027. [Google Scholar] [CrossRef]
- Oshiro, C.; Kagara, N.; Naoi, Y.; Shimoda, M.; Shimomura, A.; Maruyama, N.; Shimazu, K.; Kim, S.J.; Noguchi, S. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat. 2015, 150, 299–307. [Google Scholar] [CrossRef]
- Otsuji, K.; Sasaki, T.; Tanaka, A.; Kunita, A.; Ikemura, M.; Matsusaka, K.; Tada, k.; Masashi Fukayama, M.; Seto, Y. Use of droplet digital PCR for quantitative and automatic analysis of the HER2 status in breast cancer patients. Breast Cancer Res. Treat. 2017, 162, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Tantiwetrueangdet, A.; Panvichian, R.; Wongwaisayawan, S.; Sueangoen, N.; Lertsithichai, P. Droplet digital PCR using HER2/EIF2C1 ratio for detection of HER2 amplification in breast cancer tissues. Med. Oncol. 2018, 35, 149. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Board, R.E.; Wardley, A.M.; Dixon, J.M.; Armstrong, A.C.; Howell, S.; Renshaw, L.; Donald, E.; Greystoke, A.; Ranson, M.; Hughes, A.; et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res. Treat. 2010, 120, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Inao, T.; Sueta, A.; Fujiwara, S.; Omoto, Y.; Hirotaka Iwase, H. Prognostic role of PIK 3 CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci. 2015, 106, 1582–1589. [Google Scholar] [CrossRef]
- Baselga, J.; Im, S.-A.; Iwata, H.; Clemons, M.; Ito, Y.; Awada, A.; Chia, S.; Jagiello-Gruszfeld, A.; Pistilli, B.; Tseng, L.M.; et al. Abstract S6–01: PIK3CA status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2–advanced breast cancer (BC): First results from the randomized, phase III BELLE-2 trial. Cancer Res. 2016. [Google Scholar] [CrossRef]
- Jelovac, D.; Beaver, J.A.; Balukrishna, S.; Wong, H.Y.; Toro, P.V.; Cimino-Mathews, A.; Cimino-Mathews, A.; Argani, P.; Stearns, V.; Jacobs, L.; et al. A PIK3CA mutation detected in plasma from a patient with synchronous primary breast and lung cancers. Hum. Pathol. 2014, 45, 880–883. [Google Scholar] [CrossRef]
- Perkins, G.; Lu, H.; Garlan, F.; Taly, V. Droplet-based digital PCR: Application in cancer research. Exp. Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar]
- Litwin, M.S.; Tan, H.-J. The diagnosis and treatment of prostate cancer: A review. JAMA 2017, 317, 2532–2542. [Google Scholar] [CrossRef]
- Danila, D.C.; Samoila, A.; Patel, C.; Schreiber, N.; Herkal, A.; Anand, A.; Bastos, D.; Heller, G.; Fleisher, M.; Scher, H.I. Clinical validity of detecting circulating tumor cells by AdnaTest assay compared to direct detection of tumor mRNA in stabilized whole blood, as a biomarker predicting overall survival for metastatic castration-resistant prostate cancer patients. Cancer J. 2016, 22, 315–320. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D.; et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Goldkorn, A.; Ely, B.; Tangen, C.M.; Tai, Y.C.; Xu, T.; Li, H.; Twardowski, P.; Veldhuizen, P.J.; Agarwal, N.; Carducci, M.A.; et al. Circulating tumor cell telomerase activity as a prognostic marker for overall survival in SWOG 0421: A phase III metastatic castration resistant prostate cancer trial. Int. J. Cancer. 2015, 136, 1856–1862. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Logothetis, C.J.; Basch, E.; Molina, A.; Fizazi, K.; North, S.A.; Chi, K.N.; Jones, R.J.; Goodman, O.B.; Mainwarning, P.N.; Sternberg, C.N.; et al. Effect of abiraterone acetate and prednisone compared with placebo and prednisone on pain control and skeletal-related events in patients with metastatic castration-resistant prostate cancer: Exploratory analysis of data from the COU-AA-301 randomised trial. Lancet Oncol. 2012, 13, 1210–1217. [Google Scholar]
- Wyatt, A.W.; Azad, A.A.; Volik, S.V.; Annala, M.; Beja, K.; McConeghy, B.; Haegert, A.; Warner, E.W.; Mo, F.; Brahmbhatt, S.; et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016, 2, 1598–1606. [Google Scholar] [CrossRef]
- Kong, D.; Sethi, S.; Li, Y.; Chen, W.; Sakr, W.A.; Heath, E. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. Prostate 2015, 75, 161–174. [Google Scholar] [CrossRef]
- Uo, T.; Plymate, S.R.; Sprenger, C.C. The potential of AR-V7 as a therapeutic target. Exp. Opin. Ther. Targ. 2018, 22, 201–216. [Google Scholar] [CrossRef]
- Arnold, M.; Holterhues, C.; Hollestein, L.; Coebergh, J.; Nijsten, T.; Pukkala, E.; Holleczek, B.; Tryggvadóttir, L.; Comber, H.; Bento, M.J.; et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. J. Eur. Acad. Dermatol. Venereol. 2014, 8, 1170–1178. [Google Scholar] [CrossRef]
- Lee, R.J.; Gremel, G.; Marshall, A.; Myers, K.; Fisher, N.; Dunn, J.; Dhomen, N.; Corrie, P.G.; Middleton, M.R.; Lorigan, P.; et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann. Oncol. 2017, 29, 490–496. [Google Scholar] [CrossRef]
- Gonzalez-Cao, M.; Mayo-de-las-Casas, C.; Molina-Vila, M.A.; De Mattos-Arruda, L.; Muñoz-Couselo, E.; Manzano, J.L.; Cortes, J.; Berros, J.P.; Drozdowskyj, A.; Sanmamed, M.; et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015, 25, 486–495. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, A.C.; Warburton, L.; Al-Ogaili, Z.; Celliers, L.; Calapre, L.; Pereira, M.R.; Khattak, M.A.; Meniawy, T.M.; Millward, M.; Ziman, M.; et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. BMC Cancer 2018, 18, 726. [Google Scholar] [CrossRef] [PubMed]
- Valpione, S.; Gremel, G.; Mundra, P.; Middlehurst, P.; Galvani, E.; Girotti, M.R.; Lee, R.J.; Garner, G.; Dhomen, N.; Lorigan, P.C.; et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur. J. Cancer 2018, 88, 1–9. [Google Scholar] [CrossRef]
- Bidard, F.C.; Madic, J.; Mariani, P.; Piperno-Neumann, S.; Rampanou, A.; Servois, V.; Cassoux, N.; Desjardins, L.; Milder, M.; Vaucher, I.; et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 2014, 134, 1207–1213. [Google Scholar] [CrossRef] [PubMed]

| Molecular Detection Techniques | Properties |
|---|---|
| quantitative polymerase chain reaction (qPCR) | qPCR, is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). qPCR is regarded as the ‘gold standard’ in the quantitative analysis of nucleic acids, be it DNA, RNA or micro-RNA molecules. qPCR has high sensitivity, robustness, good reproducibility, broad dynamic quantification range, and very importantly, affordability. |
| Safe-Sequencing System (Safe-SeqS) | Safe-SeqS is a Unique Molecular Identifier (UMI) approach to detect rare variants. Safe-SeqS assigns a UMI to each template molecule and amplifies each uniquely tagged template molecule to create UMI families. The abundance of each UMI can be used to distinguish between rare mutations and technical errors, as well as to correct for PCR amplification bias |
| CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) | CAPP-Seq is a next-generation sequencing based method used to quantify circulating DNA in cancer. CAPP-Seq is an economical and ultrasensitive method and could be routinely applied clinically to detect and monitor diverse malignancies, thus facilitating personalized cancer therapy. |
| Digital PCR (dPCR) | dPCR is a modification of the qPCR method that can be employed to quantify precisely defined nucleic acid targets. The technique is based on the concept of limiting dilutions, which involves the partitioning of a PCR reaction into multiple sub-reactions such that each sub-reaction either contains none or one or more DNA targets. Following thermal cycling, reactions are classified as either positive (target detected) or negative (no target detected), hence providing the basis for a digital output format. By determining the proportion of empty partitions, Poissonian statistics can be applied and the initial number of target molecules present can be estimated. |
| Copy number alterations (CNAs) | CNA are somatic changes in chromosome structure that result in gains or losses in copies of DNA sections in somatic tissue, and are prevalent in many cancers. CNA has facilitated the discovery of tumor suppressor genes and oncogenes. Microarray-based CNA assays designed to detect these chromosomes copy number alterations on a high-resolution, genome-wide scale have emerged as a key technology in the genomic era. |
| Whole-genome sequencing (WGS) | WGS is a comprehensive method for analyzing entire genomes. Whole genome sequencing WGS has revolutionized the biosciences and proven to be essential and invaluable to the identification of gene functions and their involvement in disease. The feasibility of WGS analysis is under the support of next generation sequencing (NGS) technologies, which require substantial computational and biomedical resources to acquire and analyze large and complex sequence data. |
| Whole-exome sequencing (WES) | WES is a genomic technique for sequencing all of the protein-coding regions of genes in a genome (known as the exome). WES provides coverage of more than 95% of the exons, which harbor the majority of the large genetic variants and single nucleotide polymorphisms (SNPs) associated with human disease phenotypes. WES strategy starts by narrowing down the details of variants to be studied by filtering against databases such as HapMap, from the approximately 3.5 million SNPs identified in the human genome project. This focus enables a simpler way for discovery and validation of causative genes and common and rare variants. |
| Tagged-Amplicon deep sequencing (TAm-Seq) | TAM-Seq allows targeted sequencing of entire genes to detect mutations in ctDNA. TAM-Seq is based on a multiplex pre-amplification of tiling short amplicons with target-specific primers and initial eminent of the target regions is performed followed by a selective amplification in individual (singleplex) PCRs in order to exclude non-specific products |
| Next-Generation Sequencing (NGS) | Digital PCR (dPCR) | |
|---|---|---|
| % Sensitivity | 0.1 | 0.02 |
| Plasma volume (mL) | 4 | 2 |
| Results (week) | 2 | 1 |
| Coverage | High Mutations; Copy-Number Variation; Fusions | Restricted Point Mutations |
| Advantages | Multiple genes Quantitative | High Sensitivity Quantitative |
| Disadvantages | High Cost | Restricted Gene Coverage |
| Pitfalls of Liquid Biopsy |
|---|
| Lack of standard and convenient techniques |
| Test variability and assay sensitivity and specificity |
| Lack of consensus in technical approaches of choice |
| Absence of information concerning histology or proliferation index |
| Absence of information concerning tumor microenvironment |
| Difficulty in systematically performing immunocytochemistry and in situ hybridation circulating tumor cell (CTCs) |
| Technological problems in identifying mutations in genes of interest in circulating tumor DNA (ctDNA) |
| Potentially miss biomarkers expressed in the tumor |
| Many studies retrospective have been performed to prove the clinical utility |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Lázaro, D.; García Hernández, J.L.; Caballero García, A.; Caballero del Castillo, A.; Villaverde Hueso, M.; Cruz-Hernández, J.J. Clinical Perspective and Translational Oncology of Liquid Biopsy. Diagnostics 2020, 10, 443. https://doi.org/10.3390/diagnostics10070443
Fernández-Lázaro D, García Hernández JL, Caballero García A, Caballero del Castillo A, Villaverde Hueso M, Cruz-Hernández JJ. Clinical Perspective and Translational Oncology of Liquid Biopsy. Diagnostics. 2020; 10(7):443. https://doi.org/10.3390/diagnostics10070443
Chicago/Turabian StyleFernández-Lázaro, Diego, Juan Luis García Hernández, Alberto Caballero García, Aurora Caballero del Castillo, María Villaverde Hueso, and Juan Jesús Cruz-Hernández. 2020. "Clinical Perspective and Translational Oncology of Liquid Biopsy" Diagnostics 10, no. 7: 443. https://doi.org/10.3390/diagnostics10070443
APA StyleFernández-Lázaro, D., García Hernández, J. L., Caballero García, A., Caballero del Castillo, A., Villaverde Hueso, M., & Cruz-Hernández, J. J. (2020). Clinical Perspective and Translational Oncology of Liquid Biopsy. Diagnostics, 10(7), 443. https://doi.org/10.3390/diagnostics10070443







