A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of Low Viral Load Clinical Specimen Mimics
2.3. Preparation of High Viral Load Contrived Clinical Specimen to Test the Effect of VTM on RT-qPCR
2.4. Automated Sample Preparation
2.5. Rapid Extraction
2.6. Using Sous Vide Immersion Heaters to Create a Portable and Low-Cost Alternative to Circulating Water Baths Used in Laboratories
2.7. Automation Using a One-Servo Device
2.8. RT-qPCR Reaction Setup
2.9. Rapid PCR with Water Baths
2.10. End-Point RT-PCR Fluorescence Analysis
2.11. Ethics Statement
3. Results and Discussion
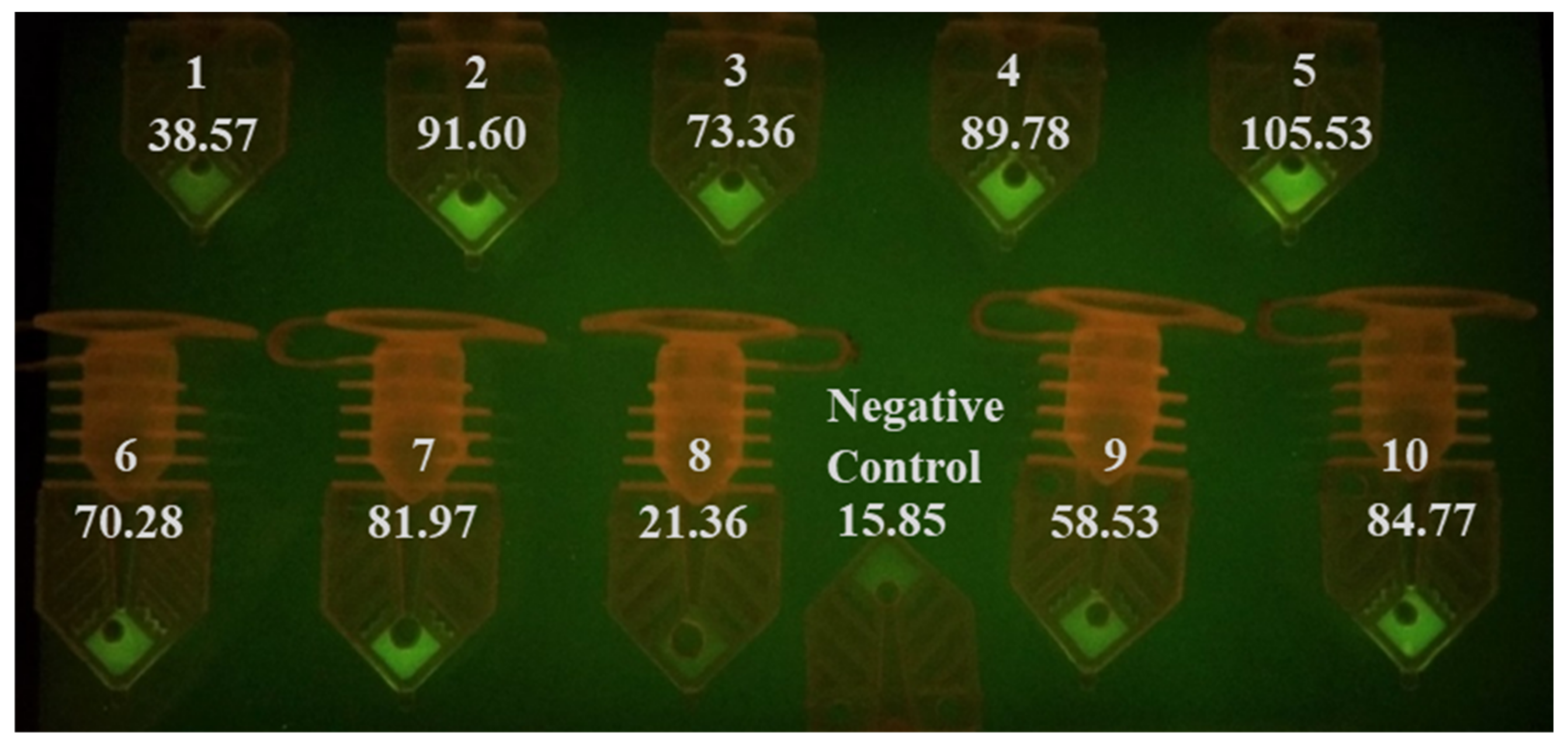
3.1. RT-qPCR Analysis of Ten COVID-19 Positive Clinical Samples Using Commercial Real-Time Thermal Cycler
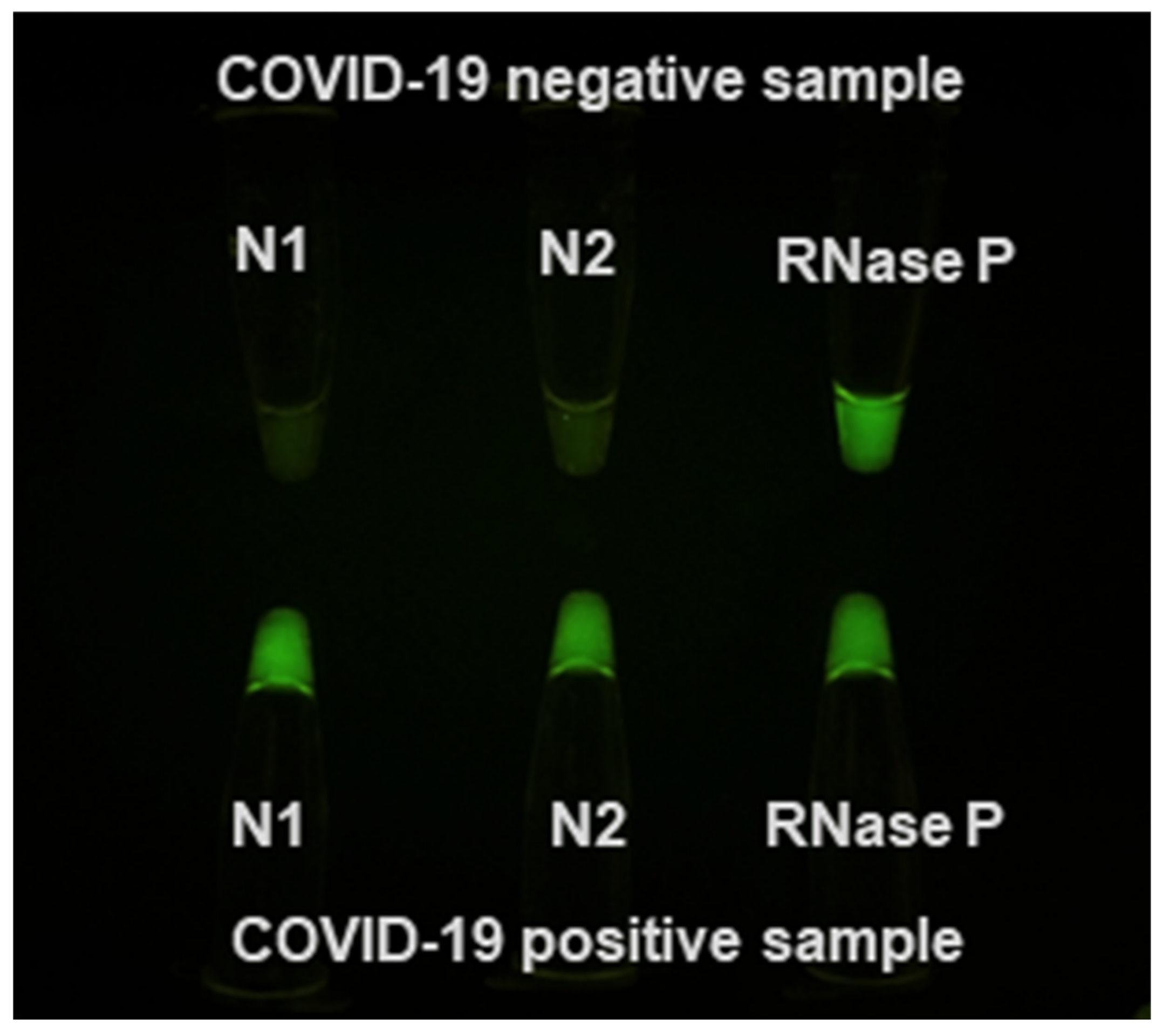
3.2. Rapid RT-PCR Detection of SARS-CoV-2 (N1 target) in Unprocessed Clinical Samples Using Water Baths
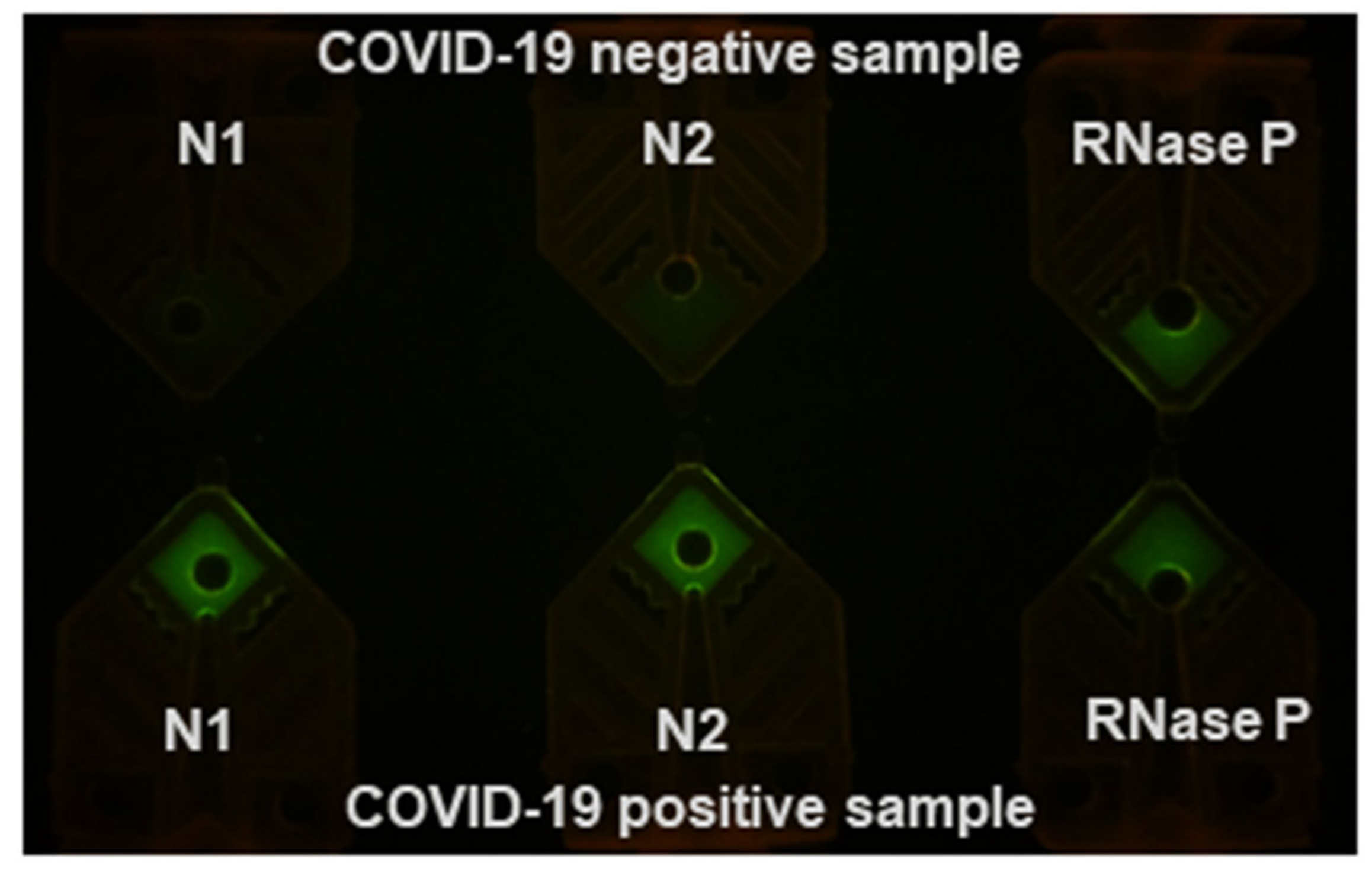
3.3. Rapid RT-PCR Detection of SARS-CoV-2 with N1, N2, and RNase P Reactions Using Water Baths
3.4. Rapid RT-PCR Detection of SARS-CoV-2 in Heat-Inactivated Clinical Samples
3.5. RT-PCR Detection of SARS-CoV-2 with A 4-Minute Extraction Protocol
3.6. Limitation Statement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Xie, C.; Jiang, L.; Huang, G.; Pu, H.; Gong, B.; Lin, H.; Ma, S.; Chen, X.; Long, B.; Si, G.; et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020, 93, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Alcoba-Florez, J.; González-Montelongo, R.; Íñigo-Campos, A.; De Artola, D.G.-M.; Gil-Campesino, H.; Ciuffreda, L.; Valenzuela-Fernández, A.; Flores, C.; The Microbiology Technical Support Team. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int. J. Infect. Dis. 2020, 97, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Bruce, E.A.; Huang, M.-L.W.; Perchetti, G.A.; Tighe, S.; Laaguiby, P.; Hoffman, J.J.; Gerrard, D.L.; Nalla, A.K.; Wei, Y.; Greninger, A.L.; et al. DIRECT RT-qPCR DETECTION OF SARS-CoV-2 RNA FROM PATIENT NASOPHARYNGEAL SWABS WITHOUT AN RNA EXTRACTION STEP 2020. bioRxiv 2020. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Muenker, M.C.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 1–7. [Google Scholar] [CrossRef]
- Luo, Z.; Ang, M.J.Y.; Chan, S.Y.; Yi, Z.; Goh, Y.Y.; Yan, S.; Tao, J.; Liu, K.; Li, X.; Zhang, H.; et al. Combating the Coronavirus Pandemic: Early Detection, Medical Treatment, and a Concerted Effort by the Global Community. Research 2020, 2020, 1–35. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Stittleburg, V.; Pond, R.; Saklawi, Y.; Sahoo, M.K.; Babiker, A.; Hussaini, L.; Kraft, C.S.; Pinsky, B.A.; Anderson, E.J.; et al. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1633–1635. [Google Scholar] [CrossRef]
- Millington, A.L.; Houskeeper, J.A.; Quackenbush, J.F.; Trauba, J.M.; Wittwer, C.T. The kinetic requirements of extreme qPCR. Biomol. Detect. Quantif. 2019, 17, 100081. [Google Scholar] [CrossRef]
- Wong, G.; Wong, I.; Chan, K.; Hsieh, Y.; Wong, S. A Rapid and Low-Cost PCR Thermal Cycler for Low Resource Settings. PLoS ONE 2015, 10, e0131701. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Wong, P.-Y.; Yu, P.; Hardick, J.; Wong, K.-Y.; Wilson, S.A.; Wu, T.; Hui, Z.; Gaydos, C.; Wong, S. A Rapid and Low-Cost PCR Thermal Cycler for Infectious Disease Diagnostics. PLoS ONE 2016, 11, e0149150. [Google Scholar] [CrossRef] [PubMed]
- Beltran-Pavez, C.; Marquez, C.L.; Munoz, G.; Valiente-Echeverría, F.; Gaggero, A.; Soto-Rifo, R.; Barriga, G.P. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wee, S.K.; Sivalingam, S.P.; Yap, E.P.H. Rapid Direct Nucleic Acid Amplification Test without RNA Extraction for SARS-CoV-2 Using a Portable PCR Thermocycler. Genes 2020, 11, 664. [Google Scholar] [CrossRef]
- Wei, S.; Kohl, E.; Djandji, A.; Morgan, S.; Whittier, S.; Mansukhani, M.M.; Hod, E.A.; D’Alton, M.; Suh, Y.; Williams, Z. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. medRxiv 2020. [Google Scholar] [CrossRef]
- Arumugam, A.; Wong, S. The Potential Use of Unprocessed Sample for RT-qPCR Detection of COVID-19 without an RNA Extraction Step. bioRxiv 2020. [Google Scholar] [CrossRef]
- Smyrlaki, I.; Ekman, M.; Lentini, A.; Vondracek, M.; Papanicoloau, N.; Aarum, J.; Safari, H.; Muradrasoli, S.; Albert, J.; Högberg, B.; et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCR. medRxiv 2020. [Google Scholar] [CrossRef]
- Brown, J.R.; Atkinson, L.; Shah, D.; Harris, K.A. Validation of an extraction-free RT-PCR protocol for detection of SARS-CoV2 RNA. medRxiv 2020. [Google Scholar] [CrossRef]
- Hasan, M.R.; Mirza, F.; Al-Hail, H.; Sundararaju, S.; Xaba, T.; Iqbal, M.; Alhussain, H.; Yassine, H.M.; Perez-Lopez, A.; Tang, P. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS ONE 2020, 15, e0236564. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Han, M.S.; Seong, M.-W.; Kim, N.; Shin, S.; Cho, S.I.; Park, H.; Kim, T.S.; Park, S.S.; Choi, E.H. Viral RNA Load in Mildly Symptomatic and Asymptomatic Children with COVID-19, Seoul. Emerg. Infect. Dis. 2020, 26, 2497–2499. [Google Scholar] [CrossRef]
- Becker, D.; Sandoval, E.; Amin, A.; De Hoff, P.; Diets, A.; Leonetti, N.; Lim, Y.W.; Elliott, C.; Laurent, L.C.; Grzymski, J.; et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv 2020. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, S.; Yang, Z.; Guan, W.; Liu, D.; Lin, Z.; Zhang, Y.; Xu, Z.; Liu, X.; Li, Y. SARS-CoV-2 Viral Load in Clinical Samples from Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2020, 201, 1435–1438. [Google Scholar] [CrossRef] [PubMed]
- Bullard, J.; Dust, K.; Funk, D.; Strong, J.E.; Alexander, D.; Garnett, L.; Boodman, C.; Bello, A.; Hedley, A.; Schiffman, Z.; et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Lv, T. Prolonged SARS-CoV-2 RNA shedding: Not a rare phenomenon. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Van Kampen, J.J.A.; Van De Vijver, D.A.; Fraaij, P.L.; Haagmans, B.L.; Lamers, M.M.; Okba, N.; Akker, J.P.V.D.; Endeman, H.; Gommers, D.A.; Cornelissen, J.J.; et al. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): Duration and key determinants. medRxiv 2020. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- The COVID-19 Investigation Team Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat. Med. 2020, 26, 861–868. [CrossRef]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Buchan, B.W.; Hoff, J.S.; Gmehlin, C.G.; Perez, A.; Faron, M.L.; Munoz-Price, L.S.; Ledeboer, N.A. Distribution of SARS-CoV-2 PCR Cycle Threshold Values Provide Practical Insight Into Overall and Target-Specific Sensitivity Among Symptomatic Patients. Am. J. Clin. Pathol. 2020, 4, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Chan, K.; Coen, M.; Hardick, J.; Gaydos, C.A.; Wong, K.-Y.; Smith, C.; Wilson, S.A.; Vayugundla, S.P.; Wong, S. Low-Cost 3D Printers Enable High-Quality and Automated Sample Preparation and Molecular Detection. PLoS ONE 2016, 11, e0158502. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Weaver, S.C.; Wong, P.-Y.; Lie, S.; Wang, E.; Guerbois, M.; Vayugundla, S.P.; Wong, S. Rapid, Affordable and Portable Medium-Throughput Molecular Device for Zika Virus. Sci. Rep. 2016, 6, 38223. [Google Scholar] [CrossRef]
- Selvaraju, S.B.; Selvarangan, R. Evaluation of Three Influenza A and B Real-Time Reverse Transcription-PCR Assays and a New 2009 H1N1 Assay for Detection of Influenza Viruses. J. Clin. Microbiol. 2010, 48, 3870–3875. [Google Scholar] [CrossRef]
- Fry, A.M.; Chittaganpitch, M.; Baggett, H.C.; Peret, T.C.T.; Dare, R.K.; Sawatwong, P.; Thamthitiwat, S.; Areerat, P.; Sanasuttipun, W.; Fischer, J.; et al. The Burden of Hospitalized Lower Respiratory Tract Infection due to Respiratory Syncytial Virus in Rural Thailand. PLoS ONE 2010, 5, e15098. [Google Scholar] [CrossRef]
- CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Centers for Disease, Control Prevention, Division of Viral Diseases. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 4 April 2020).
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1657–1665. [Google Scholar] [CrossRef]
- Jung, Y.J.; Park, G.-S.; Moon, J.H.; Ku, K.B.; Beak, S.-H.; Kim, S.; Park, E.C.; Park, D.; Lee, J.-H.; Byeon, C.W.; et al. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Nalla, A.K.; Casto, A.M.; Huang, M.-L.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2020, 58, 00557–00520. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brackney, D.; Wang, J.; Kalinich, C.C.; Ott, I.M.; Kudo, E.; Lu, P.; Venkataraman, A.; Tokuyama, M.; Moore, A.J.; et al. SalivaDirect: Simple and sensitive molecular diagnostic test for SARS-CoV-2 surveillance. medRxiv 2020. [Google Scholar] [CrossRef]



| Primers | Sequence (5′–3′) | Annealing Temperature | Product Size |
|---|---|---|---|
| 2019–nCoV_N1–F 2019–nCoV_N1–R 2019–nCoV_N1–Probe | 5′–GACCCCAAAATCAGCGAAAT–3′ 5′–TCTGGTTACTGCCAGTTGAATCTG–3′ 5′–ACCCCGCATTACGTTTGGTGGACC–3′ | 55 °C | 71 bp |
| 2019–nCoV_N2–F 2019–nCoV_N2–R 2019–nCoV_N2–Probe | 5′–TTACAAACATTGGCCGCAAA–3′ 5′–GCGCGACATTCCGAAGAA–3′ 5′–ACAATTTGCCCCCAGCGCTTCAG–3′ | 55 °C | 67 bp |
| InfA Forward InfA Reverse InfA Probe | 5′–GACCRATCCTGTCACCTCTGAC–3′ 5′–AGGGCATTYTGGACAAAKCGTCTA–3′ 5′–TGCAGTCCTCGCTCACTGGGCACG–3′ | 55 °C | 106 bp |
| InfB Forward InfB Reverse InfB Probe | 5′–TCCTCAACTCACTCTTCGAGCG–3′ 5′–CGGTGCTCTTGACCAAATTGG–3′ 5′–CCAATTCGAGCAGCTGAAACTGCGGTG–3′ | 55 °C | 102 bp |
| RSV Forward RSV Reverse RSV Probe | 5′–GGCAAATATGGAAACATACGTGAA–3′ 5′–TCTTTTTCTAGGACATTGTAYTGAACAG–3′ 5′–CTGTGTATGTGGAGCCTTCGTGAAGCT–3′ | 55 °C | 84 bp |
| RNaseP Forward RNaseP Reverse RNaseP Probe | 5′–AGATTTGGACCTGCGAGCG–3′ 5′–GAGCGGCTGTCTCCACAAGT–3′ 5′–TTCTGACCTGAAGGCTCTGCGCG–3′ | 55 °C | 65 bp |
| Process | Temperature | Time | Cycles |
|---|---|---|---|
| UNG incubation | 25 °C | 2 min | 1 |
| RT | 50 °C | 15 min | |
| Polymerase activation | 95 °C | 2 min | |
| PCR | 95 °C | 3 s | 50 |
| 55 °C | 30 s |
| Process | Temperature | Time | Cycles |
|---|---|---|---|
| RT | 53 °C | 90 s | 1 |
| Polymerase activation | 95 °C | 30 s | |
| PCR | 95 °C | 6 s | 40 or 45 |
| 55 °C | 9 s |
| Sample Number | Media | Threshold Cycle Values with Extracted Templates (n = 2) | Threshold Cycle Values with Unprocessed Samples (n = 1) |
|---|---|---|---|
| AI001 | UTM | 33.2 | 30.4 |
| AI002 | M6 VTM | 23.0 | 23.4 |
| AI003 | UTM | 30.1 | 31.9 |
| AI004 | UTM | 24.9 | 26.3 |
| AI005 | M6 VTM | 23.8 | 23.4 |
| AI006 | M6 VTM | 27.7 | 24.6 |
| AI007 | M6 VTM | 29.1 | 28.1 |
| AI008 | UTM | 34.6 | 35.9 |
| AI009 | UTM | 33.8 | 33.4 |
| AI010 | M4 VTM | 25.9 | 24.9 |
| Negative Control | VTM | N/A | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arumugam, A.; Faron, M.L.; Yu, P.; Markham, C.; Wu, M.; Wong, S. A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings. Diagnostics 2020, 10, 739. https://doi.org/10.3390/diagnostics10100739
Arumugam A, Faron ML, Yu P, Markham C, Wu M, Wong S. A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings. Diagnostics. 2020; 10(10):739. https://doi.org/10.3390/diagnostics10100739
Chicago/Turabian StyleArumugam, Arunkumar, Matthew L. Faron, Peter Yu, Cole Markham, Michelle Wu, and Season Wong. 2020. "A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings" Diagnostics 10, no. 10: 739. https://doi.org/10.3390/diagnostics10100739
APA StyleArumugam, A., Faron, M. L., Yu, P., Markham, C., Wu, M., & Wong, S. (2020). A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings. Diagnostics, 10(10), 739. https://doi.org/10.3390/diagnostics10100739







