Functional Characterization of the FNT Family Nitrite Transporter of Marine Picocyanobacteria
Abstract
:1. Introduction
2. Results
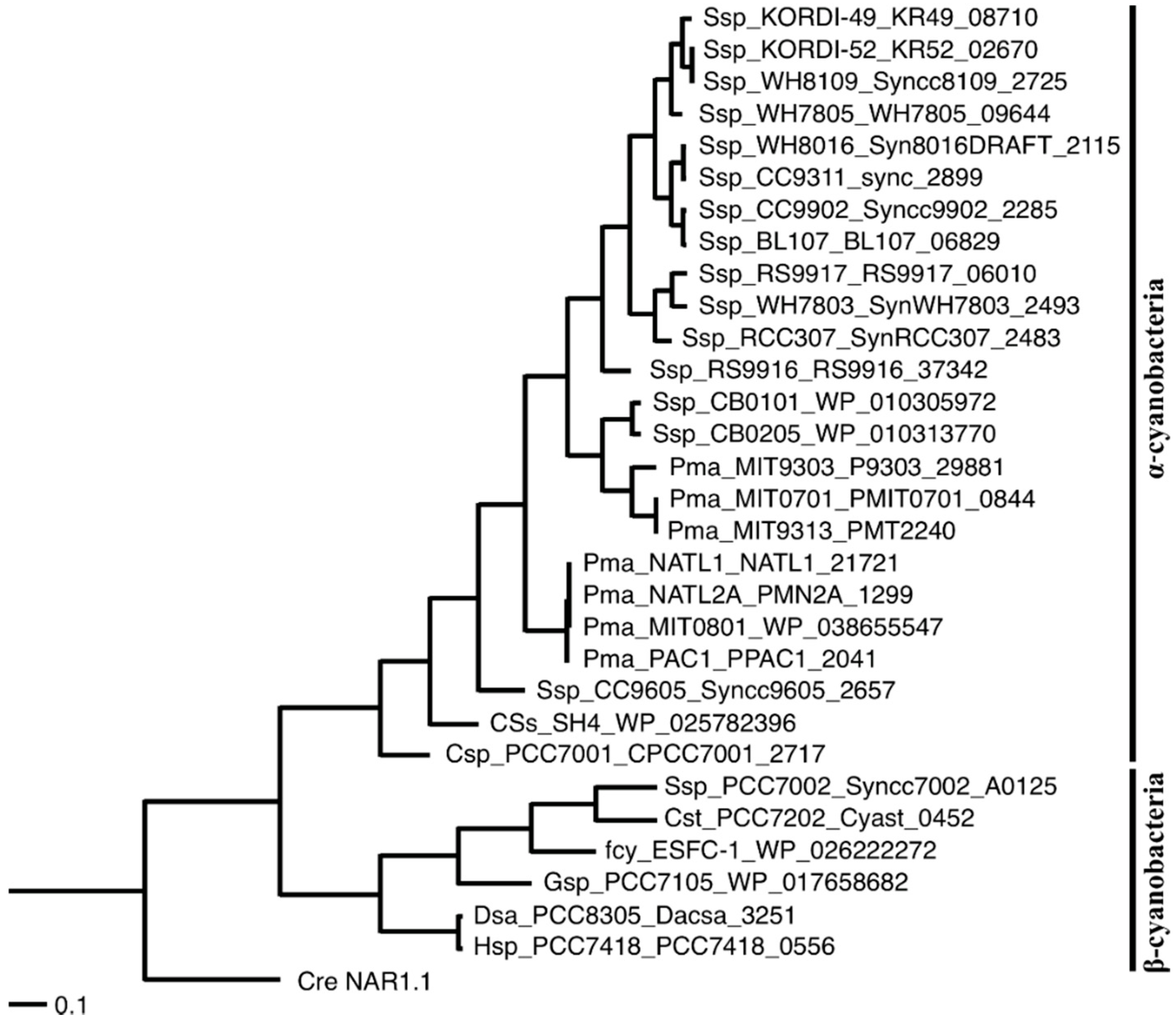
2.1. Distribution of the Gene Encoding an FNT Family Protein in Cyanobacteria
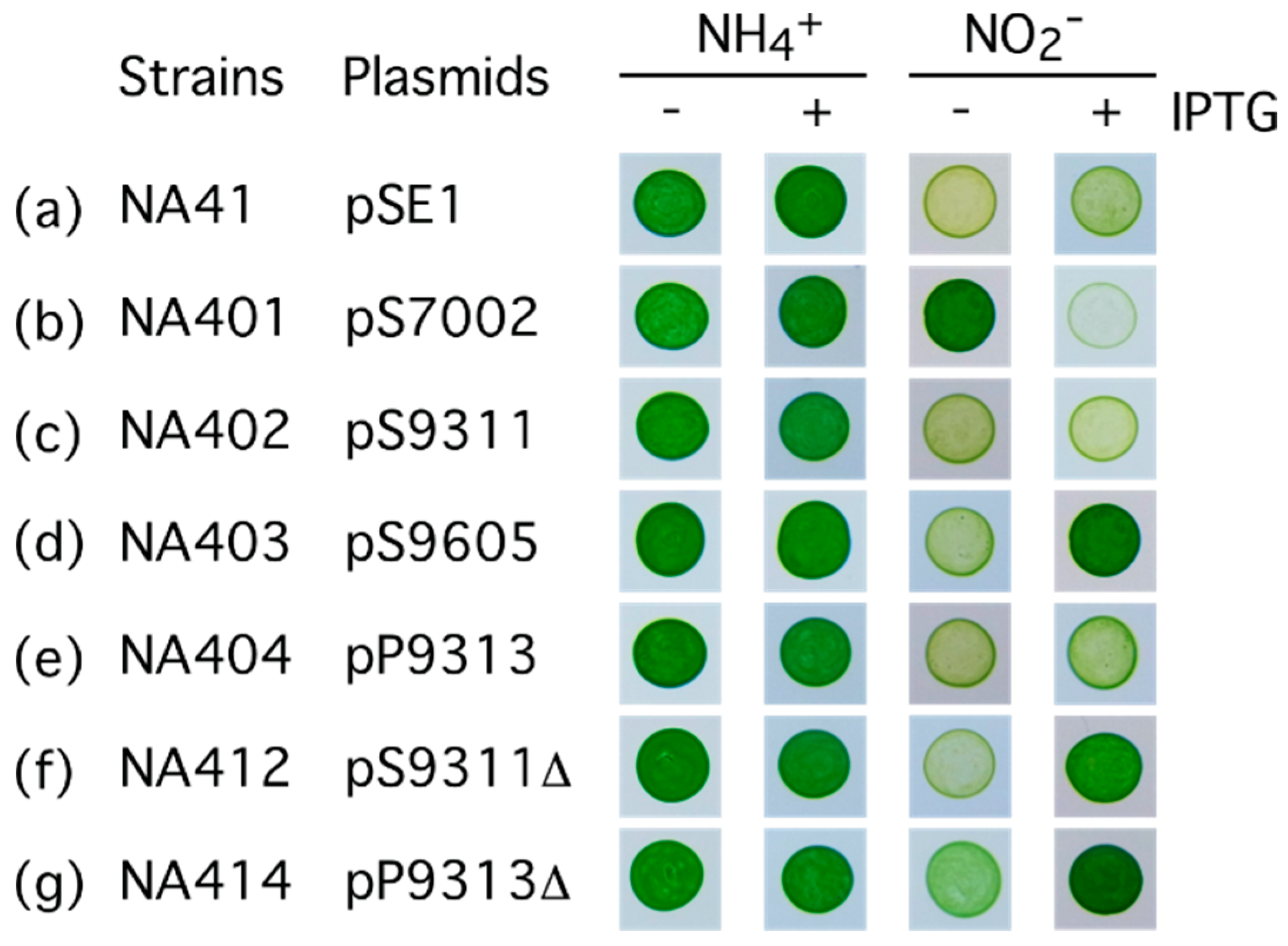
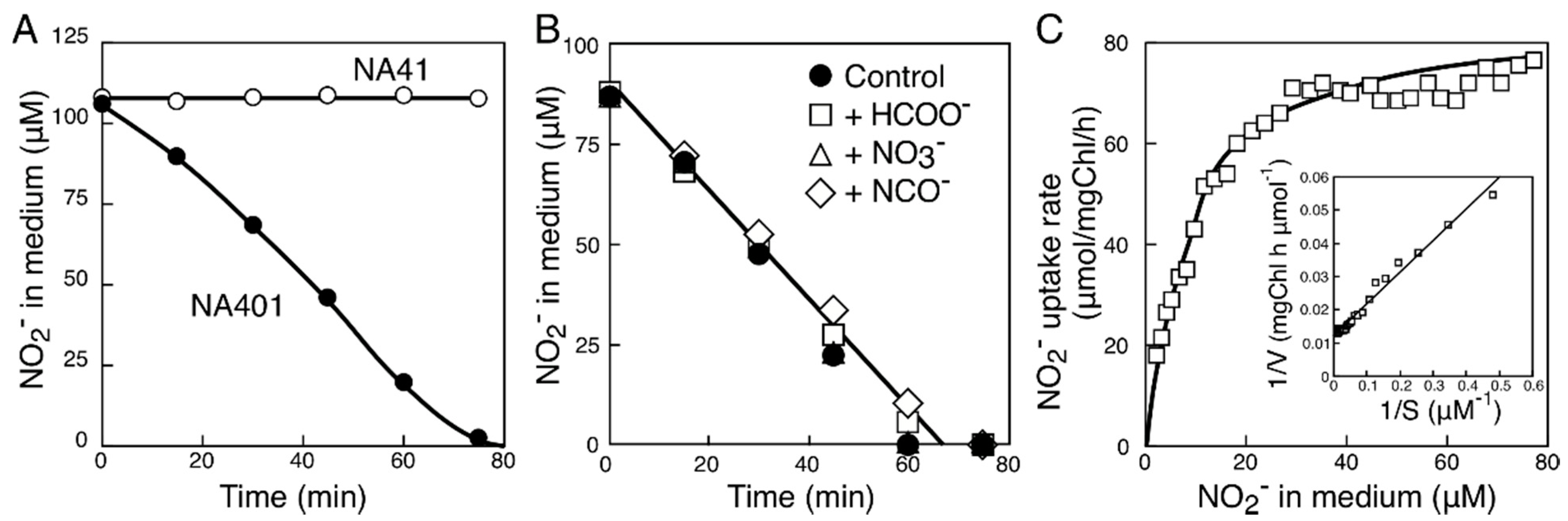
2.2. Characterization of the FNT Family Transporter from Synechococcus sp. Strain PCC 7002
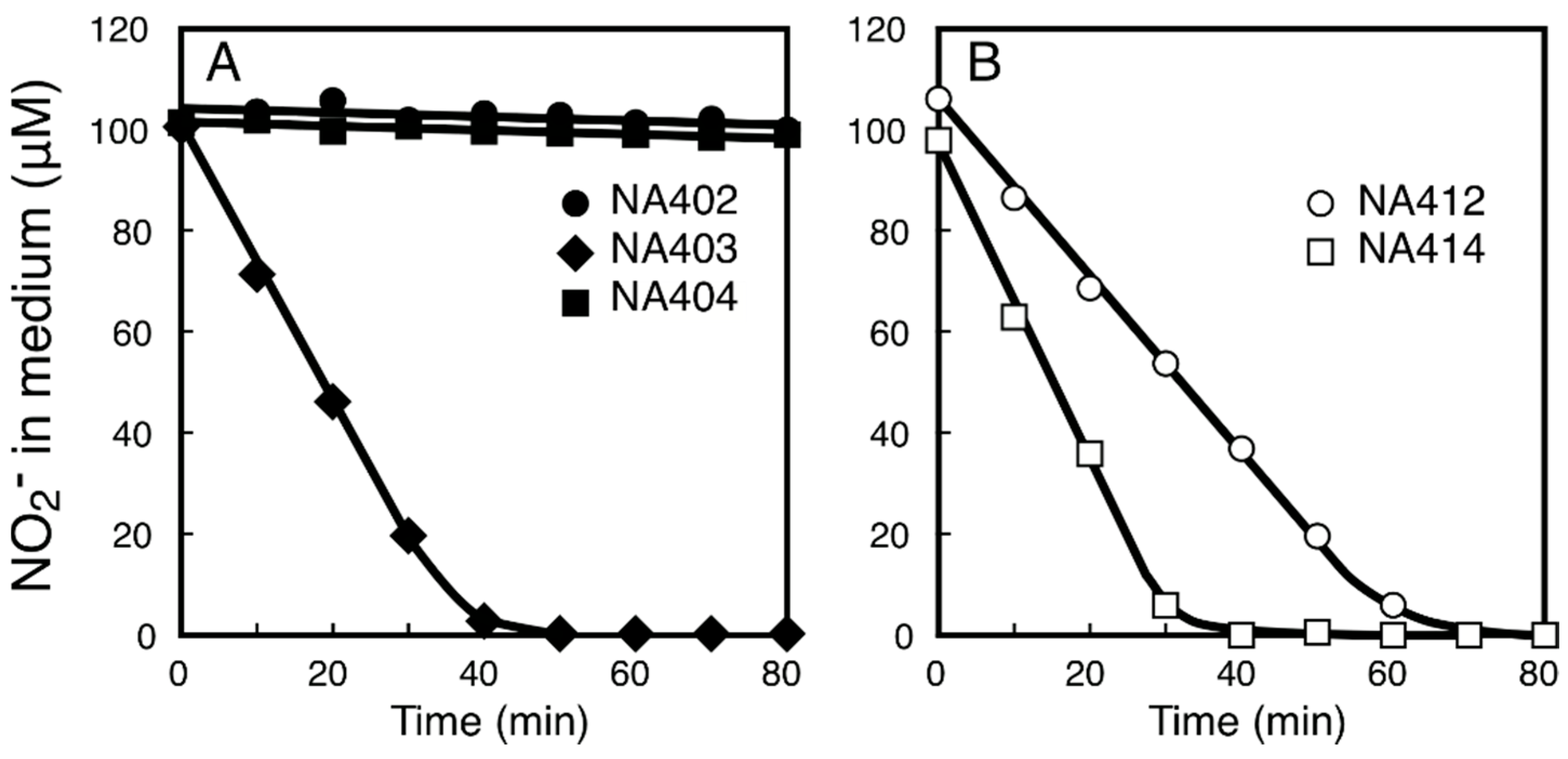
2.3. Characterization of the NitM Proteins from α-Cyanobacteria
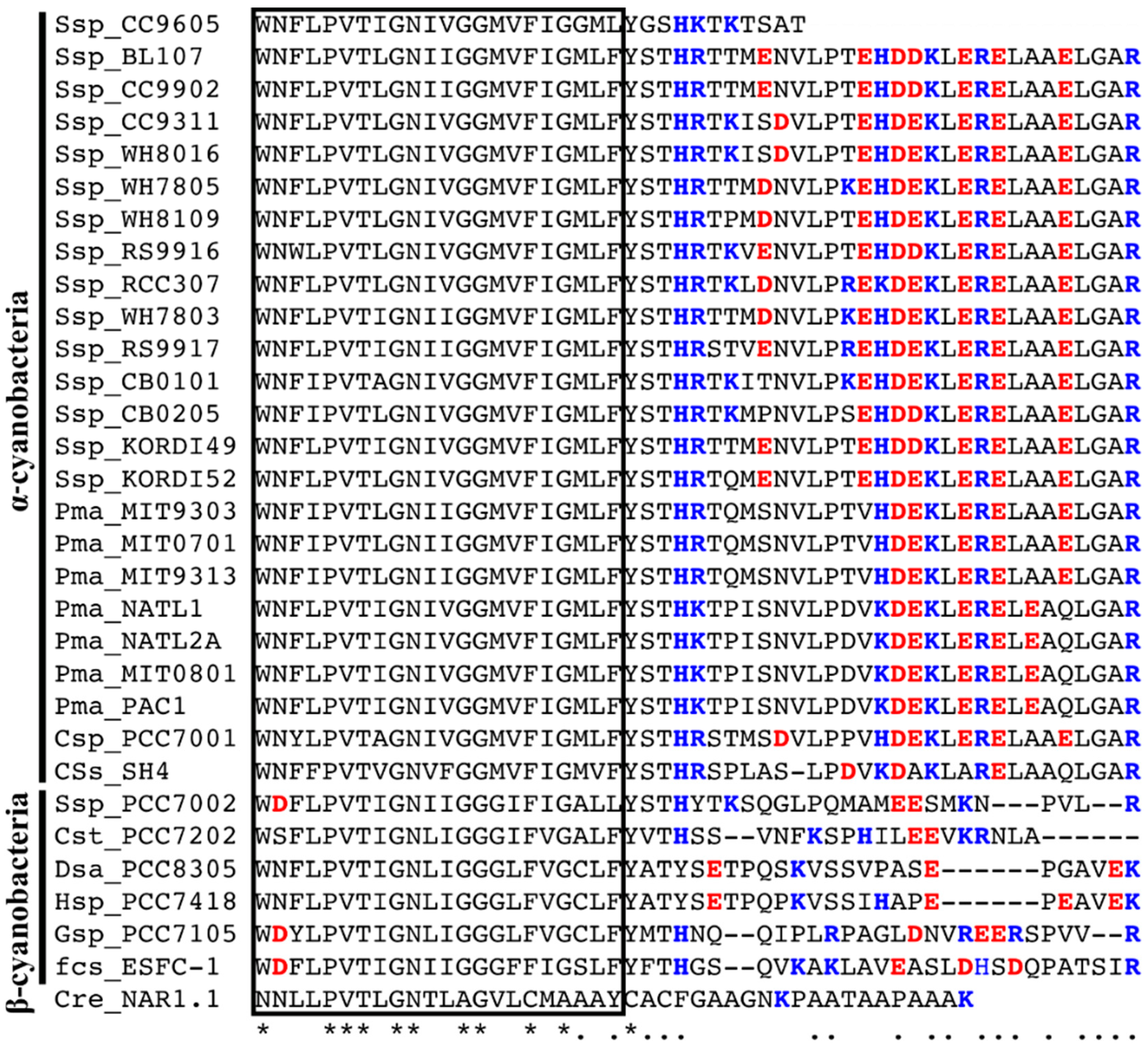
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| SPc | Synechococcus elongatus strain PCC 7942 cured of the pUH24 plasmid, wild type | [34] |
| NA4 | SPc ∆nrtABCD ∆cynABD::CmR, lacking the genes encoding an ABC-type nitrate/nitrite transporter and an ABC-type cyanate/nitrite transporter | [24] |
| NA41 | NA4 harboring pSE1 | [24] |
| NA401 | NA4 harboring pS7002 | This study |
| NA402 | NA4 harboring pS9311 | This study |
| NA403 | NA4 harboring pS9605 | This study |
| NA404 | NA4 harboring pP9313 | This study |
| NA412 | NA4 harboring pS9311∆ | This study |
| NA414 | NA4 harboring pP9313∆ | This study |
| Plasmids | ||
| pSE1 | KmR, Synechococcus shuttle expression vector | [23] |
| pS7002 | pSE1 derivative encoding nitM of Synechococcus sp. strain PCC 7002 | This study |
| pS9311 | pSE1 derivative encoding nitM of Synechococcus sp. strain CC9311 | This study |
| pS9605 | pSE1 derivative encoding nitM of Synechococcus sp. strain CC9605 | This study |
| pP9313 | pSE1 derivative encoding nitM of Prochlorococcus marinus strain MIT 9313 | This study |
| pS9311∆ | pSE1 derivative encoding nitM of Synechococcus sp. strain CC9311 lacking the C-terminal region (21 a.a.) | This study |
| pP9313∆ | pSE1 derivative encoding nitM of Prochlorococcus marinus strain MIT 9313 lacking the C-terminal region (21 a.a.) | This study |
4.2. Expression of Plasmid-Encoded Proteins in S. elongatus Strain PCC 7942
4.3. Measurements of Nitrite Uptake
4.4. Other Methods
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Capone, D.G. Marine nitrogen fixation: What’s the fuss? Curr. Opin. Microbiol. 2001, 4, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Zehr, J.P.; Ward, B.B. Nitrogen cycling in the ocean: New perspectives on processes and paradigms. Appl. Environ. Microbiol. 2002, 68, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Zubkov, M.V.; Fuchs, B.M.; Tarran, G.A.; Burkill, P.H.; Amann, R. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl. Environ. Microbiol. 2003, 69, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Rees, A.P.; Woodward, E.M.S.; Joint, I. Concentrations and uptake of nitrate and ammonium in the Atlantic Ocean. Deep Sea Res. II 2006, 53, 1649–1665. [Google Scholar] [CrossRef]
- Dugdale, R.C.; Goering, J.J. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol. Oceanogr. 1967, 12, 196–206. [Google Scholar] [CrossRef]
- Moore, J.K.; Doney, S.C.; Kleypas, J.A.; Glover, D.M.; Fung, I.Y. An intermediate complexity marine ecosystem model for the global domain. Deep Sea Res. II 2002, 49, 403–462. [Google Scholar] [CrossRef]
- Liu, H.; Campbell, L.; Landry, M.R.; Nolla, H.A.; Brown, S.L.; Constantinou, J. Prochlorococcus and Synechococcus growth rates and contributions to production in the Arabian sea during the 1995 Southwest and Northeast Monsoons. Deep Sea Res. II 1998, 45, 2327–2352. [Google Scholar] [CrossRef]
- Shiozaki, T.; Furuya, K.; Kodama, T.; Takeda, S. Contribution of N2 fixation to new production in the western North Pacific Ocean along 155°E. Mar. Ecol. Prog. Ser. 2009, 377, 19–32. [Google Scholar] [CrossRef]
- Webpage of Japan Meteorological Agency. Available online: http://www.data.jma.go.jp/gmd/kaiyou/db/vessel_obs/hq/2006spr/137e/index_line.php?id=chl (accessed on 5 February 2015).
- Webpage of Japan Meteorological Agency. Available online: http://www.data.jma.go.jp/gmd/kaiyou/db/vessel_obs/hq/2006spr/137e/index_line.php?id=no2 (accessed on 5 February 2015).
- Moore, L.R.; Post, A.F.; Rocap, G.; Chisholm, S.W. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 2002, 47, 989–996. [Google Scholar] [CrossRef]
- Rocap, G.; Larimer, F.W.; Lamerdin, J.; Malfatti, S.; Chain, P.; Ahlgren, N.A.; Arellano, A.; Coleman, M.; Hauser, L.; Hess, W.R.; et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 2003, 424, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Martiny, A.C.; Kathuria, S.; Berube, P.M. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc. Natl. Acad. Sci. USA 2009, 106, 10787–10792. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Hanson, D.T.; Price, G.D. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 2002, 29, 407–416. [Google Scholar] [CrossRef]
- Omata, T.; Ohmori, M.; Arai, N.; Ogawa, T. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc. Natl. Acad. Sci. USA 1989, 86, 6612–6616. [Google Scholar] [CrossRef] [PubMed]
- Omata, T. Cloning and characterization of the nrtA gene that encodes a 45-kDa protein involved in nitrate transport in the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1991, 32, 151–157. [Google Scholar]
- Omata, T.; Andriesse, X.; Hirano, A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC7942. Mol. Gen. Genet. 1993, 236, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Inoue-Sakamoto, K.; Bryant, D.A. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 1999, 181, 7363–7372. [Google Scholar] [PubMed]
- Rexach, J.; Fernández, E.; Galván, A. The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell 2000, 12, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, W.; Siddiqi, Y.; Symington, V.F.; Kinghorn, J.R.; Unkles, S.E.; Glass, A.D. Nitrite transport is mediated by the nitrite-specific high-affinity NitA transporter and by nitrate transporters NrtA, NrtB in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Clegg, S.; Yu, F.; Griffiths, L.; Cole, J.A. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: Two nitrate and three nitrite transporters. Mol. Microbiol. 2002, 44, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Omata, T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 1997, 272, 3036–3041. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Omata, T. Nitrite transport activity of the ABC-type cyanate transporter of the cyanobacterium Synechococcus elongatus. J. Bacteriol. 2009, 191, 3265–3272. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Bryant, D.A. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Aichi, M.; Yoshihara, S.; Yamashita, M.; Maeda, S.; Nagai, K.; Omata, T. Characterization of the nitrate-nitrite transporter of the major facilitator superfamily (the nrtP gene product) from the cyanobacterium Nostoc punctiforme strain ATCC 29133. Biosci. Biotechnol. Biochem. 2006, 70, 2682–2689. [Google Scholar] [CrossRef] [PubMed]
- Lara, C.; Romero, J.M.; Guerrero, M.G. Regulated nitrate transport in the cyanobacterium Anacystis nidulans. J. Bacteriol. 1987, 169, 4376–4378. [Google Scholar] [PubMed]
- Kobayashi, M.; Rodríguez, R.; Lara, C.; Omata, T. Involvement of the C-terminal domain of an ATP-binding subunit in the regulation of the ABC-type nitrate/nitrite transporter of the cyanobacterium Synechococcus sp. strain PCC 7942. J. Biol. Chem. 1997, 272, 27197–27201. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Takatani, N.; Tanigawa, M.; Omata, T. Posttranslational regulation of nitrate assimilation in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2005, 187, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Okamura, M.; Kobayashi, M.; Omata, T. Nitrite-specific active transport system of the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1998, 180, 6761–6763. [Google Scholar] [PubMed]
- Chang, Y.; Takatani, N.; Aichi, M.; Maeda, S.; Omata, T. Evaluation of the effects of PII deficiency and the toxicity of PipX on growth characteristics of the PII-less mutant of the cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2013, 54, 1504–1514. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Flores, E.; Herrero, A.; Houmard, J.; Tandeau de Marsac, N. A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998, 427, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Takatani, N.; Omata, T. Effects of PII deficiency on expression of the genes involved in ammonium utilization in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2006, 47, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Kuhlemeier, C.J.; Thomas, A.A.; van der Ende, A.; van Leen, R.W.; Borrias, W.E.; van den Hondel, C.A.; van Arkel, G.A. A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid 1983, 10, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, I.; Horie, N.; Sugiyama, T.; Omata, T. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J. Bacteriol. 1995, 177, 290–296. [Google Scholar] [PubMed]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Snell, F.D.; Snell, C.T. Colorimetric Methods of Analysis; Van Nostrand: New York, NY, USA, 1949; Volume 2, pp. 802–807. [Google Scholar]
- Mackinney, G. Absorption of light by chlorophyll solutions. J. Biol. Chem. 1941, 140, 315–322. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maeda, S.-i.; Murakami, A.; Ito, H.; Tanaka, A.; Omata, T. Functional Characterization of the FNT Family Nitrite Transporter of Marine Picocyanobacteria. Life 2015, 5, 432-446. https://doi.org/10.3390/life5010432
Maeda S-i, Murakami A, Ito H, Tanaka A, Omata T. Functional Characterization of the FNT Family Nitrite Transporter of Marine Picocyanobacteria. Life. 2015; 5(1):432-446. https://doi.org/10.3390/life5010432
Chicago/Turabian StyleMaeda, Shin-ichi, Akio Murakami, Hisashi Ito, Ayumi Tanaka, and Tatsuo Omata. 2015. "Functional Characterization of the FNT Family Nitrite Transporter of Marine Picocyanobacteria" Life 5, no. 1: 432-446. https://doi.org/10.3390/life5010432
APA StyleMaeda, S.-i., Murakami, A., Ito, H., Tanaka, A., & Omata, T. (2015). Functional Characterization of the FNT Family Nitrite Transporter of Marine Picocyanobacteria. Life, 5(1), 432-446. https://doi.org/10.3390/life5010432





