Automatic Annotation of Narrative Radiology Reports
Abstract
1. Introduction
2. Materials and Methods
2.1. Radiology Reports and Labeling
2.2. Data Preprocessing
| Algorithm 1:ClusterForms(, , ) |
forw in if for k in = if if return |
2.3. Symbolic Text Classification
2.3.1. Logistic Regression
2.3.2. Support Vector Machines
2.3.3. Random Forests
2.3.4. Naïve Bayes
2.4. Semantic Text Classification
2.4.1. Word Embeddings
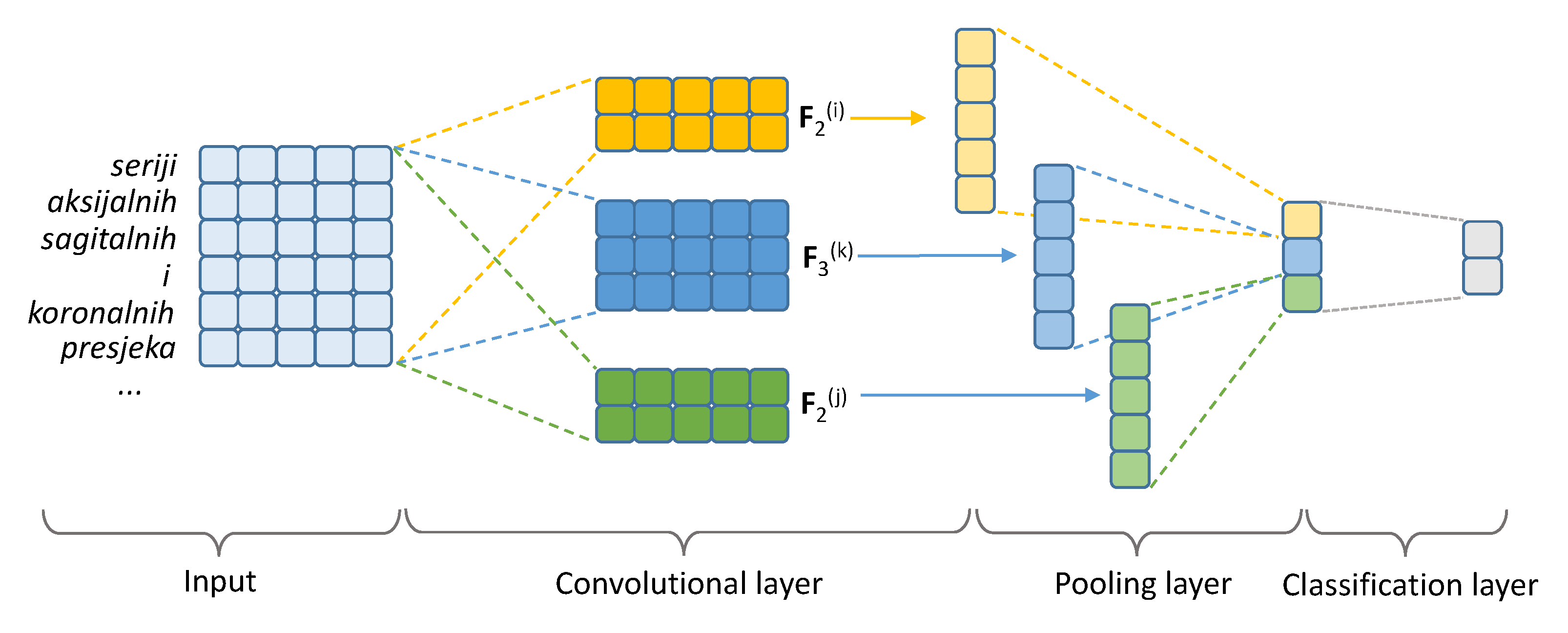
2.4.2. Convolutional Neural Network
2.5. Experimental Setup and Model Evaluation
3. Results and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Spasić, I.; Livsey, J.; Keane, J.A.; Nenadić, G. Text mining of cancer-related information: Review of current status and future directions. Int. J. Med. Informatics 2014, 83, 605–623. [Google Scholar] [CrossRef] [PubMed]
- Meystre, S.M.; Savova, G.K.; Kipper-Schuler, K.C.; Hurdle, J.F. Extracting information from textual documents in the electronic health record: A review of recent research. Yearb. Med. Inform. 2008, 8, 358–375. [Google Scholar] [CrossRef]
- Zweigenbaum, P.; Demner-Fushman, D.; Yu, H.; Cohen, K.B. Frontiers of biomedical text mining: Current progress. Brief. Bioinform. 2007, 8, 358–375. [Google Scholar] [CrossRef] [PubMed]
- Demner-Fushman, D.; Chapman, W.W.; McDonald, C.J. What can natural language processing do for clinical decision support? J. Biomed. Inform. 2009, 42, 760–772. [Google Scholar] [CrossRef]
- Pestian, J.P.; Brew, C.; Matykiewicz, P.; Hovermale, D.J.; Johnson, N.; Cohen, K.B.; Duch, W. A shared task involving multi-label classification of clinical free text. In Biological, Translational, and Clinical Language Processing; Association for Computational Linguistics: Stroudsburg, PA, USA, 2007; pp. 97–104. [Google Scholar]
- Karimi, S.; Dai, X.; Hassanzadeh, H.; Nguyen, A. Automatic Diagnosis Coding of Radiology Reports: A Comparison of Deep Learning and Conventional Classification Methods. In BioNLP 2017; Association for Computational Linguistics: Stroudsburg, PA, USA, 2017; pp. 328–332. [Google Scholar] [CrossRef]
- Lucini, F.R.S.; Fogliatto, F.C.; da Silveira, G.J.L.; Neyeloff, J.; Anzanello, M.J.; de, S.; Kuchenbecker, R.D.; Schaan, B. Text mining approach to predict hospital admissions using early medical records from the emergency department. Int. J. Med. Inform. 2017, 100, 1–8. [Google Scholar] [CrossRef]
- Koopman, B.; Zuccon, G.; Nguyen, A.; Bergheim, A.; Grayson, N. Automatic ICD-10 classification of cancers from free-text death certificates. Int. J. Med. Inform. 2015, 84, 956–965. [Google Scholar] [CrossRef]
- Botsis, T.; Nguyen, M.D.; Woo, E.J.; Markatou, M.; Ball, R. Text mining for the Vaccine Adverse Event Reporting System: Medical text classification using informative feature selection. J. Am. Med. Inform. Assoc. 2011, 18, 631–638. [Google Scholar] [CrossRef]
- Conway, M.; Doan, S.; Kawazoe, A.; Collier, N. Classifying disease outbreak reports using n-grams and semantic features. Int. J. Med. Inform. 2009, 78, 47–58. [Google Scholar] [CrossRef]
- Torii, M.; Yin, L.; Nguyen, T.; Mazumdar, C.T.; Liu, H.; Hartley, D.M.; Nelson, N.P. An exploratory study of a text classification framework for Internet-based surveillance of emerging epidemics. Int. J. Med. Inform. 2011, 80, 56–66. [Google Scholar] [CrossRef]
- Byrd, R.J.; Steinhubl, S.R.; Sun, J.; Ebadollahi, S.; Stewart, W.F. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int. J. Med. Inform. 2014, 83, 983–992. [Google Scholar] [CrossRef]
- Adams, D.Z.; Gruss, R.; Abrahams, A.S. Automated discovery of safety and efficacy concerns for joint & muscle pain relief treatments from online reviews. Int. J. Med. Inform. 2017, 100, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Seol, J.W.; Yi, W.; Choi, J.; Lee, K.S. Causality patterns and machine learning for the extraction of problem-action relations in discharge summaries. Int. J. Med. Inform. 2017, 98, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Kieseberg, P.; Weippl, E.; Tjoa, A.M. Current Advances, Trends and Challenges of Machine Learning and Knowledge Extraction: From Machine Learning to Explainable AI. In Machine Learning and Knowledge Extraction, CD-MAKE 2018, Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11015, pp. 1–8. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef]
- Holzinger, A.; Carrington, A.; Müller, H. Measuring the Quality of Explanations: The System Causability Scale (SCS). KI-Künstliche Intell. 2020, 1–6. [Google Scholar] [CrossRef]
- Köse, C.; Gençalioğlu, O.; Şevik, U. An automatic diagnosis method for the knee meniscus tears in MR images. Expert Syst. Appl. 2009, 36, 1208–1216. [Google Scholar] [CrossRef]
- Oka, H.; Muraki, S.; Akune, T.; Mabuchi, A.; Suzuki, T.; Yoshida, H.; Yamamoto, S.; Nakamura, K.; Yoshimura, N.; Kawaguchi, H. Fully automatic quantification of knee osteoarthritis severity on plain radiographs. Osteoarthr. Cartil. 2008, 16, 1300–1306. [Google Scholar] [CrossRef]
- Shamir, L.; Ling, S.M.S.; Scott, W.W.; Bos, A.; Orlov, N.; MacUra, T.J.; Eckley, D.M.; Ferrucci, L.; Goldberg, I.G. Knee X-ray image analysis method for automated detection of osteoarthritis. IEEE Trans. Biomed. Eng. 2009, 56, 407–415. [Google Scholar] [CrossRef]
- Štajduhar, I.; Mamula, M.; Miletić, D.; Ünal, G. Semi-Automated Detection of Anterior Cruciate Ligament Injury from MRI. Comput. Methods Programs Biomed. 2017, 140, 151–164. [Google Scholar] [CrossRef]
- Fripp, J.; Crozier, S.; Warfield, S.K.; Ourselin, S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans. Med. Imaging 2010, 29, 55–64. [Google Scholar] [CrossRef]
- Vincent, G.; Wolstenholme, C.; Scott, I.; Bowes, M. Fully automatic segmentation of the knee joint using active appearance models. In Medical Image Analysis for the Clinic: A Grand Challenge; CreateSpace: Scotts Valley, CA, USA, 2010; pp. 224–230. [Google Scholar]
- Jnawali, K.; Arbabshirani, M.R.; Ulloa, A.E.; Rao, N.; Patel, A.A. Automatic classification of radiological report for intracranial hemorrhage. In Proceedings of the 2019 IEEE 13th International Conference on Semantic Computing (ICSC), Newport Beach, CA, USA, 30 January–1 February 2019; pp. 187–190. [Google Scholar]
- Mujtaba, G.; Shuib, L.; Idris, N.; Hoo, W.L.; Raj, R.G.; Khowaja, K.; Shaikh, K.; Nweke, H.F. Clinical text classification research trends: Systematic literature review and open issues. Expert Syst. Appl. 2019, 116, 494–520. [Google Scholar] [CrossRef]
- Wood, D.A.; Lynch, J.; Kafiabadi, S.; Guilhem, E.; Busaidi, A.A.; Montvila, A.; Varsavsky, T.; Siddiqui, J.; Gadapa, N.; Townend, M.; et al. Automated Labelling using an Attention model for Radiology reports of MRI scans (ALARM). arXiv 2020, arXiv:2002.06588. [Google Scholar]
- Chen, M.C.; Ball, R.L.; Yang, L.; Moradzadeh, N.; Chapman, B.E.; Larson, D.B.; Langlotz, C.P.; Amrhein, T.J.; Lungren, M.P. Deep learning to classify radiology free-text reports. Radiology 2018, 286, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, I.; Ling, Y.; Chen, M.C.; Hasan, S.A.; Langlotz, C.P.; Moradzadeh, N.; Chapman, B.; Amrhein, T.; Mong, D.; Rubin, D.L.; et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif. Intell. Med. 2019, 97, 79–88. [Google Scholar] [CrossRef]
- Luo, Y. Recurrent neural networks for classifying relations in clinical notes. J. Biomed. Inform. 2017, 72, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.; Chokshi, F.H.; Lee, T.; Choi, J.D. Classification of radiology reports using neural attention models. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 4363–4370. [Google Scholar]
- Kim, Y. Convolutional Neural Networks for Sentence Classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1746–1751. [Google Scholar]
- Zhang, X.; Zhao, J.; LeCun, Y. Character-level convolutional networks for text classification. In Proceedings of the 28th International Conference on Neural Information Processing Systems (NIPS), Montreal, QC, Canada, 7–12 December 2014; pp. 649–657. [Google Scholar]
- Conneau, A.; Schwenk, H.; LeCun, Y.; Barrault, L. Very deep convolutional networks for text classification. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, Valencia, Spain, 3–7 April 2017; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2017; pp. 1107–1116. [Google Scholar]
- Šnajder, J.; Bašić, B.D.; Tadić, M. Automatic acquisition of inflectional lexica for morphological normalisation. Inf. Process. Manag. 2008, 44, 1720–1731. [Google Scholar] [CrossRef]
- Salton, G.; Buckley, C. Term-weighting approaches in automatic text retrieval. Inf. Process. Manag. 1988, 24, 513–523. [Google Scholar] [CrossRef]
- Manning, C.D.; Raghavan, P.; Schütze, H. Introduction to Information Retrieval; Cambridge University Press: Cambridge, UK, 2008; p. 506. [Google Scholar] [CrossRef]
- Vapnik, V.; Guyon, I.; Hastie, T. Support vector machines. Mach. Learn 1995, 20, 273–297. [Google Scholar]
- Platt, J. Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines; Technical Report; Microsoft Research: Redmond, WA, USA, 1998. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Domingos, P.; Pazzani, M. On the Optimality of the Simple Bayesian Classifier under Zero-One Loss. Mach. Learn. 1997, 29, 103–130. [Google Scholar] [CrossRef]
- Joachims, T. Text categorization with Support Vector Machines: Learning with many relevant features. In European Conference on Machine Learning; Springer: Berlin/Heidelberg, Germany, 1998; pp. 137–142. [Google Scholar] [CrossRef]
- Yang, Y. An Evaluation of Statistical Approaches to Text Categorization. Inf. Retr. 1999, 1, 69–90. [Google Scholar] [CrossRef]
- Mikolov, T.; Corrado, G.; Chen, K.; Dean, J. Efficient Estimation of Word Representations in Vector Space. CoRR 2013, abs/1301.3, 1–12. [Google Scholar] [CrossRef]
- Kiros, R.; Zhu, Y.; Salakhutdinov, R.R.; Zemel, R.; Urtasun, R.; Torralba, A.; Fidler, S. Skip-thought vectors. In Proceedings of the 28th International Conference on Neural Information Processing Systems (NIPS), Montreal, QC, Canada, 7–12 December 2014; pp. 3294–3302. [Google Scholar]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching word vectors with subword information. Trans. Assoc. Comput. Linguist. 2017, 5, 135–146. [Google Scholar] [CrossRef]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, (Long and Short Papers), Minneapolis, MN, USA, 2–7 June 2019; Volume 1, pp. 4171–4186. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems (NIPS), Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Ljubešić, N.; Erjavec, T. hrWaC and slWaC: Compiling web corpora for Croatian and Slovene. In International Conference on Text, Speech and Dialogue; Springer: Berlin/Heidelberg, Germany, 2011; pp. 395–402. [Google Scholar]
| Clinical Condition | Count | Observation |
|---|---|---|
| arthrosis | 820 | grade I degenerative changes, chondromalacia, pseudocyst, popliteal cyst |
| injury | 738 | patella fracture, partial ACL rupture, sprain, meniscus tear, contusion |
| degenerative disease | 160 | osteoarthritis |
| clean | 79 | lateral meniscus intact, cartilage preserved, patella positioned normally |
| inflammatory disease | 76 | osteomyelitis, septic arthritis, chronic enthesitis, Osgood–Schlatter disease, rheumatoid arthritis, discoid meniscus |
| neoplasm | 68 | sessile osteochondroma, hemangioma, enchondroma, fibroma |
| unclear | 42 | patchy areas of edema, patellar tilt |
| multicausal disease | 32 | varicose veins, chondrocalcinosis, osteochondromatosis |
| developmental anomaly | 16 | tibial developmental defect, bipartite patella, knee recurvation |
| metabolic disease | 14 | osteoporosis |
| neoplasm-like growth | 7 | fibrous cortical defect |
| idiopathic disease | 3 | ACL mucoid degeneration |
| autoimmune disease | 2 | Henoch–Schönlein purpura |
| genetic disease | 1 | osteopetrosis |
| Clinical Conditions | Count | Clinical Conditions | Count |
|---|---|---|---|
| arthrosis, injury | 306 | arthrosis, injury, degenerative dis., neoplasm | 2 |
| injury | 281 | arthrosis, injury, degenerative dis., inflammatory dis. | 2 |
| arthrosis | 261 | arthrosis, injury, degenerative dis., multicausal dis. | 2 |
| clean | 79 | arthrosis, injury, degenerative dis., developmental anomaly | 1 |
| arthrosis, injury, degenerative dis. | 66 | arthrosis, injury, neoplasm, metabolic disease | 1 |
| arthrosis, degenerative dis. | 52 | degenerative dis., developmental anomaly | 1 |
| unclear | 41 | arthrosis, developmental anomaly | 1 |
| arthrosis, neoplasm | 20 | arthrosis, injury, degenerative dis., inflammatory dis., developmental anomaly, idiopathic dis. | 1 |
| arthrosis, inflammatory dis. | 20 | developmental anomaly | 1 |
| neoplasm | 19 | arthrosis, genetic dis. | 1 |
| inflammatory dis. | 15 | arthrosis, degenerative dis., inflammatory dis., neoplasm | 1 |
| arthrosis, injury, inflammatory dis. | 15 | arthrosis, injury, degenerative dis., neoplasm-like growth | 1 |
| arthrosis, degenerative dis., inflammatory dis. | 11 | arthrosis, degenerative dis., inflammatory dis., multicausal dis. | 1 |
| injury, neoplasm | 9 | arthrosis, injury, multicausal dis., idiopathic dis. | 1 |
| arthrosis, injury, multicausal dis. | 9 | injury, neoplasm, developmental anomaly | 1 |
| arthrosis, injury, neoplasm | 7 | arthrosis, idiopathic dis. | 1 |
| arthrosis, multicausal dis. | 6 | injury, metabolic disease | 1 |
| injury, degenerative dis. | 6 | neoplasm-like growth | 1 |
| arthrosis, injury, metabolic disease | 5 | arthrosis, inflammatory dis., developmental anomaly | 1 |
| arthrosis, injury, developmental anomaly | 5 | arthrosis, degenerative dis., inflammatory dis., developmental anomaly | 1 |
| injury, inflammatory dis. | 4 | injury, neoplasm-like growth | 1 |
| arthrosis, metabolic disease | 4 | degenerative dis. | 1 |
| arthrosis, degenerative dis., multicausal dis. | 4 | unclear, neoplasm, neoplasm-like growth | 1 |
| arthrosis, injury, degenerative dis., metabolic disease | 3 | arthrosis, injury, neoplasm-like growth | 1 |
| arthrosis, degenerative dis., neoplasm | 3 | neoplasm-like growth, autoimmune dis. | 1 |
| injury, developmental anomaly | 3 | autoimmune dis. | 1 |
| arthrosis, inflammatory dis., neoplasm | 3 | multicausal dis., neoplasm-like growth | 1 |
| injury, multicausal dis. | 3 | inflammatory dis., multicausal dis. | 1 |
| multicausal dis. | 3 | arthrosis, injury, degenerative dis., neoplasm, multicausal dis. | 1 |
| uznapredovala (10), uznapredovalog (13), uznapredovalih (17), uznapredovalnim (1), uznapredovalim (5), uznapredovalu (2), uznapredovale (74), uznapredovalosti (1) uznapredovalom (5), uznapredovali (32), uznapredovao (1) |
| posteromedijalni (1), posteromedijalnog (1), posteromedijalno (9), posteromedijalnome (2) |
| infrapatelarnoj (1), infrapatelarno (29), infrapatelarnom (3), infrapatelano (1), infrapatelarnog (33), infrapatelarne (1), infrapatelanog (3) |
| ekstenzorni (1), ekstenzije (1), ekstenzora (1), ekstenziji (1), ekstenzornom (1), ekstenzivne (1), ekstenzija (1), ekstenzorne (1), ekstenzivan (1), ekstenzivno (2), ekstenzijom (5) |
| Class | NB | RF | LR | SVM | CNN |
|---|---|---|---|---|---|
| clean | 24.3 | 25.3 | 47.4 | 57.1 | 39.5 |
| unclear | 8.7 | 14.8 | 7.5 | 8.7 | 4.5 |
| arthrosis | 83.5 | 89.3 | 89.0 | 88.9 | 93.3 |
| injury | 81.3 | 82.3 | 88.3 | 87.7 | 89.2 |
| degenerative dis. | 26.4 | 94.5 | 94.8 | 72.5 | 95.9 |
| inflammatory dis. | 7.1 | 65.6 | 67.7 | 36.4 | 52.3 |
| neoplasm | 16.2 | 46.0 | 55.4 | 51.6 | 75.9 |
| multicausal dis. | 0.0 | 27.9 | 34.1 | 6.1 | 0.0 |
| developmental an. | 0.0 | 31.6 | 38.1 | 0.0 | 0.0 |
| metabolic dis. | 0.0 | 69.6 | 69.6 | 0.0 | 13.3 |
| Macro avg. | 24.8 | 54.7 | 59.2 | 40.9 | 46.4 |
| Micro avg. | 73.8 | 82.2 | 84.6 | 82.1 | 86.7 |
| Model | Class | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clean | Unclear | Arthrosis | Injury | Degenerative Dis. | Inflammatory Dis. | Neoplasm | Multicausal Dis. | Developmental An. | Metabolic Dis. | Macro Average | Micro Average | |
| Classification accuracy | ||||||||||||
| NB | 95.7 | 96.8 | 75.2 | 75.5 | 87.5 | 93.9 | 95.2 | 97.5 | 98.8 | 98.9 | 91.5 | 91.5 |
| RF | 95.0 | 96.5 | 86.2 | 79.7 | 98.7 | 96.7 | 95.8 | 97.6 | 99.0 | 99.5 | 94.5 | 94.4 |
| LR | 96.1 | 96.2 | 85.5 | 86.3 | 98.8 | 96.8 | 96.2 | 97.9 | 99.0 | 99.5 | 95.2 | 95.2 |
| SVM | 96.8 | 96.8 | 85.0 | 85.3 | 94.2 | 95.2 | 96.5 | 97.6 | 98.8 | 98.9 | 94.5 | 94.5 |
| CNN | 96.0 | 96.8 | 91.0 | 87.2 | 99.0 | 95.9 | 97.8 | 97.4 | 98.8 | 99.0 | 95.9 | 95.9 |
| Precision | ||||||||||||
| NB | 90.0 | 66.7 | 74.4 | 74.5 | 48.3 | 37.5 | 100 | 0.0 | 0.0 | 0.0 | 49.1 | 73.7 |
| RF | 47.8 | 36.4 | 91.7 | 85.3 | 98.0 | 85.4 | 71.9 | 54.5 | 100 | 88.9 | 76.0 | 89.0 |
| LR | 69.7 | 20.0 | 88.9 | 89.7 | 98.0 | 86.0 | 70.5 | 77.8 | 80.0 | 88.9 | 77.0 | 88.7 |
| SVM | 82.4 | 66.7 | 86.8 | 87.1 | 87.6 | 81.8 | 96.0 | 100 | 0.0 | 0.0 | 68.8 | 87.0 |
| CNN | 77.3 | 100 | 91.6 | 89.5 | 97.4 | 85.3 | 91.7 | 0.0 | 0.0 | 100 | 63.3 | 90.8 |
| Recall | ||||||||||||
| NB | 14.1 | 4.7 | 95.2 | 89.5 | 18.1 | 3.9 | 8.8 | 0.0 | 0.0 | 0.0 | 23.4 | 74.0 |
| RF | 17.2 | 9.3 | 86.9 | 79.5 | 91.3 | 53.2 | 33.8 | 18.8 | 18.8 | 57.1 | 46.6 | 76.4 |
| LR | 35.9 | 4.7 | 89.1 | 86.9 | 91.9 | 55.8 | 45.6 | 21.9 | 25.0 | 57.2 | 51.4 | 80.8 |
| SVM | 43.8 | 4.7 | 91.0 | 88.3 | 61.9 | 23.4 | 35.3 | 3.1 | 0.0 | 0.0 | 35.2 | 77.7 |
| CNN | 26.6 | 2.3 | 95.1 | 89.0 | 94.4 | 37.7 | 64.7 | 0.0 | 0.0 | 7.1 | 41.7 | 83.0 |
| score | ||||||||||||
| NB | 24.3 | 8.7 | 83.5 | 81.3 | 26.4 | 7.1 | 16.2 | 0.0 | 0.0 | 0.0 | 24.8 | 73.8 |
| RF | 25.3 | 14.8 | 89.3 | 82.3 | 94.5 | 65.6 | 46.0 | 27.9 | 31.6 | 69.6 | 54.7 | 82.2 |
| LR | 47.4 | 7.5 | 89.0 | 88.3 | 94.8 | 67.7 | 55.4 | 34.1 | 38.1 | 69.6 | 59.2 | 84.6 |
| SVM | 57.1 | 8.7 | 88.9 | 87.7 | 72.5 | 36.4 | 51.6 | 6.1 | 0.0 | 0.0 | 40.9 | 82.1 |
| CNN | 39.5 | 4.5 | 93.3 | 89.2 | 95.9 | 52.3 | 75.9 | 0.0 | 0.0 | 13.3 | 46.4 | 86.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krsnik, I.; Glavaš, G.; Krsnik, M.; Miletić, D.; Štajduhar, I. Automatic Annotation of Narrative Radiology Reports. Diagnostics 2020, 10, 196. https://doi.org/10.3390/diagnostics10040196
Krsnik I, Glavaš G, Krsnik M, Miletić D, Štajduhar I. Automatic Annotation of Narrative Radiology Reports. Diagnostics. 2020; 10(4):196. https://doi.org/10.3390/diagnostics10040196
Chicago/Turabian StyleKrsnik, Ivan, Goran Glavaš, Marina Krsnik, Damir Miletić, and Ivan Štajduhar. 2020. "Automatic Annotation of Narrative Radiology Reports" Diagnostics 10, no. 4: 196. https://doi.org/10.3390/diagnostics10040196
APA StyleKrsnik, I., Glavaš, G., Krsnik, M., Miletić, D., & Štajduhar, I. (2020). Automatic Annotation of Narrative Radiology Reports. Diagnostics, 10(4), 196. https://doi.org/10.3390/diagnostics10040196






