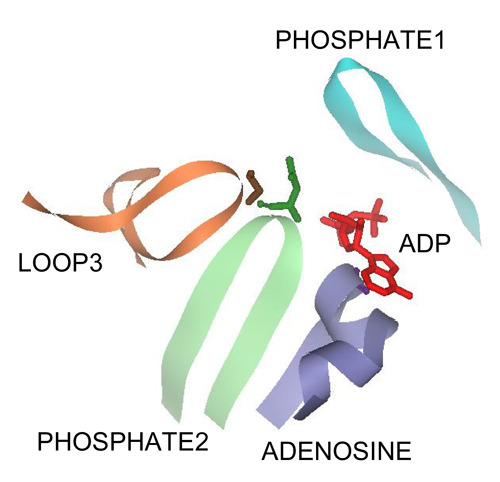
The Role of Active Site Residues in ATP Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Site-Directed Mutagenesis
2.3. Heterologous Production and Purification of MtACK Enzymes
2.4. Enzymatic Assay for ACK Activity
3. Results and Discussion
3.1. The PHOSPHATE2, ADENOSINE, and LOOP3 Regions of Methanosarcina ACK Are Highly Conserved
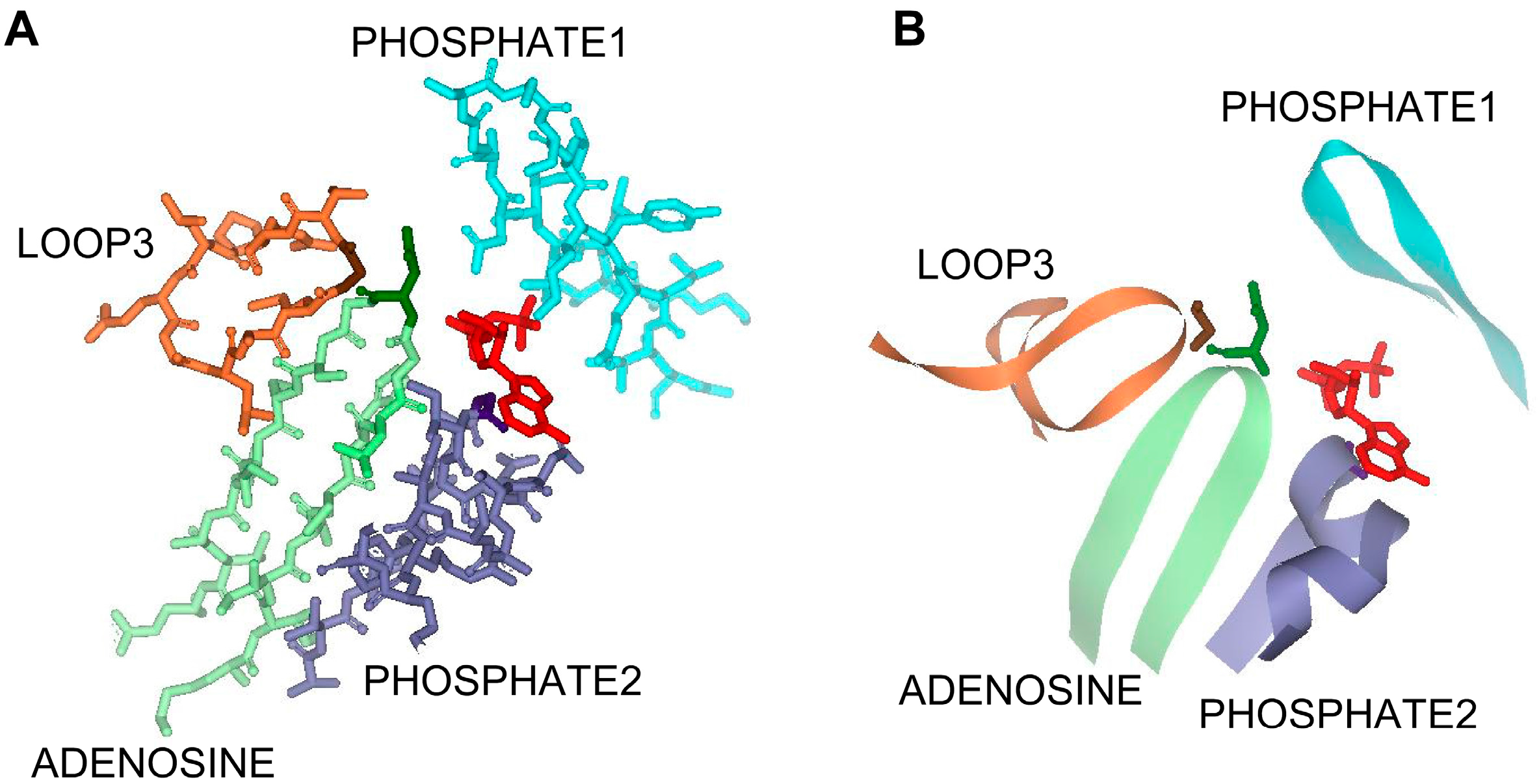
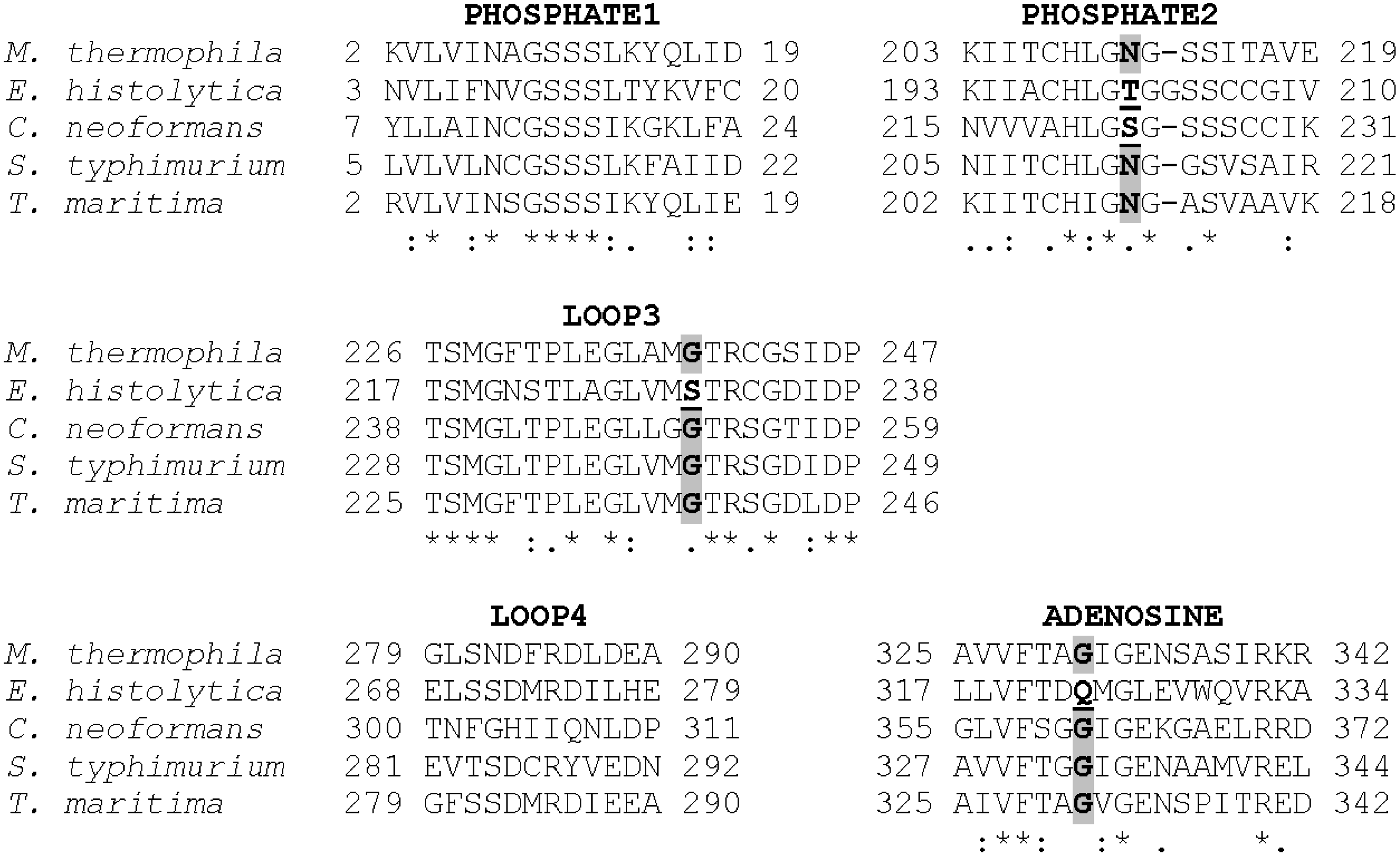
3.2. Asn211, Gly239, and Gly331 Influence NTP Preference and Utilization
| Enzyme | Specific Activity a (mol·min−1·mg−1) | Percent Activity Relative to That Observed with 10 mM ATP b | |||||
|---|---|---|---|---|---|---|---|
| ATP | CTP | GTP | TTP | UTP | ITP | ||
| Wild type | 883 ± 13 | 100.0 ± 1.4 | 50.0 ± 0.4 | 85.2 ± 1.9 | 64.5 ± 0.7 | 54.8 ± 0.5 | 80.0 ± 0.5 |
| Asn211Ala | 145 ± 1.6 | 100.0 ± 1.1 | 56.4 ± 0.6 | 65.5 ± 1.1 | 78.6 ± 2.5 | 43.0 ± 0.3 | 80.0 ± 1.1 |
| Asn211Ser | 4.4 ± 0.04 | 100.0 ± 1.0 | 56.9 ± 0.6 | 65.7 ± 0.5 | 73.9 ± 0.4 | 43.7 ± 0.2 | 83.8 ± 0.7 |
| Asn211Thr | 20 ± 0.1 | 100.0 ± 0.6 | 55.7 ± 0.2 | 62.1 ± 0.9 | 95.7 ± 0.5 | 28.4 ± 0.3 | 75.4 ± 1.4 |
| Gly239Ala | 61 ± 1.7 | 100.0 ± 2.8 | 90.9 ± 1.0 | 64.3 ± 0.6 | 142.0 ± 0.9 | 60.2 ± 1.5 | 51.8 ± 0.5 |
| Gly239Ser | 4.6 ± 0.06 | 100.0 ± 1.2 | 93.7 ± 1.0 | 72.1 ± 0.7 | 163.4 ± 1.6 | 72.8 ± 0.7 | 56.2 ± 0.4 |
| Gly331Ala | 58 ± 1.0 | 100.0 ± 1.6 | 9.7 ± 0.5 | 49.0 ± 0.4 | 8.0 ± 0.3 | 4.4 ± 0.3 | 62.5 ± 0.1 |
| Gly331Gln | 7.5 ± 0.05 | 100.0 ± 0.6 | 11.8 ± 0.9 | 46.6 ± 0.5 | 8.1 ± 0.6 | 5.1 ± 0.4 | 69.0 ± 0.5 |
| Enzyme | NTP | Km (mM) | kcat (s−1) | kcat/Km (s−1·mM−1) | Km Acetate a (mM) |
|---|---|---|---|---|---|
| WT | ATP | 4.0 ± 0.3 | 715 ± 10 | 180 ± 9 | 14.3 ± 0.5 |
| - | CTP | 3.2 ± 0.4 | 460 ± 28 | 148 ± 13 | - |
| - | GTP | 7.2 ± 0.9 | 571 ± 22 | 80 ± 7 | - |
| - | TTP | 2.7 ± 0.1 | 540 ± 9 | 202 ± 2 | - |
| - | UTP | 2.7 ± 0.3 | 415 ± 19 | 152 ± 7 | - |
| - | ITP | 4.7 ± 0.2 | 742 ± 10 | 158 ± 5 | - |
| Gly331Ala | ATP | 10.2 ± 0.4 | 88 ± 1.8 | 8.9 ± 0.18 | 601 ± 8 |
| - | CTP | 15.7 ± 1.6 | 17 ± 0.8 | 1.1 ± 0.07 | - |
| - | GTP | 5.3 ± 0.2 | 67 ± 1.0 | 12.6 ± 0.40 | - |
| - | TTP | 11.2 ± 0.5 | 20 ± 0.5 | 1.8 ± 0.05 | - |
| - | UTP | * | 43 ± 8.1 | ND | - |
| - | ITP | 10.7 ± 0.2 | 84 ± 0.4 | 7.8 ± 0.17 | - |
| Gly331Gln | ATP | 18.4 ± 0.6 | 16 ± 0.2 | 0.9 ± 0.02 | ** |
| - | CTP | 11.1 ± 0.7 | 2.1 ± 0.05 | 0.2 ± 0.01 | - |
| - | GTP | 7.4 ± 0.4 | 8.6 ± 0.15 | 1.2 ± 0.04 | - |
| - | TTP | 12.1 ± 1.0 | 2.8 ± 0.10 | 0.2 ± 0.01 | - |
| - | UTP | 14.2 ± 0.5 | 2.8 ± 0.03 | 0.2 ± 0.01 | - |
| - | ITP | 10.1 ± 0.3 | 11 ± 0.1 | 1.0 ± 0.02 | - |
| Asn211Ala | ATP | 5.9 ± 0.5 | 374 ± 13 | 63.0 ± 5.7 | 165 ± 20 |
| Asn211Ser | ATP | 6.8 ± 0.6 | 13 ± 0.8 | 1.9 ± 0.1 | 140 ± 5 |
| Asn211Thr | ATP | 7.1 ± 0.3 | 32 ± 0.7 | 4.5 ± 0.2 | 470 ± 40 |
| Gly239Ala | ATP | 1.7 ± 0.1 | 77 ± 2.1 | 45.7 ± 2.6 | 113 ± 13.4 |
| Gly239Ser | ATP | 1.8 ± 0.1 | 6.8 ± 0.05 | 3.9 ± 0.19 | 94 ± 3.0 |
3.3. Substrate Specificity in MtACK versus Other ACKs
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bork, P.; Sander, C.; Valencia, A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 7290–7294. [Google Scholar]
- Buss, K.A.; Cooper, D.R.; Ingram-Smith, C.; Ferry, J.G.; Sanders, D.A.; Hasson, M.S. Urkinase: Structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 2001, 183, 680–686. [Google Scholar] [PubMed]
- Hurley, J.H. The sugar kinase/heat shock protein 70/actin superfamily: Implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Enzymology of the fermentation of acetate to methane by Methanosarcina thermophila. Biofactors 1997, 6, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Lessner, D.J.; Li, L.; Li, Q.; Rejtar, T.; Andreev, V.P.; Reichlen, M.; Hill, K.; Moran, J.J.; Karger, B.L.; Ferry, J.G. An unconventional pathway for reduction of CO2 to methane in co-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. USA 2006, 103, 17921–17926. [Google Scholar] [CrossRef] [PubMed]
- Rother, M.; Metcalf, W.W. Anaerobic growth of Methanosarcina acetivorans C2a on carbon monoxide: An unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. USA 2004, 101, 16929–16934. [Google Scholar] [CrossRef] [PubMed]
- Blattler, W.A.; Knowles, J.R. Stereochemical course of phosphokinases. The use of adenosine [gamma-(s)-16O,17O,18O] triphosphate and the mechanistic consequences for the reactions catalyzed by glycerol kinase, hexokinase, pyruvate kinase, and acetate kinase. Biochemistry 1979, 18, 3927–3933. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.D.; Gorrell, A.; Ferry, J.G. Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 2002, 277, 22547–22552. [Google Scholar] [CrossRef] [PubMed]
- Gorrell, A.; Lawrence, S.H.; Ferry, J.G. Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 2005, 280, 10731–10742. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.D.; Iyer, P.P.; Ferry, J.G. Site-directed mutational analysis of active site residues in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 2001, 276, 45059–45064. [Google Scholar] [CrossRef] [PubMed]
- Gorrell, A.; Ferry, J.G. Investigation of the Methanosarcina thermophila acetate kinase mechanism by fluorescence quenching. Biochemistry 2007, 46, 14170–14176. [Google Scholar] [CrossRef] [PubMed]
- Thaker, T.M.; Tanabe, M.; Fowler, M.L.; Preininger, A.M.; Ingram-Smith, C.; Smith, K.S.; Iverson, T.M. Crystal structures of acetate kinases from the eukaryotic pathogens Entamoeba histolytica and Cryptococcus neoformans. J. Struct. Biol. 2013, 181, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Fowler, M.L.; Ingram-Smith, C.; Smith, K.S. Novel pyrophosphate-forming acetate kinase from the protist Entamoeba histolytica. Eukaryot. Cell 2012, 11, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Singh-Wissmann, K.; Ingram-Smith, C.; Miles, R.D.; Ferry, J.G. Identification of essential glutamates in the acetate kinase from Methanosarcina thermophila. J. Bacteriol. 1998, 180, 1129–1134. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Aceti, D.J.; Ferry, J.G. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. Evidence for regulation of synthesis. J. Biol. Chem. 1988, 263, 15444–15448. [Google Scholar] [PubMed]
- Lipmann, F.; Tuttle, L.C. A specific micromethod for determination of acyl phosphates. J. Biol. Chem. 1945, 159, 21–28. [Google Scholar]
- Rose, I.A.; Grunberg-Manago, M.; Korey, S.F.; Ochoa, S. Enzymatic phosphorylation of acetate. J. Biol. Chem. 1954, 211, 737–756. [Google Scholar] [PubMed]
- Latimer, M.T.; Ferry, J.G. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J. Bacteriol. 1993, 175, 6822–6829. [Google Scholar] [PubMed]
- Ingram-Smith, C.; Barber, R.D.; Ferry, J.G. The role of histidines in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 2000, 275, 33765–33770. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Gorrell, A.; Lawrence, S.H.; Iyer, P.; Smith, K.; Ferry, J.G. Characterization of the acetate binding pocket in the Methanosarcina thermophila acetate kinase. J. Bacteriol. 2005, 187, 2386–2394. [Google Scholar] [CrossRef] [PubMed]
- Singh-Wissmann, K.; Miles, R.D.; Ingram-Smith, C.; Ferry, J.G. Identification of essential arginines in the acetate kinase from Methanosarcina thermophila. Biochemistry 2000, 39, 3671–3677. [Google Scholar] [CrossRef] [PubMed]
- Chittori, S.; Savithri, H.S.; Murthy, M.R. Structural and mechanistic investigations on Salmonella typhimurium acetate kinase (AckA): Identification of a putative ligand binding pocket at the dimeric interface. BMC Struct. Biol. 2012, 12. [Google Scholar] [CrossRef]
- Bork, P.; Sander, C.; Valencia, A. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993, 2, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, A.; Murata, K.; Kawai, S. Structural and mutational analysis of amino acid residues involved in ATP specificity of Escherichia coli acetate kinase. J. Biosci. Bioeng. 2014, 118, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef] [PubMed]
- Glenn, K.; Ingram-Smith, C.; Smith, K.S. Biochemical and kinetic characterization of xylulose 5-phosphate/fructose 6-phosphate phosphoketolase 2 (Xfp2) from Cryptococcus neoformans. Eukaryot. Cell 2014, 13, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Ingram-Smith, C.; Martin, S.R.; Smith, K.S. Acetate kinase: Not just a bacterial enzyme. Trends Microbiol. 2006, 14, 249–253. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingram-Smith, C.; Wharton, J.; Reinholz, C.; Doucet, T.; Hesler, R.; Smith, K. The Role of Active Site Residues in ATP Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase. Life 2015, 5, 861-871. https://doi.org/10.3390/life5010861
Ingram-Smith C, Wharton J, Reinholz C, Doucet T, Hesler R, Smith K. The Role of Active Site Residues in ATP Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase. Life. 2015; 5(1):861-871. https://doi.org/10.3390/life5010861
Chicago/Turabian StyleIngram-Smith, Cheryl, Jeffrey Wharton, Christian Reinholz, Tara Doucet, Rachel Hesler, and Kerry Smith. 2015. "The Role of Active Site Residues in ATP Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase" Life 5, no. 1: 861-871. https://doi.org/10.3390/life5010861
APA StyleIngram-Smith, C., Wharton, J., Reinholz, C., Doucet, T., Hesler, R., & Smith, K. (2015). The Role of Active Site Residues in ATP Binding and Catalysis in the Methanosarcina thermophila Acetate Kinase. Life, 5(1), 861-871. https://doi.org/10.3390/life5010861