Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Blood Culture Criteria
2.3. Statistical Analysis
2.4. Experimental Methodology
3. Results
3.1. Patient Characteristics
3.2. Feature Selection and Risk Classification
3.3. Subgroup Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nelson, D.S.; Walsh, K.; Fleisher, G.R. Spectrum and frequency of pediatric illness presenting to a general community hospital emergency department. Pediatrics 1992, 90, 5–10. [Google Scholar] [PubMed]
- Massin, M.M.; Montesanti, J.; Gerard, P.; Lepage, P. Spectrum and frequency of illness presenting to a pediatric emergency department. Acta Clin. Belg. 2006, 61, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Baraff, L.J. Management of fever without source in infants and children. Ann. Emerg. Med. 2000, 36, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, S.E.; Moons, K.G.; Derksen-Lubsen, G.; Grobbee, D.E.; Moll, H.A. Predicting serious bacterial infection in young children with fever without apparent source. Acta Paediatr. 2001, 90, 1226–1232. [Google Scholar] [CrossRef]
- Bass, J.W.; Steele, R.W.; Wittler, R.R.; Weisse, M.E.; Bell, V.; Heisser, A.H.; Brien, J.H.; Fajardo, J.E.; Wasserman, G.M.; Vincent, J.M.; et al. Antimicrobial treatment of occult bacteremia: A multicenter cooperative study. Pediatr. Infect. Dis. J. 1993, 12, 466–473. [Google Scholar] [CrossRef]
- Woods-Hill, C.Z.; Fackler, J.; Nelson McMillan, K.; Ascenzi, J.; Martinez, D.A.; Toerper, M.F.; Voskertchian, A.; Colantuoni, E.; Klaus, S.A.; Levin, S.; et al. Association of a Clinical Practice Guideline with Blood Culture Use in Critically Ill Children. JAMA Pediatr. 2017, 171, 157–164. [Google Scholar] [CrossRef]
- Parikh, K.; Davis, A.B.; Pavuluri, P. Do we need this blood culture? Hosp. Pediatr. 2014, 4, 78–84. [Google Scholar] [CrossRef]
- Thuler, L.C.; Jenicek, M.; Turgeon, J.P.; Rivard, M.; Lebel, P.; Lebel, M.H. Impact of a false positive blood culture result on the management of febrile children. Pediatr. Infect. Dis. J. 1997, 16, 846–851. [Google Scholar] [CrossRef]
- Min, H.; Park, C.S.; Kim, D.S.; Kim, K.H. Blood culture contamination in hospitalized pediatric patients: A single institution experience. Korean J. Pediatr. 2014, 57, 178–185. [Google Scholar] [CrossRef]
- Self, W.H.; Mickanin, J.; Grijalva, C.G.; Grant, F.H.; Henderson, M.C.; Corley, G.; Blaschke Ii, D.G.; McNaughton, C.D.; Barrett, T.W.; Talbot, T.R.; et al. Reducing blood culture contamination in community hospital emergency departments: A multicenter evaluation of a quality improvement intervention. Acad. Emerg. Med. 2014, 21, 274–282. [Google Scholar] [CrossRef]
- Hall, R.T.; Domenico, H.J.; Self, W.H.; Hain, P.D. Reducing the blood culture contamination rate in a pediatric emergency department and subsequent cost savings. Pediatric 2013, 131, e292–e297. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, P.H. Blood culture contamination: A clinical and financial burden. Infect. Control. Hosp. Epidemiol. 2013, 34, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Segal, G.S.; Chamberlain, J.M. Resource utilization and contaminated blood cultures in children at risk for occult bacteremia. Arch. Pediatr. Adolesc. Med. 2000, 154, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Baraff, L.J.; Bass, J.W.; Fleisher, G.R.; Klein, J.O.; McCracken, G.H., Jr.; Powell, K.R.; Schriger, D.L. Practice guideline for the management of infants and children 0 to 36 months of age with fever without source. Agency for Health Care Policy and Research. Ann. Emerg. Med. 1993, 22, 1198–1210. [Google Scholar] [CrossRef]
- Pulliam, P.N.; Attia, M.W.; Cronan, K.M. C-reactive protein in febrile children 1 to 36 months of age with clinically undetectable serious bacterial infection. Pediatric 2001, 108, 1275–1279. [Google Scholar] [CrossRef]
- van Rossum, A.M.; Wulkan, R.W.; Oudesluys-Murphy, A.M. Procalcitonin as an early marker of infection in neonates and children. Lancet Infect. Dis. 2004, 4, 620–630. [Google Scholar] [CrossRef]
- Rey, C.; Los Arcos, M.; Concha, A.; Medina, A.; Prieto, S.; Martinez, P.; Prieto, B. Procalcitonin and C-reactive protein as markers of systemic inflammatory response syndrome severity in critically ill children. Intensive Care Med. 2007, 33, 477–484. [Google Scholar] [CrossRef]
- Andreola, B.; Bressan, S.; Callegaro, S.; Liverani, A.; Plebani, M.; Da Dalt, L. Procalcitonin and C-reactive protein as diagnostic markers of severe bacterial infections in febrile infants and children in the emergency department. Pediatr. Infect. Dis. J. 2007, 26, 672–677. [Google Scholar] [CrossRef]
- Bradley, J.S.; Byington, C.L.; Shah, S.S.; Alverson, B.; Carter, E.R.; Harrison, C.; Kaplan, S.L.; Mace, S.E.; McCracken, G.H., Jr.; Moore, M.R.; et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: Clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin. Infect. Dis. 2011, 53, e25–e76. [Google Scholar] [CrossRef]
- Shah, S.S.; Dugan, M.H.; Bell, L.M.; Grundmeier, R.W.; Florin, T.A.; Hines, E.M.; Metlay, J.P. Blood cultures in the emergency department evaluation of childhood pneumonia. Pediatr. Infect. Dis. J. 2011, 30, 475–479. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Towns, M.L.; Quartey, S.M.; Mirrett, S.; Reimer, L.G.; Parmigiani, G.; Reller, L.B. The clinical significance of positive blood cultures in the 1990s: A prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1997, 24, 584–602. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P. Blood culture contamination: Persisting problems and partial progress. J. Clin. Microbiol. 2003, 41, 2275–2278. [Google Scholar] [CrossRef]
- Pavlovsky, M.; Press, J.; Peled, N.; Yagupsky, P. Blood culture contamination in pediatric patients: Young children and young doctors. Pediatr. Infect. Dis. J. 2006, 25, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Pien, B.C.; Sundaram, P.; Raoof, N.; Costa, S.F.; Mirrett, S.; Woods, C.W.; Reller, L.B.; Weinstein, M.P. The clinical and prognostic importance of positive blood cultures in adults. Am. J. Med. 2010, 123, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.; Bulloch, B.; Smith, M. Prevalence of occult bacteremia in children aged 3 to 36 months presenting to the emergency department with fever in the postpneumococcal conjugate vaccine era. Acad. Emerg. Med. Off. J. Soc. Acad. Emerg. Med. 2009, 16, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Gomez, B.; Hernandez-Bou, S.; Garcia-Garcia, J.J.; Mintegi, S. Bacteraemia Study Working Group from the Infectious Diseases Working Group, S.S.O.P.E. Bacteremia in previously healthy children in emergency departments: Clinical and microbiological characteristics and outcome. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2015, 34, 453–460. [Google Scholar] [CrossRef]
- Sard, B.; Bailey, M.C.; Vinci, R. An analysis of pediatric blood cultures in the postpneumococcal conjugate vaccine era in a community hospital emergency department. Pediatr. Emerg. Care 2006, 22, 295–300. [Google Scholar] [CrossRef]
- Mintegi, S.; Benito, J.; Sanchez, J.; Azkunaga, B.; Iturralde, I.; Garcia, S. Predictors of occult bacteremia in young febrile children in the era of heptavalent pneumococcal conjugated vaccine. Eur. J. Emerg. Med. Off. J. Eur. Soc. Emerg. Med. 2009, 16, 199–205. [Google Scholar] [CrossRef]
- Irwin, A.D.; Drew, R.J.; Marshall, P.; Nguyen, K.; Hoyle, E.; Macfarlane, K.A.; Wong, H.F.; Mekonnen, E.; Hicks, M.; Steele, T.; et al. Etiology of childhood bacteremia and timely antibiotics administration in the emergency department. Pediatrics 2015, 135, 635–642. [Google Scholar] [CrossRef]
- Strand, C.L.; Wajsbort, R.R.; Sturmann, K. Effect of iodophor vs iodine tincture skin preparation on blood culture contamination rate. JAMA 1993, 269, 1004–1006. [Google Scholar] [CrossRef]
- Jaye, D.L.; Waites, K.B. Clinical applications of C-reactive protein in pediatrics. Pediatr. Infect. Dis. J. 1997, 16, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Du Clos, T.W. Function of C-reactive protein. Ann. Med. 2000, 32, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Danino, D.; Rimon, A.; Scolnik, D.; Grisaru-Soen, G.; Glatstein, M. Does extreme leukocytosis predict serious bacterial infections in infants in the post-pneumococcal vaccine era? The experience of a large, tertiary care pediatric hospital. Pediatr. Emerg. Care 2015, 31, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.Y.; Cho, H.R.; Lee, J.S.; Ryu, J.M.; Lee, J. High Concentration of C-Reactive Protein Is Associated With Serious Bacterial Infection in Previously Healthy Children Aged 3 to 36 Months With Fever and Extreme Leukocytosis. Pediatr. Emerg. Care 2017. [Google Scholar] [CrossRef]
- Shaoul, R.; Lahad, A.; Tamir, A.; Lanir, A.; Srugo, I. C reactive protein (CRP) as a predictor for true bacteremia in children. Med. Sci. Monit. 2008, 14, CR255–CR261. [Google Scholar]
- Buendia, J.A.; Sanchez-Villamil, J.P.; Urman, G. Cost-effectiveness of diagnostic strategies of severe bacterial infection in infants with fever without a source. Biomed. Rev. Del. Inst. Nac. De Salud 2016, 36, 406–414. [Google Scholar] [CrossRef]
- Weiss, G.; Goodnough, L.T. Anemia of chronic disease. New Engl. J. Med. 2005, 352, 1011–1023. [Google Scholar] [CrossRef]
- Jansson, L.T.; Kling, S.; Dallman, P.R. Anemia in children with acute infections seen in a primary care pediatric outpatient clinic. Pediatr. Infect. Dis. 1986, 5, 424–427. [Google Scholar] [CrossRef]
- Ballin, A.; Lotan, A.; Serour, F.; Ovental, A.; Boaz, M.; Senecky, Y.; Rief, S. Anemia of acute infection in hospitalized children-no evidence of hemolysis. J. Pediatr. Hematol. /Oncol. 2009, 31, 750–752. [Google Scholar] [CrossRef]
- Nemeth, E.; Ganz, T. Anemia of inflammation. Hematol. Oncol. Clin. N. Am. 2014, 28, 671–681. [Google Scholar] [CrossRef]
- Zierk, J.; Arzideh, F.; Rechenauer, T.; Haeckel, R.; Rascher, W.; Metzler, M.; Rauh, M. Age- and sex-specific dynamics in 22 hematologic and biochemical analytes from birth to adolescence. Clin. Chem. 2015, 61, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Lanzkowsky, P. Lanzkowsky’s Manual of Pediatric Hematology and Oncology; Elsevier: Boston, MA, USA, 2016. [Google Scholar]
- Bleeker, J.S.; Hogan, W.J. Thrombocytosis: Diagnostic evaluation, thrombotic risk stratification, and risk-based management strategies. Thrombosis 2011, 2011, 536062. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.R.; Petersen, N.J.; Gardner, T.J.; Hamill, R.J.; Trautner, B.W. Etiology of thrombocytosis in a general medicine population: Analysis of 801 cases with emphasis on infectious causes. J. Clin. Med. Res. 2012, 4, 415–423. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schattner, A.; Kadi, J.; Dubin, I. Reactive thrombocytosis in acute infectious diseases: Prevalence, characteristics and timing. Eur. J. Intern. Med. 2019, 63, 42–45. [Google Scholar] [CrossRef]
- Zeidler, C.; Kanz, L.; Hurkuck, F.; Rittmann, K.L.; Wildfang, I.; Kadoya, T.; Mikayama, T.; Souza, L.; Welte, K. In vivo effects of interleukin-6 on thrombopoiesis in healthy and irradiated primates. Blood 1992, 80, 2740–2745. [Google Scholar] [CrossRef]
- Bass, D.A.; Gonwa, T.A.; Szejda, P.; Cousart, M.S.; DeChatelet, L.R.; McCall, C.E. Eosinopenia of acute infection: Production of eosinopenia by chemotactic factors of acute inflammation. J. Clin. Invest. 1980, 65, 1265–1271. [Google Scholar] [CrossRef]
- Lipkin, W.I. Eosinophil counts in bacteremia. Arch. Intern. Med. 1979, 139, 490–491. [Google Scholar] [CrossRef]
- Weinberg, A.G.; Rosenfeld, C.R.; Manroe, B.L.; Browne, R. Neonatal blood cell count in health and disease. II. Values for lymphocytes, monocytes, and eosinophils. J. Pediatr. 1985, 106, 462–466. [Google Scholar] [CrossRef]
- Setterberg, M.J.; Newman, W.; Potti, A.; Smego, R.A., Jr. Utility of eosinophil count as predictor of bacteremia. Clin. Infect. Dis. 2004, 38, 460–461. [Google Scholar] [CrossRef]
- Terradas, R.; Grau, S.; Blanch, J.; Riu, M.; Saballs, P.; Castells, X.; Horcajada, J.P.; Knobel, H. Eosinophil count and neutrophil-lymphocyte count ratio as prognostic markers in patients with bacteremia: A retrospective cohort study. PLoS ONE 2012, 7, e42860. [Google Scholar] [CrossRef] [PubMed]
- Zeretzke, C.M.; McIntosh, M.S.; Kalynych, C.J.; Wylie, T.; Lott, M.; Wood, D. Reduced use of occult bacteremia blood screens by emergency medicine physicians using immunization registry for children presenting with fever without a source. Pediatr. Emerg. Care 2012, 28, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Klein-Kremer, A.; Goldman, R.D. Return visits to the emergency department among febrile children 3 to 36 months of age. Pediatr. Emerg. Care 2011, 27, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.H.; Kim, J.H.; Lee, J.Y.; Kim, Y.J.; Sohn, C.H.; Lim, K.S.; Kim, W.Y. Low utility of blood culture in pediatric community-acquired pneumonia: An observational study on 2705 patients admitted to the emergency department. Medicine 2017, 96, e7028. [Google Scholar] [CrossRef] [PubMed]
- McCulloh, R.J.; Koster, M.P.; Yin, D.E.; Milner, T.L.; Ralston, S.L.; Hill, V.L.; Alverson, B.K.; Biondi, E.A. Evaluating the use of blood cultures in the management of children hospitalized for community-acquired pneumonia. PLoS ONE 2015, 10, e0117462. [Google Scholar] [CrossRef]
- Gendrel, D.; Bohuon, C. Procalcitonin as a marker of bacterial infection. Pediatr. Infect. Dis. J. 2000, 19, 679–687. [Google Scholar] [CrossRef]
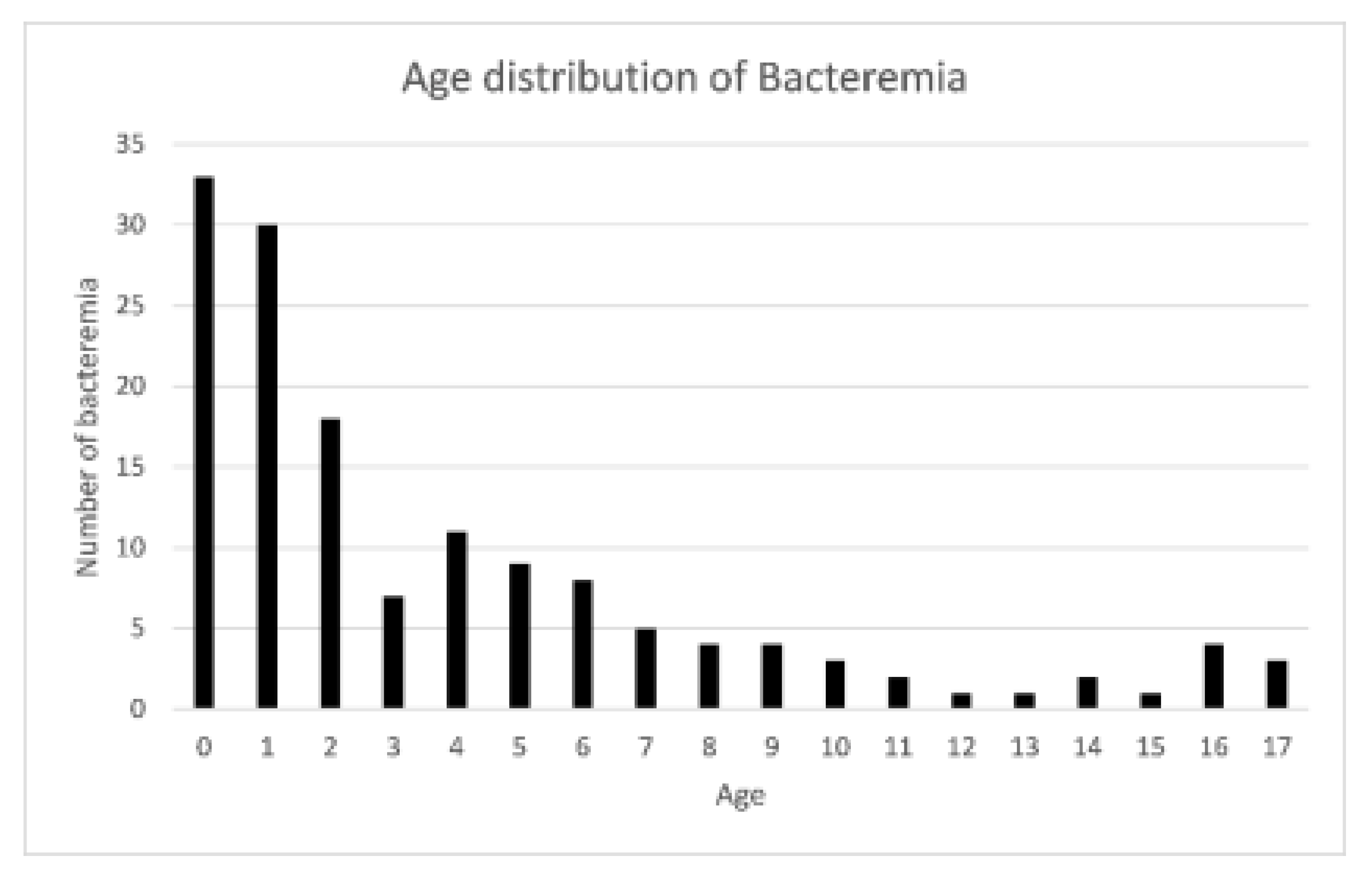
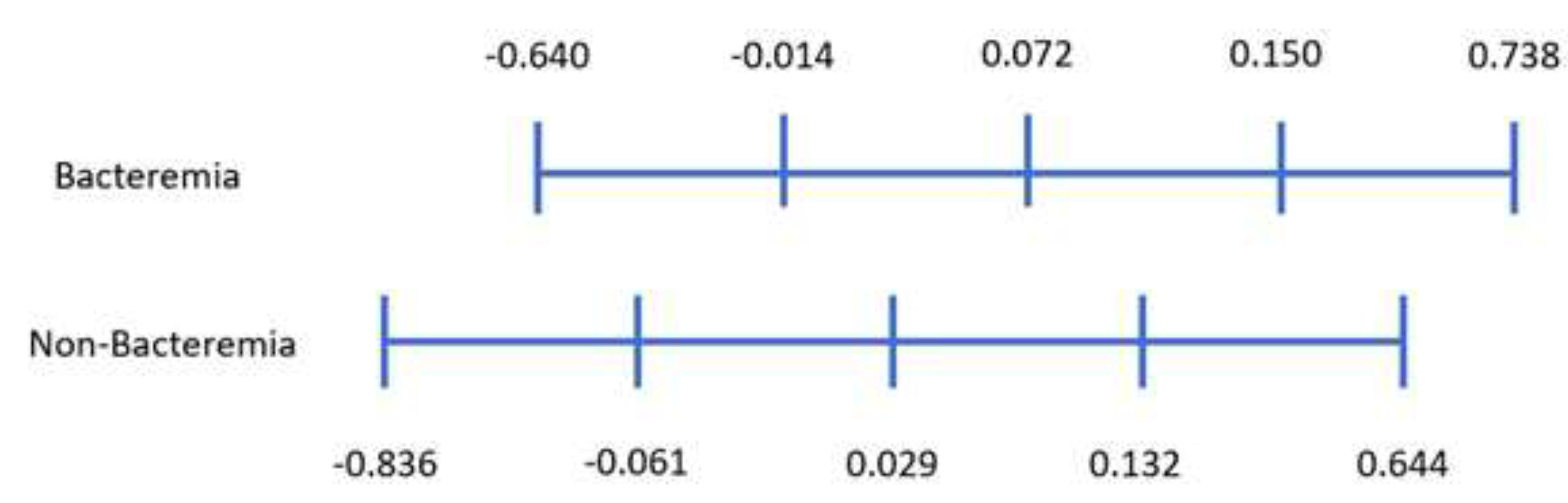
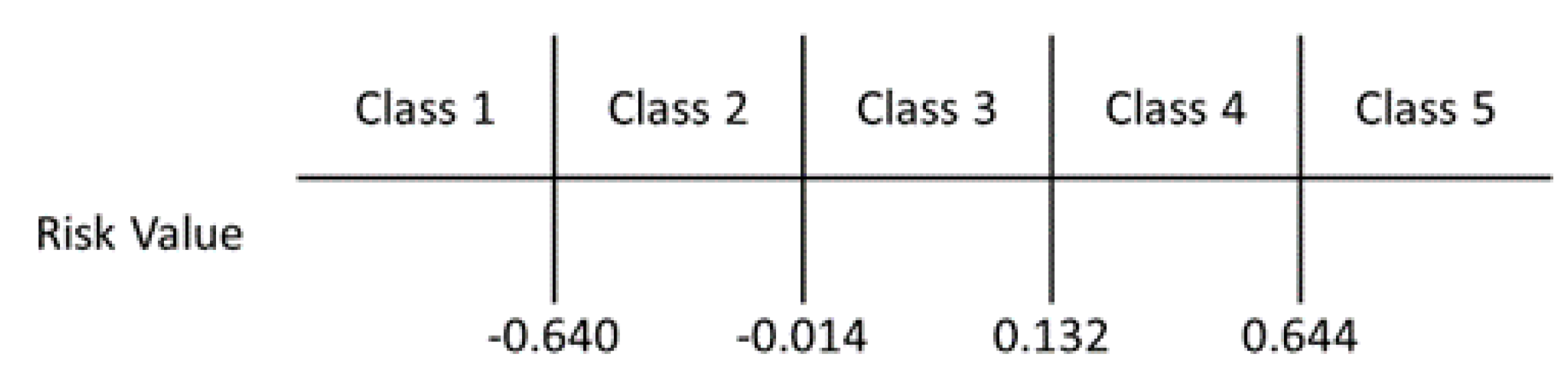
| Variable | Bacteremia | Non-Bacteremia | p-Value |
|---|---|---|---|
| Demographics | N = 146 | N = 16,821 | |
| Age | 3.85 ± 4.50 | 5.17 ± 3.97 | <0.001 |
| Gender = Male | 80 (54.79%) | 9308 (55.34%) | 0.896 |
| Gender = Female | 66 (45.21%) | 7513 (44.66%) | |
| Laboratory Values | |||
| WBC (103/μL) | 12.14 ± 6.92 | 11.00 ± 7.33 | 0.062 |
| Neutrophil (%) | 56.35 ± 24.04 | 62.06 ± 19.67 | 0.005 |
| Lymphocyte (%) | 28.93 ± 19.18 | 26.64 ± 16.37 | 0.152 |
| Band (%) | 0.76 ± 1.86 | 0.34 ± 1.54 | 0.007 |
| Monocyte (%) | 7.54 ± 5.38 | 7.80 ± 3.98 | 0.561 |
| Eosinophil (%) | 0.69 ± 1.28 | 0.93 ± 1.57 | 0.067 |
| Basophil (%) | 0.22 ± 0.33 | 0.24 ± 0.36 | 0.501 |
| ANC (103/μL) | 8.57 ± 3.60 | 9.36 ± 2.95 | 0.009 |
| Hemoglobin (g/dL) | 11.68 ± 1.63 | 12.27 ± 1.35 | <0.001 |
| MCV (fL) | 80.51 ± 7.50 | 79.76 ± 6.66 | 0.179 |
| MCH (pg) | 27.19 ± 3.02 | 26.95 ± 2.55 | 0.339 |
| MCHC (g/L) | 33.71 ± 1.38 | 33.75 ± 1.02 | 0.646 |
| Platelet (103/μL) | 279.28 ± 119.06 | 255.50 ± 93.04 | 0.017 |
| AST (U/L) | 42.88 ± 36.44 | 37.45 ± 39.94 | 0.101 |
| ALT (U/L) | 27.75 ± 28.15 | 22.35 ± 34.04 | 0.056 |
| CRP (mg/L) | 53.59 ± 68.24 | 36.46 ± 52.04 | 0.003 |
| Variable | Minimum–Maximum | (Mean ± SD) | ||||||
|---|---|---|---|---|---|---|---|---|
| AGE | −0.0092 | – | 0.0369 | ( | 0.0155 | ± | 0.0084 | ) |
| WBC | −0.1275 | – | −0.0323 | ( | −0.0822 | ± | 0.0176 | ) |
| CRP | 0.0020 | – | 0.0059 | ( | 0.0039 | ± | 0.0008 | ) |
| Hemoglobin | −0.4393 | – | 0.3613 | ( | 0.0119 | ± | 0.1562 | ) |
| MCV | −0.7722 | – | −0.2038 | ( | −0.5010 | ± | 0.1216 | ) |
| MCH | 0.4870 | – | 2.1123 | ( | 1.4471 | ± | 0.3296 | ) |
| MCHC | −1.7403 | – | −0.3591 | ( | −1.0818 | ± | 0.2826 | ) |
| Platelet | −0.0002 | – | 0.0012 | ( | 0.0004 | ± | 0.0003 | ) |
| Lymphocyte | −0.0116 | – | 0.0002 | ( | −0.0053 | ± | 0.0026 | ) |
| AST | −0.0048 | – | 0.0106 | ( | 0.0008 | ± | 0.0026 | ) |
| ALT | −0.0044 | – | 0.0129 | ( | 0.0032 | ± | 0.0033 | ) |
| RBC | −1.6189 | – | 0.6399 | ( | −0.6277 | ± | 0.4279 | ) |
| Band | 0.0003 | – | 0.2040 | ( | 0.0758 | ± | 0.0364 | ) |
| Monocyte | −0.0715 | – | −0.0263 | ( | −0.0491 | ± | 0.0082 | ) |
| Eosinophil | −0.2581 | – | −0.1520 | ( | −0.1999 | ± | 0.0215 | ) |
| Basophil | −0.1549 | – | 0.5387 | ( | 0.1284 | ± | 0.1594 | ) |
| ANC | 0.0731 | – | 0.2211 | ( | 0.1424 | ± | 0.0271 | ) |
| Segment+Band | −0.0282 | – | −0.0152 | ( | −0.0212 | ± | 0.0032 | ) |
| Variable. | OR | OR (95% CI) | Number of p-Value < 0.05 | ||||
|---|---|---|---|---|---|---|---|
| AGE | 1.0156 | ( | 0.9632 | – | 1.0685 | ) | 0 |
| WBC | 0.9212 | ( | 0.8239 | – | 1.0184 | ) | 6 |
| CRP | 1.0039 | ( | 1.0008 | – | 1.0069 | ) | 88 |
| Hemoglobin | 1.0243 | ( | 0.4922 | – | 2.3101 | ) | 0 |
| MCV | 0.6104 | ( | 0.3573 | – | 1.0680 | ) | 33 |
| MCH | 4.4778 | ( | 0.8719 | – | 21.6080 | ) | 36 |
| MCHC | 0.3528 | ( | 0.1058 | – | 1.2778 | ) | 26 |
| Platelet | 1.0004 | ( | 0.9985 | – | 1.0024 | ) | 0 |
| Lymphocyte | 0.9947 | ( | 0.9819 | – | 1.0080 | ) | 0 |
| AST | 1.0008 | ( | 0.9933 | – | 1.0071 | ) | 2 |
| ALT | 1.0032 | ( | 0.9944 | – | 1.0112 | ) | 3 |
| RBC | 0.5837 | ( | 0.0609 | – | 4.5206 | ) | 0 |
| Band | 1.0795 | ( | 0.9836 | – | 1.1761 | ) | 35 |
| Monocyte | 0.9522 | ( | 0.9110 | – | 0.9920 | ) | 82 |
| Eosinophil | 0.8190 | ( | 0.6873 | – | 0.9530 | ) | 96 |
| Basophil | 1.1514 | ( | 0.6484 | – | 1.8965 | ) | 0 |
| ANC | 1.1534 | ( | 1.0014 | – | 1.3394 | ) | 43 |
| Segment+Band | 0.9790 | ( | 0.9615 | – | 0.9972 | ) | 82 |
| Variable | minimum | – | maximum | ( | Mean | ± | SD | ) |
|---|---|---|---|---|---|---|---|---|
| CRP | 0.0020 | – | 0.0088 | ( | 0.0045 | ± | 0.0010 | ) |
| MONOCYTE | −0.0689 | – | −0.0177 | ( | −0.0434 | ± | 0.0093 | ) |
| EOSINOPHIL | −0.2244 | – | −0.0950 | ( | −0.1789 | ± | 0.0263 | ) |
| Segment+Band | −0.0346 | – | −0.0112 | ( | −0.0244 | ± | 0.0044 | ) |
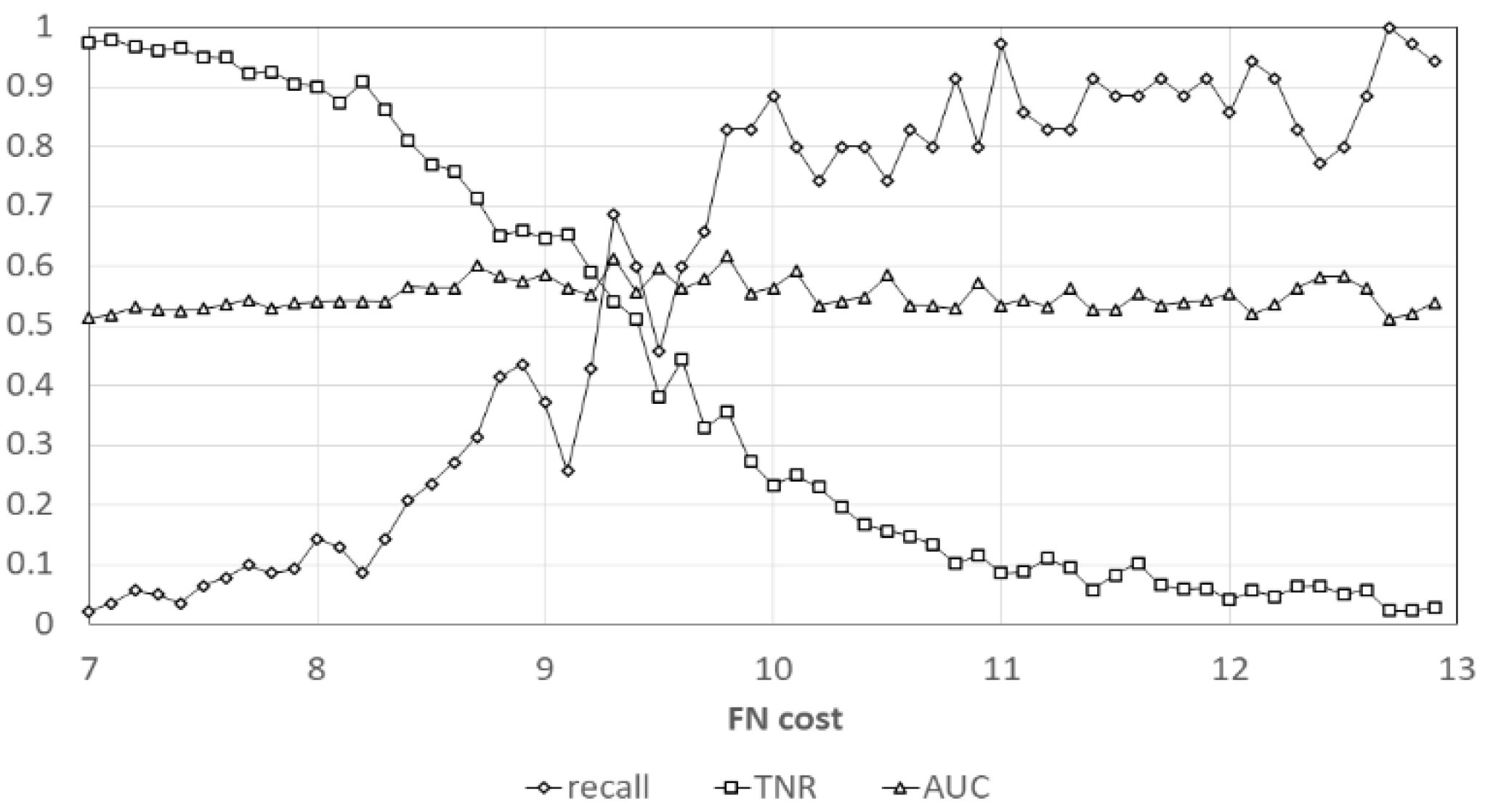
| Performance Index | Max Value |
|---|---|
| Sensitivity (recall) | 0.92 (@cost = 12) |
| Specificity (TN rate) | 0.96 (@cos = 7) |
| Positive likelihood ratio | 1.14 (@cost = 9) |
| Negative likelihood ratio | 1.25 (@cost = 11) |
| Positive predictive value | 0.013(@cost = 8) |
| Negative predictive value | 0.993 (@cost = 9) |
| AUC | 0.768 (@cost = 10) |
| Variable | Bacteremia | Non-Bacteremia | p-Value |
|---|---|---|---|
| Demographics | N = 81 | N = 5033 | |
| Age | 0.81 ± 0.78 | 1.12 ± 0.73 | <0.001 |
| Gender = Male | 43 (53.09%) | 2812 (55.87%) | 0.617 |
| Gender = Female | 38 (46.91%) | 2221 (44.13%) | |
| Laboratory Values | |||
| WBC (103/μL) | 12.76 ± 6.40 | 11.40 ± 5.98 | 0.590 |
| Neutrophil (%) | 45.63 ± 21.57 | 48.65 ± 18.75 | 0.151 |
| Lymphocyte (%) | 34.58 ± 17.99 | 37.43 ± 17.19 | 0.140 |
| Band (%) | 0.87 ± 1.88 | 0.39 ±1.64 | 0.026 |
| Monocyte (%) | 8.72 ± 6.38 | 8.91 ± 4.36 | 0.797 |
| Eosinophil (%) | 0.71 ± 1.09 | 1.07 ± 1.53 | 0.004 |
| Basophil (%) | 0.24 ± 0.40 | 0.27 ± 0.35 | 0.591 |
| ANC (103/μL) | 6.98 ± 3.20 | 7.36 ± 2.82 | 0.228 |
| Hemoglobin (g/dL) | 11.20 ± 1.49 | 11.73 ± 1.31 | <0.001 |
| MCV (fL) | 80.43 ± 8.84 | 79.07 ± 7.39 | 0.171 |
| MCH (pg) | 27.09 ± 3.25 | 26.63 ± 2.79 | 0.207 |
| MCHC (g/L) | 33.64 ± 1.05 | 33.64 ± 0.98 | 0.983 |
| Platelet (103/μL) | 296.43 ± 124.97 | 286.64 ± 112.58 | 0.438 |
| AST (U/L) | 46.43 ± 39.59 | 42.80 ± 38.53 | 0.400 |
| ALT (U/L) | 28.43 ± 25.84 | 25.26 ± 30.02 | 0.345 |
| CRP (mg/L) | 60.24 ± 71.39 | 31.47 ± 48.95 | 0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, C.-M.; Lin, C.-H.R.; Zhang, H.; Chiu, I.-M.; Cheng, C.-Y.; Yu, H.-R.; Huang, Y.-H. Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department. Diagnostics 2020, 10, 307. https://doi.org/10.3390/diagnostics10050307
Tsai C-M, Lin C-HR, Zhang H, Chiu I-M, Cheng C-Y, Yu H-R, Huang Y-H. Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department. Diagnostics. 2020; 10(5):307. https://doi.org/10.3390/diagnostics10050307
Chicago/Turabian StyleTsai, Chih-Min, Chun-Hung Richard Lin, Huan Zhang, I-Min Chiu, Chi-Yung Cheng, Hong-Ren Yu, and Ying-Hsien Huang. 2020. "Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department" Diagnostics 10, no. 5: 307. https://doi.org/10.3390/diagnostics10050307
APA StyleTsai, C.-M., Lin, C.-H. R., Zhang, H., Chiu, I.-M., Cheng, C.-Y., Yu, H.-R., & Huang, Y.-H. (2020). Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department. Diagnostics, 10(5), 307. https://doi.org/10.3390/diagnostics10050307







