Abstract
A tripartite comparison of Archaea phylogeny and taxonomy at and above the rank order is reported: (1) the whole-genome-based and alignment-free CVTree using 179 genomes; (2) the 16S rRNA analysis exemplified by the All-Species Living Tree with 366 archaeal sequences; and (3) the Second Edition of Bergey’s Manual of Systematic Bacteriology complemented by some current literature. A high degree of agreement is reached at these ranks. From the newly proposed archaeal phyla, Korarchaeota, Thaumarchaeota, Nanoarchaeota and Aigarchaeota, to the recent suggestion to divide the class Halobacteria into three orders, all gain substantial support from CVTree. In addition, the CVTree helped to determine the taxonomic position of some newly sequenced genomes without proper lineage information. A few discrepancies between the CVTree and the 16S rRNA approaches call for further investigation.
1. Introduction
Prokaryotes are the most abundant and diverse creatures on Earth. The recognition of Archaea as one of the three main domains of life [1,2] was a milestone in the development of biology and a great success of using the 16S rRNA sequences as molecular clocks for prokaryotes, as suggested by Carl Woese and coworkers [3,4]. The Second Edition of Bergey’s Manual of Systematic Bacteriology [5] (hereafter, the Manual), a magnificent work of more than 8000 pages, took 12 years (2001–2012) to complete and is being considered by many microbiologists as the best approximation to an official classification of prokaryotes [6]. As stated in the Preface to vol. 1 of the Manual, these volumes “follow a phylogenetic framework based on analysis of the nucleotide sequence of the small ribosomal subunit RNA, rather than a phenotypic structure.” However, the “congruence” of phylogeny and taxonomy on the basis of 16S rRNA sequence analysis raises a question of principle, namely the necessity of cross-verification of whether the present classification is capable of providing a natural and objective demarcation of microbial organisms.
The answer comes with the advent of the genomic era. A whole-genome-based, alignment-free, composition vector approach to prokaryotic phylogeny, called CVTree [7,8,9,10,11,12], has produced robust phylogenetic trees that agree with prokaryotic taxonomy almost at all taxonomic ranks, from domain down to genera and species, and more importantly, many apparent disagreements have disappeared, with new taxonomic revisions appearing. In fact, all published taxonomic revisions for prokaryotes with sequenced genomes have added to the agreement of CVTree with taxonomy. A recent example from the domain Archaea was the reclassification of Thermoproteus neutrophilus to Pyrobaculum neutrophilum [13].
In this paper, we study Archaea phylogeny across many phyla. This is distinct from the phylogeny of species in a narrow range of taxa, e.g., that of vertebrates (a subphylum) or human versus close relatives (a few genera). Accordingly, the phylogeny should be compared with taxonomy at large or, as Cavalier-Smith [14] put it, with “mega-classification” of prokaryotes, focusing on taxonomy of higher ranks. Although in taxonomy, the description of a newly discovered organism necessarily starts from the lower ranks, higher rank assignments are often incomplete or lacking. At present, the ranks above class are not covered by the Bacteriological Code [15,16]. The number of plausible microbial phyla may reach hundreds, and archaeal ones are among the least studied. According to the 16S rRNA analysis, the major archaeal classes and their subordinate orders have been more or less delineated. Therefore, in order to carry out the aforementioned cross-verification, we make an emphasis on higher ranks, such as phyla, classes and orders. A study using 179 Archaea genomes provides a framework for the further study of lower ranks.
2. Material and Method
Publicly available Archaea genome sequences are the material for this study. At present, more than 30,000 prokaryotic genomes have been sequenced [17], among which, about 16,000 have been annotated [18]. These numbers keep growing and make whole-genome approaches more than ever feasible.
As of the end of 2014, there were 165 Archaea genomes released on the NCBI FTP site [19]. These genomes with corresponding lineage information from NCBI taxonomy were part of the built-in database of the CVTree web servers [20,21]. A search of NCBI databases revealed 14 more archaeal genomes; these were uploaded to the web server at run time. Archaea genomes listed in the EBI Genome Pages [22] were all included. A full list of these 179 genomes with accession numbers is given in the Appendix.
A whole-genome-based phylogeny avoids the selection of sequence segments or orthologous genes. It must be alignment-free, due to the extreme diversity of prokaryotic genome size and gene content. Our way of implementing alignment-free comparison consists of using K-peptide counts in all protein products encoded in a genome to form a raw “composition vector” (CV). The raw CV components then undergo a subtraction procedure in order to diminish the background caused by neutral mutations, hence to highlight the shaping role of natural selection [23]. Using whole genomes as input data also helps to circumvent the problem of lateral gene transfer (LGT), as the latter is merely a mechanism of genome evolution together with lineage-dependent gene loss. Being a nightmare for single- or few-protein-based phylogeny, LGT may even play a positive role in whole-genome approaches, as it takes place basically in shared ecological niches [24] and among closely-related species [25]. Plasmid genomes were excluded from our input data, thus further reducing plasmid-mediated LGT. Using whole genome input and the alignment-free method also makes CVTree a parameter-free approach. In other words, given the genomes, phylogenetic trees are generated without any adjustment of the parameters or the selection of sequence segments.
As the CVTree methodology has been elucidated in many previous publications (see, e.g., [7,8,9,10,11,12]) and a web server was released twice in 2004 [26] and 2009 [20], we will not discuss the methodological aspects of CVTree here. However, it should be understood that the peptide length K, though looking like a parameter, does not function as a parameter. For a discussion on the role of K and why K = 5, 6 leads to the best results, we refer to a recent paper [27]. All CVTree figures shown in this paper were generated at K = 6. In this paper, the term CVTree is used to denote the method [7,8,9,10,11,12,27], the web server [20,21,26] and the resulting tree; see, e.g., [28].
Traditionally, a newly generated phylogenetic tree is subject to statistical re-sampling tests, such as bootstrap and jackknife. CVTree does not use sequence alignment. Consequently, there is no way to recognize informative or non-informative sites. Instead, we take all of the protein products encoded in a genome as a sampling pool for carrying out bootstrap or jackknife tests [7]. Although it was very time-consuming, CVTrees did pass these tests well [11]. However, successfully passing of statistical re-sampling tests only informs about the stability and self-consistency of the tree with respect to small variations of the input data. It is by far not a proof of the objective correctness of the tree. Direct comparison of all branchings in a tree with an independent taxonomy at all ranks would provide such a proof. The 16S rRNA phylogeny cannot be verified by Bergey’s taxonomy, as the latter follows the former. However, the agreement of branchings in CVTree with Bergey’s taxonomy would provide much stronger support to the tree, as compared to statistical tests. This is the strategy we adopt for the CVTree approach.
There are two aspects of a phylogenetic tree: the branching order (topology) and the branch lengths. Branching order is related to classification and branch length to evolution time. Calibration of branch lengths is always associated with the assumption that the mutation rate remains more or less a constant across all species represented in a tree, an assumption that cannot hold true in a large-scale phylogenetic study, like the present one. Therefore, branching order in trees is of primary concern, whereas calibration of branch lengths makes less sense. Accordingly, all figures in this paper only show the branching scheme without the indication of branch lengths and bootstrap values.
Branching order in a tree by itself does not bring about taxonomic ranks, e.g., class or order. The latter can be assigned only after comparison with a reference taxonomy, which is not a rigid framework, but a modifiable system. Though there is a dissimilarity measure in the CVTree algorithm, it is not realistic to delineate taxa by using this measure, at least for the time being. Even if defined in the future, it must be lineage dependent. For example, it cannot be expected that the same degree of dissimilarity may be used to delineate classes in all phyla. In addition, monophyly is a guiding principle in comparing branching order with taxonomy. Here, monophyly must be understood in a pragmatic way, restricted to the given set of input data and the reference taxonomy. If all genomes from a taxon appear exclusively in a tree branch, the branch is said to be monophyletic.
In order to effectively deal with several thousands of genomes in a run, we have parallelized the CVTree algorithm and moved the web server to a computer cluster with 64 cores. The new CVTree3 web server [21] is capable of producing trees with several thousands of leaves in a few minutes for a range of K-values, say for K = 3 to 7. In addition, the CVTree3 web server has the following advanced features:
- (1)
- CVTree3 is equipped with an interactive tree display, which allows collapsing or expanding the tree branches at the disposal of the user. The user may concentrate on an interested taxon by submitting an enquiry; only the neighborhood of the taxon is expanded and all of the rest collapsed properly, keeping the topology unchanged. Here, “collapsing” means replacing a whole branch by a single leaf. Usually, a collapsed branch is labeled by the name of the highest common taxon followed by the number of strains it represents. For example, <C>Methanococci{12} denotes a class-level monophyletic branch containing 12 leaves. If a taxon name is seen in two (or more) collapsed branches, such as <C>Classname{3/12} and <C>Classname{9/12}, then the taxonomically monophyletic class does not correspond to a single branch in the collapsed tree.
- (2)
- The web server reports “convergence statistics” of all tree branches, i.e., a list of all monophyletic and non-monophyletic taxa at all taxonomic ranks for every K-value. For example, the first two lines of the report read:<D>Archaea{165}− − K5K6K7−(Numerals in curly brackets tell the number of organisms present in a collapsed branch.) Therefore, the two domains Archaea and Bacteria are both well defined as monophyletic branches at K = 5 and 6. We note that in the statistics, only genomes with complete lineage information are counted. The example project referred to in this paper contained, in addition, 14 archaeal and 143 bacterial genomes with one or more “unclassified” rank in the lineage. Therefore, in total {165 + 14}= 179 Archaea and {2707+243}= 2850 Bacteria genomes were used. The {m+n}convention is useful for looking for incomplete lineages in CVTree branches.<D>Bacteria{2707} − − K5K6 − −
- (3)
- The lineage information of an organism is given in one line with labels <D>, <P>, <C>, <O>, <F>, <G> and <S>, standing for the ranks domain, phylum, class, order, family, genus and species. The sTrain label <T> does not appear in lineage information, but may be seen in a leaf. The original lineage information of the built-in genomes was taken from the NCBI taxonomy. The lineage information of user’s genomes was provided at uploading. Users are allowed to make lineage modifications and to see new statistics after doing re-collapsing.
- (4)
- When displaying a tree, the user may pull down a lineage modification window and enter a trial lineage in the form “old_lineage new_lineage”. For example, the initial lineage for <T>Caldiarchaeum_cryptofilum_OPF8_uid58601 put it in phylum Thaumarchaeota, but there is evidence that it belongs to a new phylum, Aigarchaeota, so the modification may look like:The modification line is not required to contain all ranks, but the written part must be uniquely recognizable. By submitting the lineage modification, the user performs “re-collapsing” and gets a new report of “convergence statistics”.<P>Thaumarchaeota · · · <G>Caldiarchaeum <P>Aigarchaeota · · · <G>Caldiarchaeum
- (5)
- The user may select any part of a CVTree and produce a print-quality figure in SVG, EPS, PDF or PNG format.
All of these useful features help to reveal the agreement and discrepancy of a large tree with taxonomy.
3. Outline of Archaea Taxonomy at and above the Rank Order
The taxonomy of Archaea was described in Volume 1 of the Manual, which appeared in 2001 [29], thus being somewhat outdated. Two phyla, the Crenarchaeota and the Euryarchaeota, were listed there. The Crenarchaeota contained only one class, Thermoprotei. According to the latest information provided in the List of Prokaryotic Names with Standing in Nomenclature (LPSN [30]), the class Thermoprotei contains five orders: Thermoproteales, Desulfococcales, Sulfolobales, Acidilobales and Fervidicoccales, the last two being proposed in 2009 [31] and 2010 [32], respectively. Originally, the phylum, Euryarchaeota, contained seven classes: Methanobacteria, Methanococci, Halobacteria, Thermoplasmata, Thermococci, Archaeoglobi and Methanopyri; all comprising one order, except for Methanococci, which contained three orders. Later on, in a revised roadmap of the Manual [33], the class Methanococci was left with only one order; the other two orders became part of the newly proposed class, Methanomicrobia. A third order, Methanocellales, in the last class was proposed in 2008 [34]. Very recently, there appeared a proposal [35] to divide the single-order class, Halobacteria, into three orders.
Over the past 15 years, a few new archaeal phyla have been proposed: Korarchaeota [36,37], Thaumarchaeota [38,39,40], Nanoarchaeota [41,42,43], Aigarchaeota [44], Parvarchaeota [45] and Bathyarchaeota [46]. All but the last three phyla have been listed in LPSN [30]. We will not touch on Parvarchaeota and Bathyarchaeota, due to a lack of well-annotated genome data.
The main focus of the present study is to check and compare the positions of these high-rank taxa in CVTree and to compare them with the 16S rRNA sequence analysis where some results obtained by other authors are available.
4. Results and Discussion
4.1. 16S rRNA Archaeal Phylogeny According to All-Species Living Tree
An authoritative reference to the 16S rRNA phylogeny is the All-Species Living Tree Project (LTP) [47,48,49]. LTP is an ambitious project to construct a single 16S rRNA tree based on all available type strains of hitherto named species of Archaea and Bacteria. The latest release, LTPs115 [50], of March, 2014, was based on 366 archaeal and 9905 bacterial 16S rRNA sequences. However, the 104-page PDF of the tree is hard to comprehend, especially when it comes to comparing the tree branchings with classification at various taxonomic ranks. We fetched the treeing and lineage information files LPTs115_SSU_tree.newick and LTPs115_SSU.csv from the LTP web site [50] and then collapsed the fully-fledged tree into various taxonomic ranks where possible.
We first obtained the Archaea branch containing 366 leaves and collapsed basically to the rank class without doing lineage modification (figure not shown). In fact, it was cut from the original “All-Species Living Tree” LTPs115 [50] based on all 366 archaeal and 9905 bacterial 16S rRNA sequences.
There was a line <C>Methanomicrobia{71/72} indicating that an outlier violated the monophyly of the branch. By inspecting the figure, the outlier turned out to be:
<O>Unclassified_Methanomicrobia · · · <T>HQ896499 · · · Unclassified_Methanomicrobia
It was located next to the monophyletic <C>Thermoplasmata{8}. Therefore, it does not look like an “Unclassified_Methanomicrobia”, but might be a miss-classified Thermoplasmata. Judging by its close neighborhood, we may temporarily modify the lineage to:
<C>Thermoplasmata<O>Thermoplasmatales<F>Thermoplasmataceae<G>Methanomassiliicoccus· · ·
After making the lineage modification, we get Figure 1. The branchings in Figure 1 fully agree with the taxonomy of Archaea, as outlined in Section 3, at the phylum and class ranks. In particular, the eight classes of Euryarchaeota all behave as well-defined monophyletic branches. Further more, if one expands the class Methanomicrobia, its three subordinate orders, Methanocellales{3}, Methanosarcinales{31} and Methanomicrobiales{37}, all appear as monophyletic branches (not shown in Figure 1). The definition of orders within Thermoprotei, the only class in Crenarchaeota, is somehow problematic (more on this point near the end of Subsection 4.2).
This kind of agreement should be expected, as the archaeal taxonomy is largely based on the 16S rRNA sequence analysis. However, as by design, the LTP is restricted to type strains with validly published names, one cannot check the positions of the newly proposed phyla and those strains lacking a definite lineage. The whole-genome-based CVTree approach may complement these aspects of phylogeny, since the criterion for inclusion of a strain into the tree is the availability of a sequenced genome, independent of its standing in nomenclature. In Subsection 4.3, the CVTree results are compared with 16S rRNA analyses done by other authors.
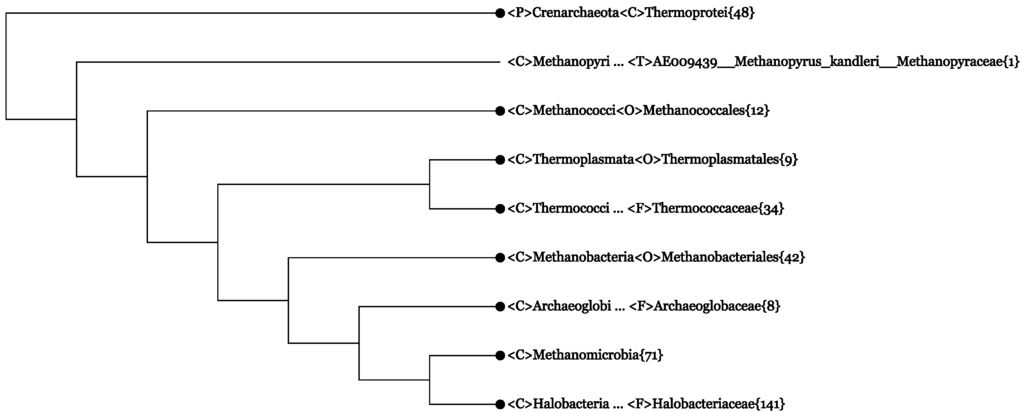
Figure 1.
The Archaea branch in the All-Species Living Tree based on 366 16S rRNA sequences. The tree has been collapsed to the rank class (<C>), and only one lineage modification has been made. Numerals in curly brackets indicate the number of sequences contained in a collapsed branch. The collapsing and lineage modification was performed by using a web server similar to CVTree3. This Living Tree Viewer is accessible to all users [51].
4.2. The Whole-Genome-Based CVTree Phylogeny
CVTrees based on 179 Archaea, 2850 Bacteria and eight Eukarya genomes were generated by using the improved version CVTree3 [21] of the web server [20]. We show the Archaea part of a big CVTree in Figure 2. When inspecting the figure, we pay more attention to the newly proposed phyla and those taxa with incomplete or suspicious lineage information.
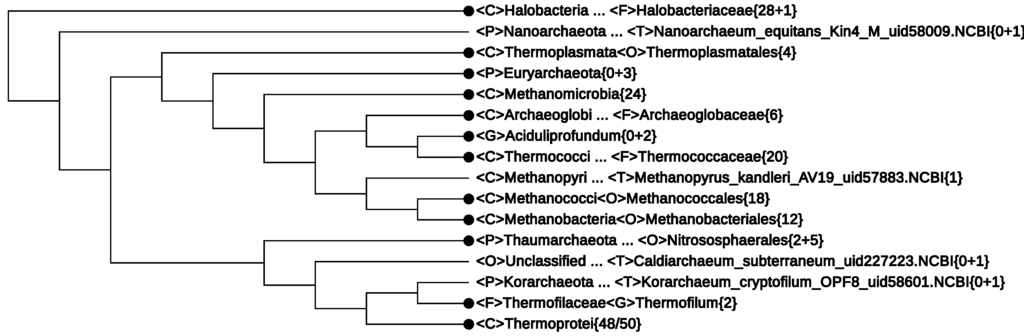
Figure 2.
The 179-genome Archaea branch of CVTree obtained by using the CVTree3 web server [21] without making lineage modifications. It has been collapsed to the rank class where possible. The branching order is to be compared with taxonomy, but does not scale the branch lengths.
In what follows, the non-monophyletic branches are summarized and possible lineage modifications are suggested.
- (1)
- The first line of Figure 2 <F>Halobacteriaceae{28+1} informs that among the 29 genomes, there was one without proper lineage information. In fact, it was Halophilic_archaeon_DL31_uid72619, a name not validly published and not following the basic rule for a binomen. Its NCBI lineage from phylum down to genus was “unclassified”. However, by expanding this line, the strain is seen to be located deeply inside the class Halobacteria (see Figure 4). As at present, the class consists of only one order, which, in turn, is made of one family [33], it is safe to assign this strain to a yet unspecified genus. This modification would yield a monophyletic branch, Halobacteria{29}.
- (2)
- The fourth line of Figure 2 <P>Euryarchaeota{0+3} represents a cluster obtained by collapsing three strains (not explicitly written in the figure):
- Thermoplasmatales_archaeon_BRNA1_uid195930, with NCBI lineage <C>Thermoplasmata<O>Unclassified<F>Unclassified;
- Candidatus_Methanomethylophilus_alvus_Mx1201_uid196597, with NCBI lineage <C>Unclassified<O>Unclassified<F>Unclassified,
- Methanomassiliicoccus_sp_Mx1_Issoire_uid207287, with NCBI lineage <C>Methanomicrobia<O>Unclassified<F>Unclassified.
If the NCBI lineage would be accepted, two of the above strains must violate the monophyly of the classes Thermoplasmata{4/5} and Methanomicrobia{24/25}. However, the fact that these three strains, taken together, make a monophyletic branch hint of the possibility to assign them to a yet unspecified class. This modification would restore the monophyly of the two classes Methanomicrobia{24} (Line 5 in Figure 2) and Thermoplasmata{4} (Line 3 in Figure 2), as seen in Figure 2. - (3)
- The newly proposed phylum, Thaumarchaeota, appears to be non-monophyletic, as an outlying strain, Candidatus Caldiarchaeum subterranum, was assigned to this phylum according to the NCBI taxonomy. The NCBI assignment might reflect its position in some phylogenetic tree based on concatenated proteins, e.g., Figure 2 in [52]. However, in the original paper reporting the discovery of this strain [44] and in recent 16S rRNA studies, e.g., [46], Candidatus Caldiarchaeum subterranum was proposed to make a new phylum, Aigarchaeota. CVTrees support the introduction of this new phylum. A lineage modification of Candidatus Caldiarchaeum subterranum from Thaumarchaeota to Aigarchaeota would lead to a monophyletic Thaumarchaeota.
- (4)
- The Candidatus genus, Aciduliprofundum, is considered a member of the DHEV2 (deep-sea hydrothermal vent euryarchaeotic 2) phylogenetic cluster. No taxonomic information was given in the original papers [53,54]. The NCBI taxonomy did not provide definite lineage information for this taxon at the class, order and family ranks. According to [53], the whole DHEV2 cluster was located close to Thermoplasmatales in a maximum-likelihood analysis of 16S rRNA sequences. A similar placement was seen in [52], where a Bayesian tree of the archaeal domain based on concatenation of 57 ribosomal proteins put a lonely Aciduliprofundum next to Thermoplasmata. However, in CVTrees, constructed for all K-values from three to nine, Aciduliprofundum is juxtaposed with the class Thermococci{18}. An observation in [54] that this organism shares a rare lipid structure with a few species from Thermococcales may hint to its possible association with the latter. If we temporarily presume a lineage:one might have a monophyletic class <C>Thermococci{20}.<C>Thermococci<O>Unclassified<F>Unclassified<G>Aciduliprofundum · · ·Since none of the 13 DHEV2 members listed in [53] have a sequenced genome so far, CVTree cannot tell the placement of the DHEV2 cluster as a whole for the time being. It remains an open problem whether DHEV2 is close to Thermoplasmata or to Thermococci or if a new class is needed to accommodate DHEV2.
- (5)
- The new phylum, Korarchaeota, violates the monophyly of the phylum, Crenarchaeota, by drawing to itself the family, Thermofilaceae. However, in an on-going study of ours (not published yet) using a much larger dataset, this violation no longer shows up; both Korarchaeota and Crenarchaeota restore their phylum status. Taking into account the fact that both Korarchaeota and Thermofilaceae are represented by single species for the time being, their placement certainly requires further study with broader sampling of genomes.However, it is worth noting that the whole lower cluster of Figure 2 supports a recent proposal for a new “TACK” superphylum [55], made of Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota.
After making all of the aforementioned lineage modifications, the resulting CVTree (not shown) looks much like Figure 2 with minor changes of some labels.
All eight classes of Euryarchaeota, as listed in Section 3, are well-defined on their own. In addition, a new class might be introduced for the three archaeons without detailed lineage information, collapsed as <P>Euryarchaeota{0+3}. The last point cannot be checked in the All-Species Living Tree without extending it to cover organisms without validly published names.
Now, it comes to inspect the orders in the single-class phylum, Crenarchaeota. There is no a priori reason to expect that 16S rRNA sequence analysis and the CVTree approach should lead to identical tree branchings. Though all being assigned to Crenarchaeota, the forty eight 16S rRNA sequences in the All-Species Living Tree and the 50 genomes in the CVTree do not belong to the same set of organisms. One can only compare those in common.
Two orders, Sulfolobales and Thermoproteales, are monophyletic in both CVTree and 16S rRNA trees, putting aside the insertion of the single-species, Korarchaeota, into Thermoproteales in CVTree. The introduction of the new orders, Acidilobales in 2009 [31] and Fervidicoccales in 2010 [32], violated the monophyly of the so-far monophyletic order, Desulfurococcales (the genus, Acidilobus, was considered part of Desulfurococcaceae before 2009). A main criterion to distinguish species of the new order from that in Desulfurococcales was indicated in [31] as acidophily, a point that might require further verification.
The CVTree results summarized above were a continuation and extension of a similar study [56] based on 62 Archaea genomes available at the beginning of 2010. The fact that, five years apart and with 117 more genomes added, the results remain consistent informs of the robustness of the CVTree approach.
4.3. Phylum Distribution in Other Phylogenies
The conclusions drawn above concerning the positions of the newly proposed phyla and organisms with uncertain lineage information cannot be directly compared with the All-Species Living Tree Project [47,48,49], as by design, LTP only includes strains with validly published names and standing in nomenclature. To this end, one must look for other published studies.
An effective way of comprehending a tree with many leaves consists of collapsing the tree branches to appropriate taxonomic ranks, as we did in Figure 1 and Figure 2. For published results of other authors, we collapsed their trees manually. Figure 3 shows four such trees collapsed to the phylum level from corresponding trees in [44] and [52]. Figure 3a is a maximum likelihood tree of concatenated SSU and LSU rRNAs using 3063 nucleotide positions; Figure 3b is a maximum likelihood tree of 45 concatenated ribosomal proteins and nine RNA polymerase subunits using 5993 aligned amino acids; and Figure 3c is a maximum likelihood tree from translation EF2 proteins based on 590 residues. All of these three subfigures were obtained by collapsing Figure 4 in [44]. Figure 3d was collapsed from a Bayesian tree based on concatenation of 67 ribosomal proteins from 89 genomes (Figure 2 in [52]).
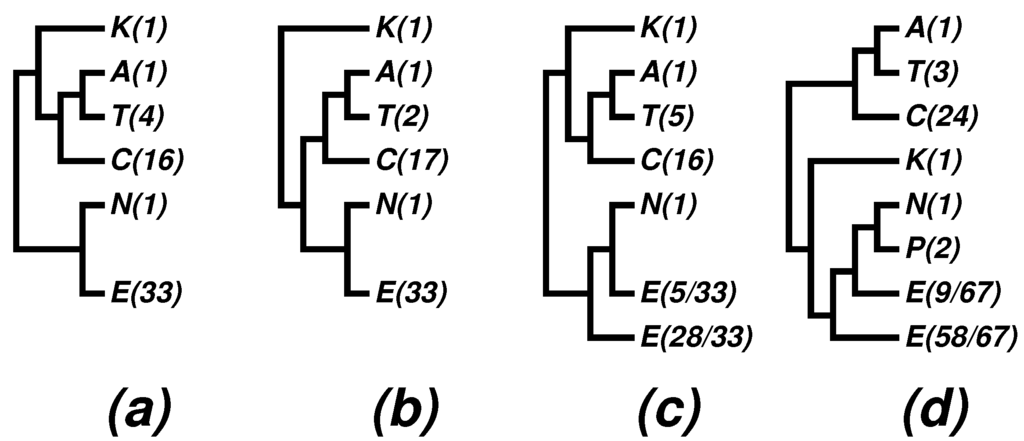
Figure 3.
Archaea trees collapsed to phyla. Abbreviations: A = Aigarchaeota, C = Crenarchaeota, E = Euryarchaeota, K = Korarchaeota, P = Parvarchaeota, N = Nanoarchaeota, T = Thaumarchaeota. (a–c) Obtained by collapsing Figure 4 in [44]; (d) obtained by collapsing Figure 2 in [52]. Numerals in parentheses indicate the number of species represented in each phylum. For details, see the text and the cited papers.
The interrelationship among phyla deduced from a limited number of representatives in a tree is subject to further changes when more data become available. In 2001, when there was only one genome from each of the bacterial phyla, Aquificae and Thermotogae, there was speculation that these phyla would make a clade [57,58]. A decade later, it was observed that, though remaining in a big cluster, many other phyla have gotten inserted in between Aquificae and Thermotogae; see, e.g., [10]. This point concerns especially the archaeal phyla with only one representative genome for the time being.
By comparing our Figure 2 with trees in Figure 3, we see:
- (1)
- The newly proposed phyla, Thaumarchaeota, Korarchaeota and Aigarchaeota, are supported in many phylogenies; especially the superphylum “TACK” is supported in most phylogenies, with “TAC” being a persistent core.
- (2)
- The nano-sized archaean symbiont, Nanoarchaeum equitans, has a highly reduced genome (490,885 bp [42]). It is the only described representative of a newly proposed phylum, Nanoarchaeota, and it cuts into the otherwise monophyletic phylum, Euryarchaeota. We note that the monophyly of Euryarchaeota was also violated by Nanoarchaeum in some 16S rRNA trees; see, e.g., Figure 4 in a 2009 paper [59], as well as (c) and (d) in Figure 3. It has been known that tiny genomes of endosymbiont microbes often tend to move towards the baseline of a tree and distort the overall picture. In fact, we have suggested skipping such tiny genomes when studying bacterial phylogeny; see, e.g., [28] and a note on the home page of the CVTree web server [20]. In the present case, we may at most say that Nanoarchaeota probably makes a separate phylum, but its cutting into Euryarchaeota might be a side effect due to the tiny size of the highly-reduced genome.
So far, we have concentrated on “mega-classification” [14] of Archaea species, mainly their taxonomy at the rank order and above. Quite recently, there appeared a proposal [35] to split the single-order class, Halobacteria, into three orders: Haloferacales, Natrialbales and Halobacteriales. In order to check whether CVTree supports this proposal or not, an expansion of the class, Halobacteria{29}, the first line in Figure 2, is given in Figure 4. Indeed, the three main branches are clearly seen in Figure 4, corresponding to the three proposed orders, except for a single genus, Halakalicoccus, which did not take a definite position, even in trees obtained by different methods in [35]. Being supported by the previous predictive power of CVTree, we anticipate that the position of Halakalicoccus in Figure 4 may better reflect the reality, a point verifiable in the future.
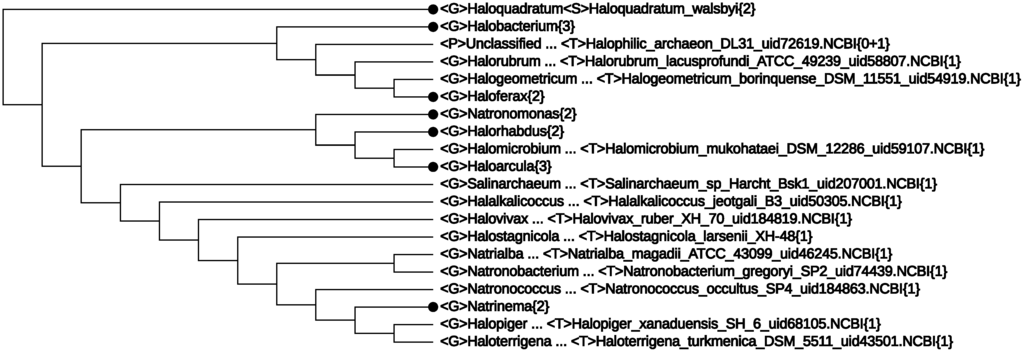
Figure 4.
The class, Halobacteria, expanded to the genus level.
5. Conclusions
The CVTree approach to prokaryotic phylogeny distinguishes itself from the 16S rRNA sequence analysis, both in the input data (genomes instead of RNA sequences) and in the methodology (K-peptide counting versus sequence alignment). The agreement of the two approaches makes the results more objective and convincing, whereas a few discrepancies call for further study. A phylogenetic study across many phyla naturally places emphasis on building a robust backbone for classification. At taxonomic rank order and above, whole-genome approaches are essentially simpler, as the only prerequisite is having the genomes at hand. Sooner or later, phylogenetic information and taxonomic placement will become by-products of genome analyses. The cost of sequencing a prokaryotic genome will drop below the average expense of carrying out conventional phenotyping experiments. To this end, a crucial factor is the availability of reliable, convenient and easy-to-use tools, such as the CVTree web server. The technique of collapsing and expanding tree branches with an interactive display, as well as automatic reporting of comparison results at all taxonomic ranks makes large-scale studies more feasible. The experience accumulated in this study on 179 archaeal strains will be instructive for carrying out similar studies on Bacteria, which would cover hundred-fold more strains.
The 16S rRNA sequence analysis will remain an indispensable tool in microbiology. The number of sequenced genomes can never catch up with that of rRNA sequences. Although the CVTree method adds more agreement than discrepancy to the 16S rRNA results, the difference between the two approaches certainly deserves in-depth scrutiny. In addition, since high resolution power at the species level and below is a prominent advantage of CVTree as compared to 16S rRNA sequence analysis [12,60], we will elaborate on this aspect in the future when the amount of sequenced archaeal genomes will have increased substantially.
Acknowledgments
The authors thank the support of the National Basic Research Program of China (973 Project Grant No. 2013CB34100) and of The State Key Laboratory of Applied Surface Physics, as well as the Department of Physics, Fudan University. An early discussion with Jiandong Sun on problems raised in this study is also gratefully acknowledged. The authors thank the three anonymous reviewers for making essential comments and suggestions to improve the manuscript.
Author Contributions
Bailin Hao designed the study and wrote the manuscript. Guanghong Zuo and Zhao Xu built and maintained the web server, collected data and carried out the calculation. Guanghong Zuo and Bailin Hao performed the analysis. All authors have read and approved the final manuscript.
Appendix: List of Genomes Used in This Study
All of the 179 genomes used in the present study are listed in the following table, together with their accession number and approximate proteome size (in 106 amino acids). The 165 genomes from the NCBI FTP site [19] come with uid numbers, but the uploaded ones appear without uid. We note that in the EBI list of Archaea [22], there are 176 species. Excluding a tiny one, 175 genomes remain. The four genomes present at NCBI, but absent at EBI, are Nos. 31, 40, 106 and 137.

Table A1.
list of genomes used in this study.
| No. | Name of Strain | Proteome Size (106AA) | Accession Number |
|---|---|---|---|
| 1 | Acidianus hospitalis W1 uid66875 | 0.62 | NC_015518 |
| 2 | Acidilobus saccharovorans 345 15 uid51395 | 0.45 | NC_014374 |
| 3 | Aciduliprofundum boonei T469 uid43333 | 0.47 | NC_013926 |
| 4 | Aciduliprofundum sp. MAR08 339 uid184407 | 0.45 | NC_019942 |
| 5 | Aeropyrum camini SY1 JCM 12091 uid222311 | 0.47 | NC_022521 |
| 6 | Aeropyrum pernix K1 uid57757 | 0.49 | NC_000854 |
| 7 | Archaeoglobus fulgidus DSM 4304 uid57717 | 0.67 | NC_000917 |
| 8 | Archaeoglobus fulgidus DSM 8774 | 0.69 | CP006577 |
| 9 | Archaeoglobus profundus DSM 5631 uid43493 | 0.48 | NC_013741 |
| 10 | Archaeoglobus sulfaticallidus PM70 1 uid201033 | 0.61 | NC_021169 |
| 11 | Archaeoglobus veneficus SNP6 uid65269 | 0.56 | NC_015320 |
| 12 | Caldisphaera lagunensis DSM 15908 uid183486 | 0.44 | NC_019791 |
| 13 | Caldivirga maquilingensis IC 167 uid58711 | 0.60 | NC_009954 |
| 14 | Candidatus Caldiarchaeum subterraneum uid227223 | 0.51 | NC_022786 |
| 15 | Candidatus Korarchaeum cryptofilum OPF8 uid58601 | 0.48 | NC_010482 |
| 16 | Candidatus Methanomethylophilus alvus Mx1201 uid196597 | 0.49 | NC_020913 |
| 17 | Candidatus Nitrosopumilus koreensis AR1 uid176129 | 0.47 | NC_018655 |
| 18 | Candidatus Nitrosopumilus sp. AR2 uid176130 | 0.49 | NC_018656 |
| 19 | Candidatus Nitrososphaera evergladensis SR1 | 0.82 | CP007174 |
| 20 | Candidatus Nitrososphaera gargensis Ga9 2 uid176707 | 0.77 | NC_018719 |
| 21 | Methanomassiliicoccus sp. Mx1 Issoire uid207287 | 0.56 | NC_021353 |
| 22 | Cenarchaeum symbiosum A uid61411 | 0.62 | NC_014820 |
| 23 | Desulfurococcus fermentans DSM 16532 uid75119 | 0.40 | NC_018001 |
| 24 | Desulfurococcus kamchatkensis 1221n uid59133 | 0.40 | NC_011766 |
| 25 | Desulfurococcus mucosus DSM 2162 uid62227 | 0.39 | NC_014961 |
| 26 | Ferroglobus placidus DSM 10642 uid40863 | 0.66 | NC_013849 |
| 27 | Ferroplasma acidarmanus fer1 uid54095 | 0.57 | NC_021592 |
| 28 | Fervidicoccus fontis Kam940 uid162201 | 0.38 | NC_017461 |
| 29 | Halalkalicoccus jeotgali B3 uid50305 | 0.83 | NC_014297 |
| 30 | Haloarcula hispanica ATCC 33960 uid72475 | 1.00 | NC_0159432 |
| 31 | Haloarcula hispanica N601 uid230920 | 0.98 | NC_0230102 |
| 32 | Haloarcula marismortui ATCC 43049 uid57719 | 0.97 | NC_0063972 |
| 33 | Halobacterium salinarum R1 uid61571 | 0.60 | NC_010364 |
| 34 | Halobacterium sp. DL1 | 0.83 | CP007060 |
| 35 | Halobacterium sp. NRC 1 uid57769 | 0.59 | NC_002607 |
| 36 | Haloferax mediterranei ATCC 33500 uid167315 | 0.84 | NC_017941 |
| 37 | Haloferax volcanii DS2 uid46845 | 0.82 | NC_013967 |
| 38 | Halogeometricum borinquense DSM 11551 uid54919 | 0.82 | NC_014729 |
| 39 | Halomicrobium mukohataei DSM 12286 uid59107 | 0.90 | NC_013202 |
| 40 | Halophilic archaeon DL31 uid72619 | 0.81 | NC_015954 |
| 41 | Halopiger xanaduensis SH 6 uid68105 | 1.05 | NC_015666 |
| 42 | Haloquadratum walsbyi C23 uid162019 | 0.77 | NC_017459 |
| 43 | Haloquadratum walsbyi DSM 16790 uid58673 | 0.78 | NC_008212 |
| 44 | Halorhabdus tiamatea SARL4B uid214082 | 0.79 | NC_021921 |
| 45 | Halorhabdus utahensis DSM 12940 uid59189 | 0.91 | NC_013158 |
| 46 | Halorubrum lacusprofundi ATCC 49239 uid58807 | 0.93 | NC_0120292 |
| 47 | Halostagnicola larsenii XH-48 | 0.78 | CP007055 |
| 48 | Haloterrigena turkmenica DSM 5511 uid43501 | 1.09 | NC_013743 |
| 49 | Halovivax ruber XH 70 uid184819 | 0.91 | NC_019964 |
| 50 | Hyperthermus butylicus DSM 5456 uid57755 | 0.45 | NC_008818 |
| 51 | Ignicoccus hospitalis KIN4 I uid58365 | 0.40 | NC_009776 |
| 52 | Ignisphaera aggregans DSM 17230 uid51875 | 0.54 | NC_014471 |
| 53 | Metallosphaera cuprina Ar 4 uid66329 | 0.54 | NC_015435 |
| 54 | Metallosphaera sedula DSM 5348 uid58717 | 0.64 | NC_009440 |
| 55 | Methanobacterium formicicum strain BRM9 | 0.67 | CP006933 |
| 56 | Methanobacterium sp. AL 21 uid63623 | 0.72 | NC_015216 |
| 57 | Methanobacterium sp. MB1 complete sequence uid231690 | 0.56 | NC_023044 |
| 58 | Methanobacterium sp. SWAN 1 uid67359 | 0.66 | NC_015574 |
| 59 | Methanobrevibacter ruminantium M1 uid45857 | 0.76 | NC_013790 |
| 60 | Methanobrevibacter smithii ATCC 35061 uid58827 | 0.56 | NC_009515 |
| 61 | Methanobrevibacter sp. AbM4 uid206516 | 0.50 | NC_021355 |
| 62 | Methanocaldococcus fervens AG86 uid59347 | 0.44 | NC_013156 |
| 63 | Methanocaldococcus infernus ME uid48803 | 0.41 | NC_014122 |
| 64 | Methanocaldococcus jannaschii DSM 2661 uid57713 | 0.48 | NC_000909 |
| 65 | Methanocaldococcus sp. JH146 | 0.47 | CP009149 |
| 66 | Methanocaldococcus sp. FS406 22 uid42499 | 0.51 | NC_013887 |
| 67 | Methanocaldococcus vulcanius M7 uid41131 | 0.49 | NC_013407 |
| 68 | Methanocella arvoryzae MRE50 uid61623 | 0.89 | NC_009464 |
| 69 | Methanocella conradii HZ254 uid157911 | 0.70 | NC_017034 |
| 70 | Methanocella paludicola SANAE uid42887 | 0.86 | NC_013665 |
| 71 | Methanococcoides burtonii DSM 6242 uid58023 | 0.69 | NC_007955 |
| 72 | Methanococcus aeolicus Nankai 3 uid58823 | 0.44 | NC_009635 |
| 73 | Methanococcus maripaludis C5 uid58741 | 0.51 | NC_009135 |
| 74 | Methanococcus maripaludis C6 uid58947 | 0.51 | NC_009975 |
| 75 | Methanococcus maripaludis C7 uid58847 | 0.51 | NC_009637 |
| 76 | Methanococcus maripaludis KA1 DNA | 0.55 | AP011526 |
| 77 | Methanococcus maripaludis OS7 DNA | 0.52 | AP011528 |
| 78 | Methanococcus maripaludis S2 uid58035 | 0.49 | NC_005791 |
| 79 | Methanococcus maripaludis X1 uid70729 | 0.51 | NC_015847 |
| 80 | Methanococcus vannielii SB uid58767 | 0.49 | NC_009634 |
| 81 | Methanococcus voltae A3 uid49529 | 0.51 | NC_014222 |
| 82 | Methanocorpusculum labreanum Z uid58785 | 0.52 | NC_008942 |
| 83 | Methanoculleus bourgensis MS2T uid171377 | 0.77 | NC_018227 |
| 84 | Methanoculleus marisnigri JR1 uid58561 | 0.72 | NC_009051 |
| 85 | Methanohalobium evestigatum Z 7303 uid49857 | 0.63 | NC_014253 |
| 86 | Methanohalophilus mahii DSM 5219 uid47313 | 0.59 | NC_014002 |
| 87 | Methanolobus psychrophilus R15 uid177925 | 0.87 | NC_018876 |
| 88 | Methanomethylovorans hollandica DSM 15978 uid184864 | 0.69 | NC_019977 |
| 89 | Methanoplanus petrolearius DSM 11571 uid52695 | 0.83 | NC_014507 |
| 90 | Methanopyrus kandleri AV19 uid57883 | 0.50 | NC_003551 |
| 91 | Methanoregula boonei 6A8 uid58815 | 0.73 | NC_009712 |
| 92 | Methanoregula formicicum SMSP uid184406 | 0.81 | NC_019943 |
| 93 | Methanosaeta concilii GP6 uid66207 | 0.84 | NC_015416 |
| 94 | Methanosaeta harundinacea 6Ac uid81199 | 0.73 | NC_017527 |
| 95 | Methanosaeta thermophila PT uid58469 | 0.51 | NC_008553 |
| 96 | Methanosalsum zhilinae DSM 4017 uid68249 | 0.61 | NC_015676 |
| 97 | Methanosarcina acetivorans C2A uid57879 | 1.42 | NC_003552 |
| 98 | Methanosarcina barkeri str Fusaro uid57715 | 1.12 | NC_007355 |
| 99 | Methanosarcina mazei Go1 uid57893 | 1.02 | NC_003901 |
| 100 | Methanosarcina mazei Tuc01 uid190185 | 0.82 | NC_020389 |
| 101 | Methanosphaera stadtmanae DSM 3091 uid58407 | 0.49 | NC_007681 |
| 102 | Methanosphaerula palustris E1 9c uid59193 | 0.82 | NC_011832 |
| 103 | Methanospirillum hungatei JF 1 uid58181 | 1.01 | NC_007796 |
| 104 | Methanothermobacter marburgensis str Marburg uid51637 | 0.49 | NC_014408 |
| 105 | Methanothermobacter thermautotrophicus str Delta H uid57877 | 0.53 | NC_000916 |
| 106 | Methanothermobacter thermautotrophicus CaT2 DNA | 0.51 | AP011952 |
| 107 | Methanothermococcus okinawensis IH1 uid51535 | 0.45 | NC_015636 |
| 108 | Methanothermus fervidus DSM 2088 uid60167 | 0.38 | NC_014658 |
| 109 | Methanotorris igneus Kol 5 uid67321 | 0.51 | NC_015562 |
| 110 | Nanoarchaeum equitans Kin4 M uid58009 | 0.15 | NC_005213 |
| 111 | Natrialba magadii ATCC 43099 uid46245 | 1.05 | NC_013922 |
| 112 | Natrinema pellirubrum DSM 15624 uid74437 | 1.06 | NC_019962 |
| 113 | Natrinema sp. J7 2 uid171337 | 1.05 | NC_018224 |
| 114 | Natronobacterium gregoryi SP2 uid74439 | 1.04 | NC_019792 |
| 115 | Natronococcus occultus SP4 uid184863 | 1.12 | NC_019974 |
| 116 | Natronomonas moolapensis 8 8 11 uid190182 | 0.82 | NC_020388 |
| 117 | Natronomonas pharaonis DSM 2160 uid58435 | 0.78 | NC_007426 |
| 118 | Nitrosopumilus maritimus SCM1 uid58903 | 0.49 | NC_010085 |
| 119 | Nitrososphaera viennensis EN76 | 0.73 | CP007536 |
| 120 | Palaeococcus pacificus DY20341 | 0.56 | CP006019 |
| 121 | Picrophilus torridus DSM 9790 uid58041 | 0.47 | NC_005877 |
| 122 | Pyrobaculum aerophilum str IM2 uid57727 | 0.66 | NC_003364 |
| 123 | Pyrobaculum arsenaticum DSM 13514 uid58409 | 0.61 | NC_009376 |
| 124 | Pyrobaculum calidifontis JCM 11548 uid58787 | 0.61 | NC_009073 |
| 125 | Pyrobaculum islandicum DSM 4184 uid58635 | 0.53 | NC_008701 |
| 126 | Pyrobaculum neutrophilum V24Sta uid58421 | 0.53 | NC_010525 |
| 127 | Pyrobaculum oguniense TE7 uid84411 | 0.71 | NC_016885 |
| 128 | Pyrobaculum sp. 1860 uid82379 | 0.73 | NC_016645 |
| 129 | Pyrococcus abyssi GE5 uid62903 | 0.54 | NC_000868 |
| 130 | Pyrococcus furiosus COM1 uid169620 | 0.57 | NC_018092 |
| 131 | Pyrococcus furiosus DSM 3638 uid57873 | 0.59 | NC_003413 |
| 132 | Pyrococcus horikoshii OT3 uid57753 | 0.55 | NC_000961 |
| 133 | Pyrococcus sp. NA2 uid66551 | 0.57 | NC_015474 |
| 134 | Pyrococcus sp. ST04 uid167261 | 0.52 | NC_017946 |
| 135 | Pyrococcus yayanosii CH1 uid68281 | 0.51 | NC_015680 |
| 136 | Pyrolobus fumarii 1A uid73415 | 0.54 | NC_015931 |
| 137 | Salinarchaeum sp. Harcht Bsk1 uid207001 | 0.91 | NC_021313 |
| 138 | Staphylothermus hellenicus DSM 12710 uid45893 | 0.46 | NC_014205 |
| 139 | Staphylothermus marinus F1 uid58719 | 0.46 | NC_009033 |
| 140 | Sulfolobus acidocaldarius DSM 639 uid58379 | 0.63 | NC_007181 |
| 141 | Sulfolobus acidocaldarius N8 uid189027 | 0.62 | NC_020246 |
| 142 | Sulfolobus acidocaldarius Ron12 I uid189028 | 0.64 | NC_020247 |
| 143 | Sulfolobus acidocaldarius SUSAZ uid232254 | 0.59 | NC_023069 |
| 144 | Sulfolobus islandicus HVE10 4 uid162067 | 0.76 | NC_017275 |
| 145 | Sulfolobus islandicus L D 8 5 uid43679 | 0.77 | NC_013769 |
| 146 | Sulfolobus islandicus L S 2 15 uid58871 | 0.76 | NC_012589 |
| 147 | Sulfolobus islandicus LAL14 1 uid197216 | 0.71 | NC_021058 |
| 148 | Sulfolobus islandicus M 14 25 uid58849 | 0.74 | NC_012588 |
| 149 | Sulfolobus islandicus M 16 27 uid58851 | 0.76 | NC_012632 |
| 150 | Sulfolobus islandicus M 16 4 uid58841 | 0.75 | NC_012726 |
| 151 | Sulfolobus islandicus REY15A uid162071 | 0.72 | NC_017276 |
| 152 | Sulfolobus islandicus Y G 57 14 uid58923 | 0.78 | NC_012622 |
| 153 | Sulfolobus islandicus Y N 15 51 uid58825 | 0.77 | NC_012623 |
| 154 | Sulfolobus solfataricus 98 2 uid167998 | 0.72 | NC_017274 |
| 155 | Sulfolobus solfataricus P2 uid57721 | 0.84 | NC_002754 |
| 156 | Sulfolobus tokodaii str 7 uid57807 | 0.76 | NC_003106 |
| 157 | Thermococcus barophilus MP uid54733 | 0.62 | NC_014804 |
| 158 | Thermococcus eurythermalis strain A501 | 0.60 | CP008887 |
| 159 | Thermococcus gammatolerans EJ3 uid59389 | 0.64 | NC_012804 |
| 160 | Thermococcus kodakarensis KOD1 uid58225 | 0.64 | NC_006624 |
| 161 | Thermococcus litoralis DSM 5473 uid82997 | 0.67 | NC_022084 |
| 162 | Thermococcus nautili strain 30 1 | 0.61 | CP007264 |
| 163 | Thermococcus onnurineus NA1 uid59043 | 0.56 | NC_011529 |
| 164 | Thermococcus sibiricus MM 739 uid59399 | 0.55 | NC_012883 |
| 165 | Thermococcus sp. 4557 uid70841 | 0.61 | NC_015865 |
| 166 | Thermococcus sp. AM4 uid54735 | 0.63 | NC_016051 |
| 167 | Thermococcus sp. CL1 uid168259 | 0.58 | NC_018015 |
| 168 | Thermococcus sp. ES1 | 0.58 | CP006965 |
| 169 | Thermofilum pendens Hrk 5 uid58563 | 0.54 | NC_008698 |
| 170 | Thermofilum sp. 1910b uid215374 | 0.52 | NC_022093 |
| 171 | Thermogladius cellulolyticus 1633 uid167488 | 0.41 | NC_017954 |
| 172 | Thermoplasma acidophilum DSM 1728 uid61573 | 0.45 | NC_002578 |
| 173 | Thermoplasma volcanium GSS1 uid57751 | 0.45 | NC_002689 |
| 174 | Thermoplasmatales archaeon BRNA1 uid195930 | 0.44 | NC_020892 |
| 175 | Thermoproteus tenax Kra 1 uid74443 | 0.55 | NC_016070 |
| 176 | Thermoproteus uzoniensis 768 20 uid65089 | 0.59 | NC_015315 |
| 177 | Thermosphaera aggregans DSM 11486 uid48993 | 0.40 | NC_014160 |
| 178 | Vulcanisaeta distributa DSM 14429 uid52827 | 0.71 | NC_014537 |
| 179 | Vulcanisaeta moutnovskia 768 28 uid63631 | 0.67 | NC_015151 |
Conflicts of Interest
The authors declare no conflict of interest.
References
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.E.; Magrum, L.J.; Balch, W.E.; Wolfe, R.S.; Woese, C.R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc. Natl. Acad. Sci. USA 1977, 74, 4537–4541. [Google Scholar] [CrossRef] [PubMed]
- Fox, G.E.; Pechman, K.R.; Woese, C.R. Comarative cataloging of 16S ribosomal ribonucleic acid: Molecular approach to procaryotic systematics. Int. J. Syst. Bacteriol. 1977, 27, 44–57. [Google Scholar] [CrossRef]
- The Bergey’s Manual Trust. In Bergey’s Manual of Systematic bacteriology, 2nd ed.; Springer: New York, NY, USA; Volumes 1∼5, pp. 2001–2012.
- Konstantinidis, K.T.; Tiedje, J.M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005, 187, 6258–6264. [Google Scholar]
- Qi, J.; Wang, B.; Hao, B. Whole genome prokaryote phylogeny without sequence alignment: A K-string composition approach. J. Mol. Evol. 2004, 58, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Qi, J. Prokaryote phylogeny without sequence alignment: From avoidance signature to composition distance. J. Bioinf. Comput. Biol. 2004, 2. [Google Scholar] [CrossRef]
- Gao, L.; Qi, J.; Sun, J.; Hao, B. Prokaryote phylogeny meets taxonomy: An exhaustive comparison of composition vector trees with systematic bacteriology. Sci. China Life Sci. 2007, 50, 587–599. [Google Scholar] [CrossRef]
- Li, Q.; Xu, Z.; Hao, B. Composition vector approach to whole-genome-based prokaryotic phylogeny: Success and foundations. J. Biotech. 2010, 149, 115–119. [Google Scholar] [CrossRef]
- Zuo, G.; Xu, Z.; Yu, H.; Hao, B. Jackknife and bootstrap tests of the composition vector trees. Genomics Proteomics Bioinform. 2010, 8, 262–267. [Google Scholar] [CrossRef]
- Hao, B. CVTrees support the Bergey’s systematics and provide high resolution at species level and below. Bull. BISMiS 2011, 2, 189–196. [Google Scholar]
- Chan, P.P.; Cozen, A.E.; Lowe, T.M. Reclassification of Thermoproteus neutrophilus Stetter and Zillig 1989 as Pyrobaculum neutrophilum comb. nov., based on phylogenetic analysis. Int. J. Syst. Evol. Microbiol. 2013, 63, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002, 52, 7–76. [Google Scholar] [PubMed]
- Lapage, S.P.; Sneath, P.H.A.; Lessel, E.F.; Skerman, V.B.D.; Seeliger, H.P.R.; Clark, W.A. International Code of Nomenclature of Bacteria: Bacteriological Code 1990; ASM Press: Washington, DC, USA, 1992. [Google Scholar]
- De Vos, P.; Trüper, H.G. Judicial Commission of the International Committee on Systematic Bacteriology. Int. J. Syst. Evol. Microbiol. 2000, 50, 2239–2244. [Google Scholar] [CrossRef]
- The GOLD (Genomes On Line Database) site. Available online: https://gold.jgi-psf.org (accessed on 12 February 2015).
- PATRIC (Pathosystems Resource Integration Center). Available online: http://particbrc.org/portal/portal/patric/Genomes (accessed on 12 February 2015).
- The NCBI FTP site. Available online: ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ (accessed on 27 February 2015).
- Xu, Z.; Hao, B. CVTree update: A newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009, 37, W174–W178. [Google Scholar] [CrossRef] [PubMed]
- The much improved CVTree3 Web Server. Available online: http://tlife.fudan.edu.cn/cvtree3/ (accessed on 25 February 2015).
- The EBI Archaea genome list. Available online: http://www.ebi.ac.uk/genomes/archaea.html (accessed on 15 February 2015).
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Pess: Cambridge, UK, 1985. [Google Scholar]
- Woese, C. The universal ancestor. Proc. Natl. Acad, Sci. USA 1998, 95, 6854–6859. [Google Scholar] [CrossRef]
- Wagner, A; de la Chaus, N. Distant horizontal gene transfer is rare for multiple families of prokaryotic insertion sequences. Mol. Genet. Genomics 2008, 280, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Luo, H.; Hao, B. CVTree: A phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004, 32, W45–W47. [Google Scholar] [CrossRef] [PubMed]
- Zuo, G.; Li, Q.; Hao, B. On K-peptide length in composition vector phylogeny of prokaryotes. Comput. Biol. Chem. 2014, 53, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Hao, B. Whole-genome based prokaryotic branches in the Tree of Life. In Darwin’s Heritage Today: Proceedings of the Darwin 200 Beijing International Conference; Long, M., Gu, H., Zhou, Z., Eds.; High Education Press: Beijing, China, 2010; pp. 102–113. [Google Scholar]
- Garrity, G.M.; Holt, J.G. Taxonomic Outline of the Archaea and Bacteria. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Boone, D.R., Castenholz, R.W., Eds.; Springer: New York, NY, USA, 2001; Volume 1, pp. 155–156. [Google Scholar]
- Parte, A.C. LPSN—list of prokaryotic names with standing in Nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Prokofeva, M.I.; Kostrikina, N.A.; Kolganova, T.V.; Tourova, T.P.; Lysenko, A.M.; Lebedinsky, A.V.; Bonch-Osmolovskaya1, F.A. Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 3116–3122. [Google Scholar] [CrossRef] [PubMed]
- Perevalova, A.A.; Bidzhieva, S.K.; Kublanov, I.V.; Hinrichs, K.-U.; Liu, X.L.; Mardanov, A.V.; Lebedinsky, A.V.; Bonch-Osmolovskaya, E.A. Fervidicoccus fontis gen. nov., sp. nov., an anaerobic, thermophilic crenarchaeote from terrestrial hot springs, and proposal of Fervidicoccaceae fam. nov. and Fervidicoccales ord. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2082–2088. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. The Revised Roadmap to the Manual. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2005; Volume 2, pp. 159–187. [Google Scholar]
- Sakai, S.; Imachi, H.; Hanada, S.; Ohashi, A.; Harada1, H.; Kamagata, Y. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage “Rice Cluster I”, and proposal of the new archaeal order Methanocellales ord. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Naushad, S.; Baker, S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: A proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov. Int. J. Syst. Evol. Microbiol. 2014. [CrossRef]
- Barns, S.M.; Delwiche, C.F.; Palmer, J.D.; Pace, N.R. Perspectives on archaeal diversity, thermophyly and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 1996, 93, 9188–9193. [Google Scholar] [CrossRef] [PubMed]
- Auchtung, T.A.; Shyndriayeva, G.; Cavanaugh, C.M. 16S rRNA phylogenetic analysis and quantification of Koarchaeota indigenous to the hot springs of Kamchatka, Russia. Extremophiles 2011, 15, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Brochier-Armanet, C.; Boussau, B.; Gribaldo, S.; Forterre, P. Mesophilic crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008, 6, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Shami, A. Molecular signatures for the Crenarchaeota and Thaumarchaeota. Antonie van Leeuwenhoek 2011, 99, 133–157. [Google Scholar] [CrossRef] [PubMed]
- Pester, M.; Schleper, C.; Wagner, M. The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 2011, 14, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Huber, H.; Hohn, M.J.; Rachel, R.; Fuchs, T.; Wimmer, V.C.; Stetter, K.O. A new phylum of Archaea represented by a nano-sized hyperthermophilic symbiont. Nature 2002, 417, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.; Hohn, M.J.; Ahel, I.; Graham, D.E.; Adams, M.D.; Barnstead, M.; Beeson, K.Y.; Bibbs, L.; Bolanos, R.; Keller, M.; et al. The genome of Nanoarchaeum equitan: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Aad. Sci. USA 2003, 100, 12984–12988. [Google Scholar] [CrossRef]
- Clingenpeel, S.; Kan, J.; Macur, R.E.; Woyke, T.; Lavalvo, D.; Carley, J.; Inskeep, W.P.; Nealson, K.; McDermott, T. Yellowstone Lake Nanoarchaeota. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Nunoura1, T.; Takaki, Y.; Kakuta, J.; Nishi, S.; Sugahara, J.; Kazama, H.; Chee, G.-J.; Hattori, M.; Kanai, A.; Atomi, H.; et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011, 39, 3204–3223. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Comolli, L.R.; Dick, G.J.; Hauser, L.J.; Haytt, D.; Dill, B.J.; Land, M.L.; VerBerkmoes, N.C.; Hettich, R.L.; Banfield, J.F. Enegmatic, ultrasmall, uncultivated Archaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8806–8811. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Xu, J.; Qin, D.; He, Y.; Xiao, X.; Wang, F. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 2014, 8, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Richter, M.; Peplies, J.; Euzéby, J.; Amann, R.; Schleifer, K.-H.; Ludwig, W.; Glöckner, F.O.; Roselló-Móra, R. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all se-quenced type strains. Syst. Appl. Microbiol. 2008, 31, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Ludwig, W.; Euzéby, J.; Amann, R.; Schleifer, K.-H.; Glöckner, F.O.; Rossweló-Móra, R. Update of the All-Species Living Tree project based on 16S and 23S rRNA sequence analysis. Syst. Appl. Microbiol. 2010, 33, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Wegener-Parfrey, L.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- LTPs115 web site. Available online: http://www.silva-arb.de/projects/livibg-tree/ (accessed on 25 November 2014).
- LVTree Viewer. Available online: http://tlife.fudan.edu.cn/lvtree/ (accessed on 25 November 2014).
- Brochier-Armanet, C.; Forterre, P.; Gribaldo, S. Phylogeny and evolution of the Archaea: One hundred genomes later. Curr. Opin. Microbiol. 2011, 14, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Reysenbach, A.-L.; Liu, Y.; Banta, A.B.; Beveridge, T.J.; Kirshtein, J.D.; Schouten, S.; Tivey, M.K.; von Damm, K.L.; Voytek, M.A. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 2006, 422, 444–447. [Google Scholar] [CrossRef]
- Schouten, S.; Baas, M.; Hopmans, E.C.; Reysenbach, A.-L.; Sinninghe Damste, J.S. Tetraether membrane lipids of Candidatus “Aciduliprofundum boonei”, a cultivated obligate thermoacidophilic euryarchaeote from deep-sea hydrothermal vents. Extremophiles 2008, 12, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Guy, L.; Ettema, T.J.G. The archaeal “TACK” superphylum and the origin of eukaryotes. Trends Micrbiol. 2011, 19, 580–587. [Google Scholar] [CrossRef]
- Sun, J.; Xu, Z.; Hao, B. Whole-genome based Archaea phylogeny and taxonomy: A composition vector approach. Chin. Sci. Bull. 2010, 55, 2323–2328. [Google Scholar] [CrossRef]
- Daubin, V.; Gouy, M.; Perriére, G. Bacterial molecular phylogeny using supertree approach. Genome Inform. 2001, 12, 155–164. [Google Scholar] [PubMed]
- Wolf, Y.I.; Rogiozin, I.B.; Grishin, N.V.; Tatusov, R.L.; Koonin, E.V. Genome tree constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 2001, 1. [Google Scholar] [CrossRef]
- Gribaldo, S.; Brochier, C. Phylogeny of prokaryotes: Does it exist and why should we care? Res. Microbiol. 2009, 160, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Zuo, G.; Hao, B.; Staley, J.R. Geographic divergence of “Sulfolobus islandicus” strains assessed by genomic analyses including electronic DNA hybridization confirms they are geovars. Antonie van Leeuwenoek 2014, 105, 431–435. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).