Genome-Wide Identification of the CBL-CIPK Gene Family in the Ice Plant and Functional Analysis of Salt Stress Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of CBL-CIPK Genes
2.2. Analysis of Physicochemical Properties of CBL-CIPK Proteins
2.3. Analysis of Conserved Domains, Motifs, and Gene Structure of CBL-CIPK Gene Family Members
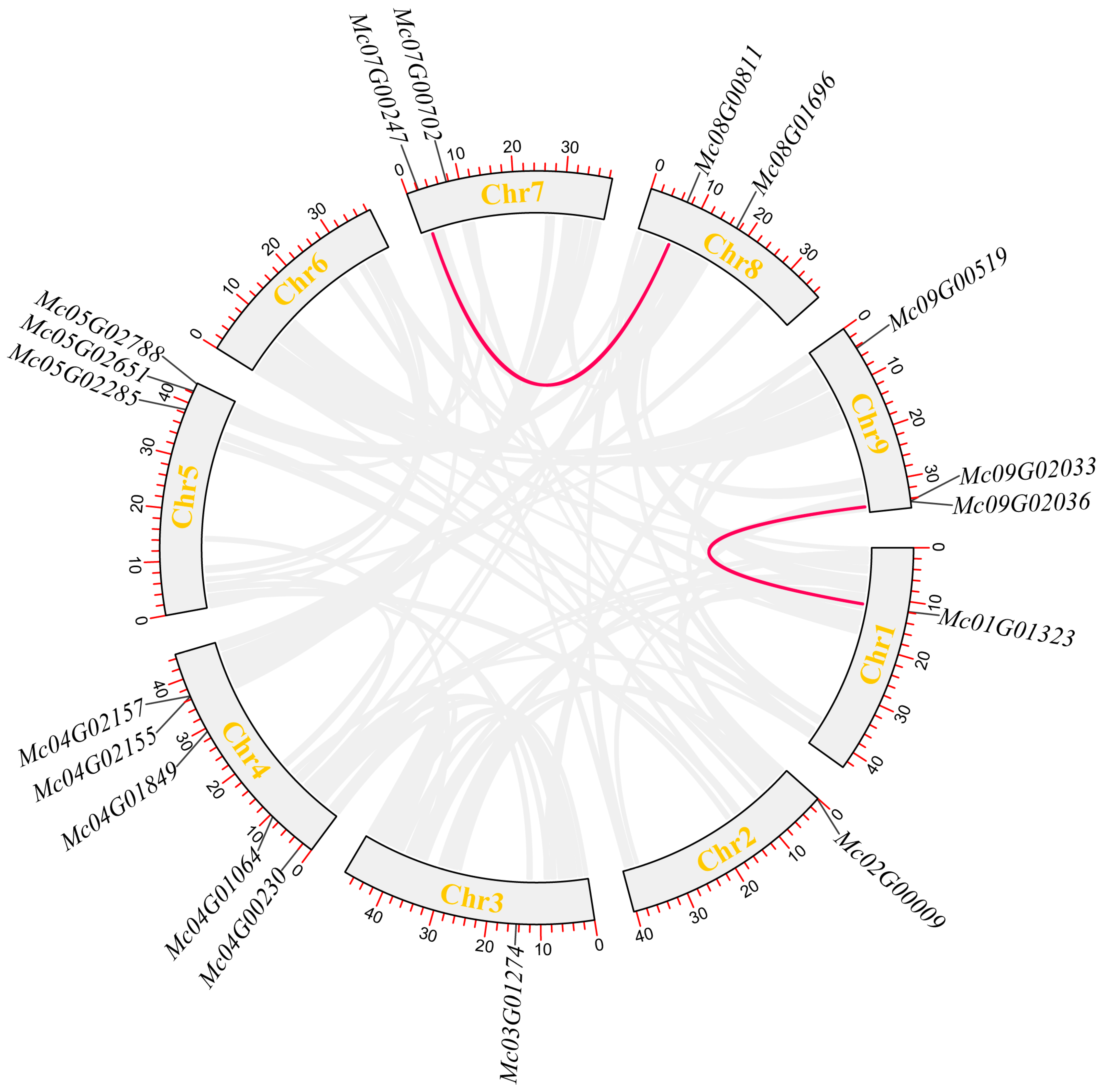
2.4. Chromosomal Distribution and Intraspecies Collinearity Analysis of Ice Plant CBL and CIPK Genes
2.5. Analysis of Cis-Acting Elements in Ice Plant CBL and CIPK Genes
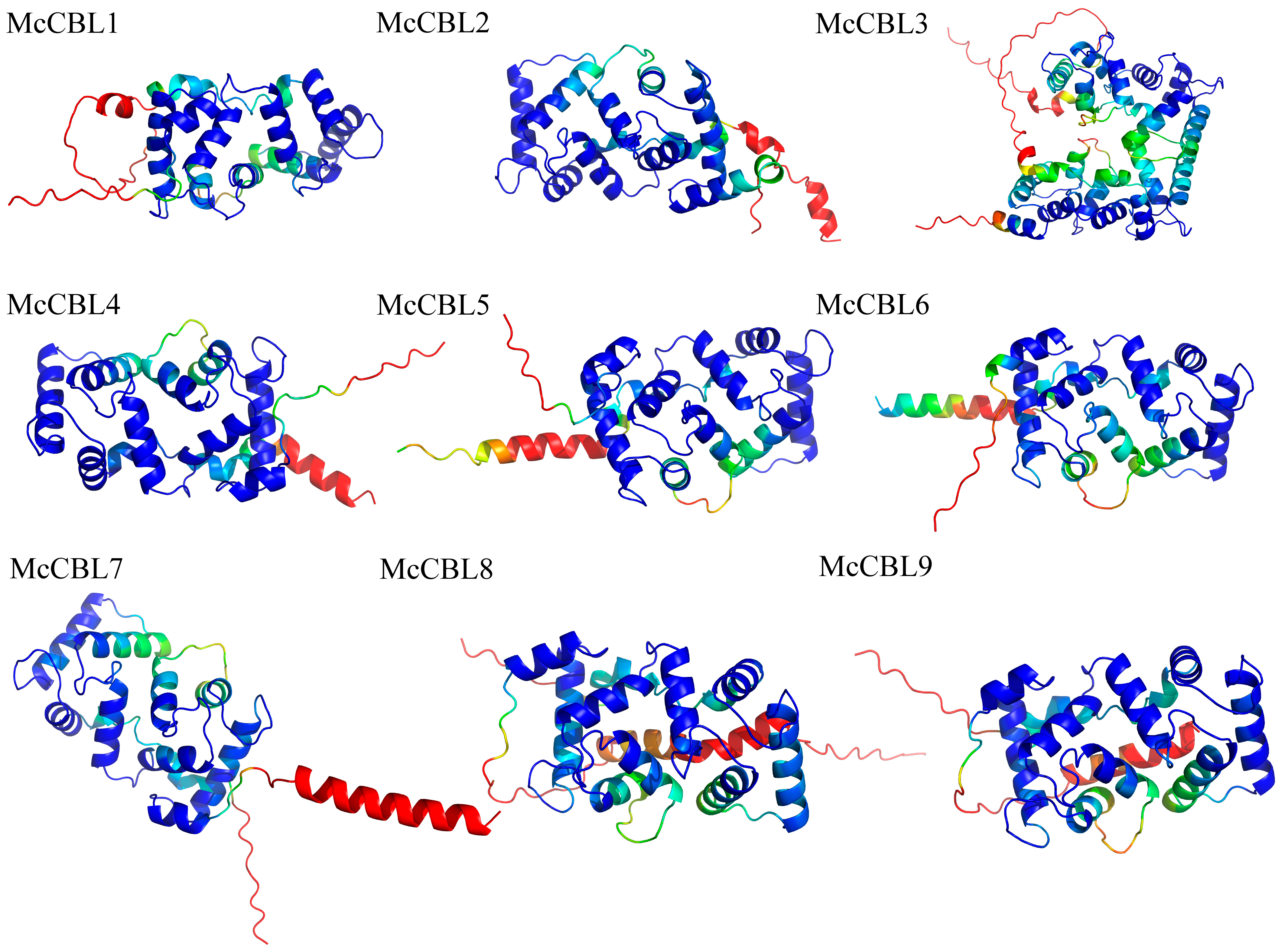
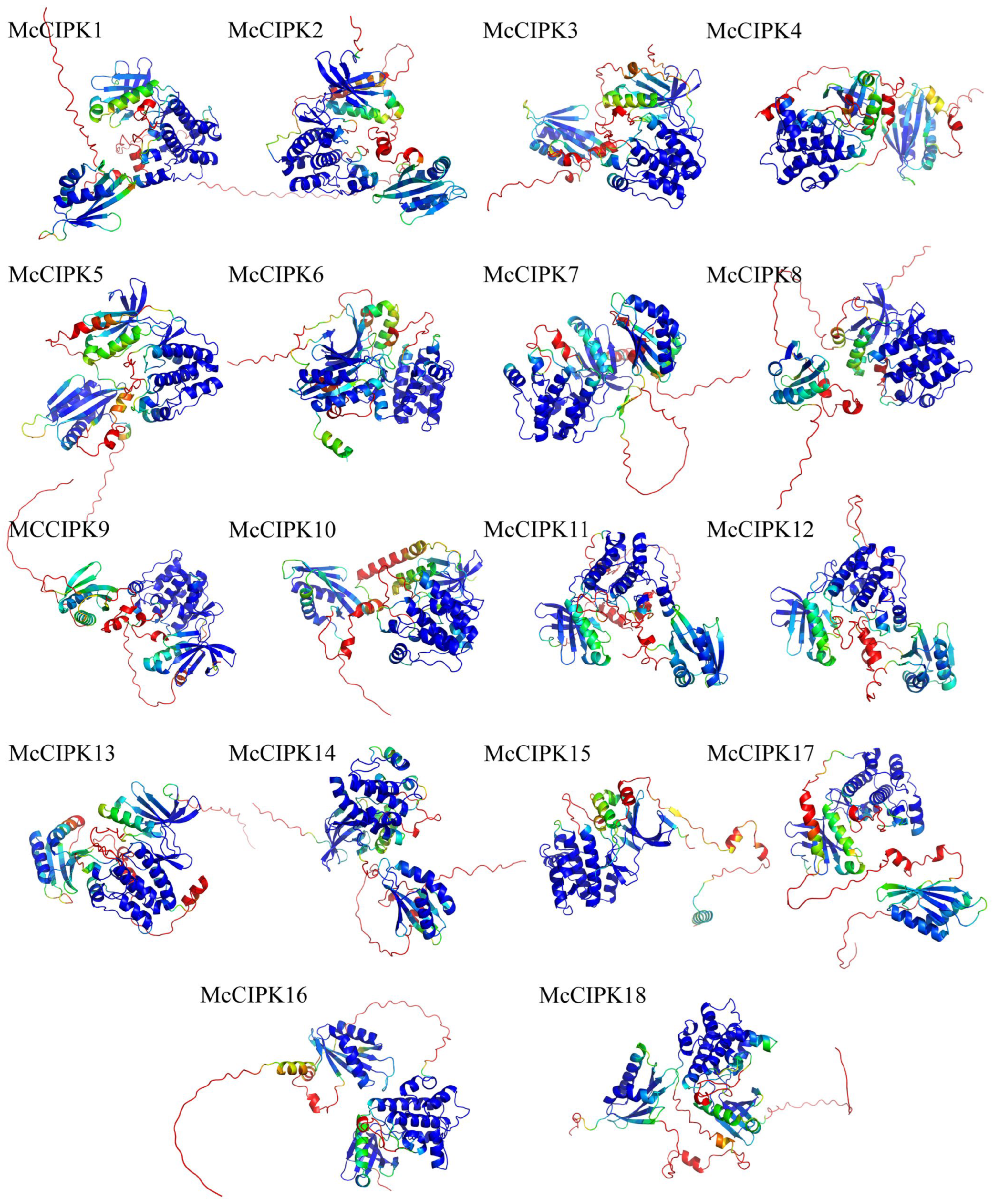
2.6. Construction of 3D Models for Ice Plant CBL-CIPK Proteins
2.7. Protein–Protein Interaction Network Analysis
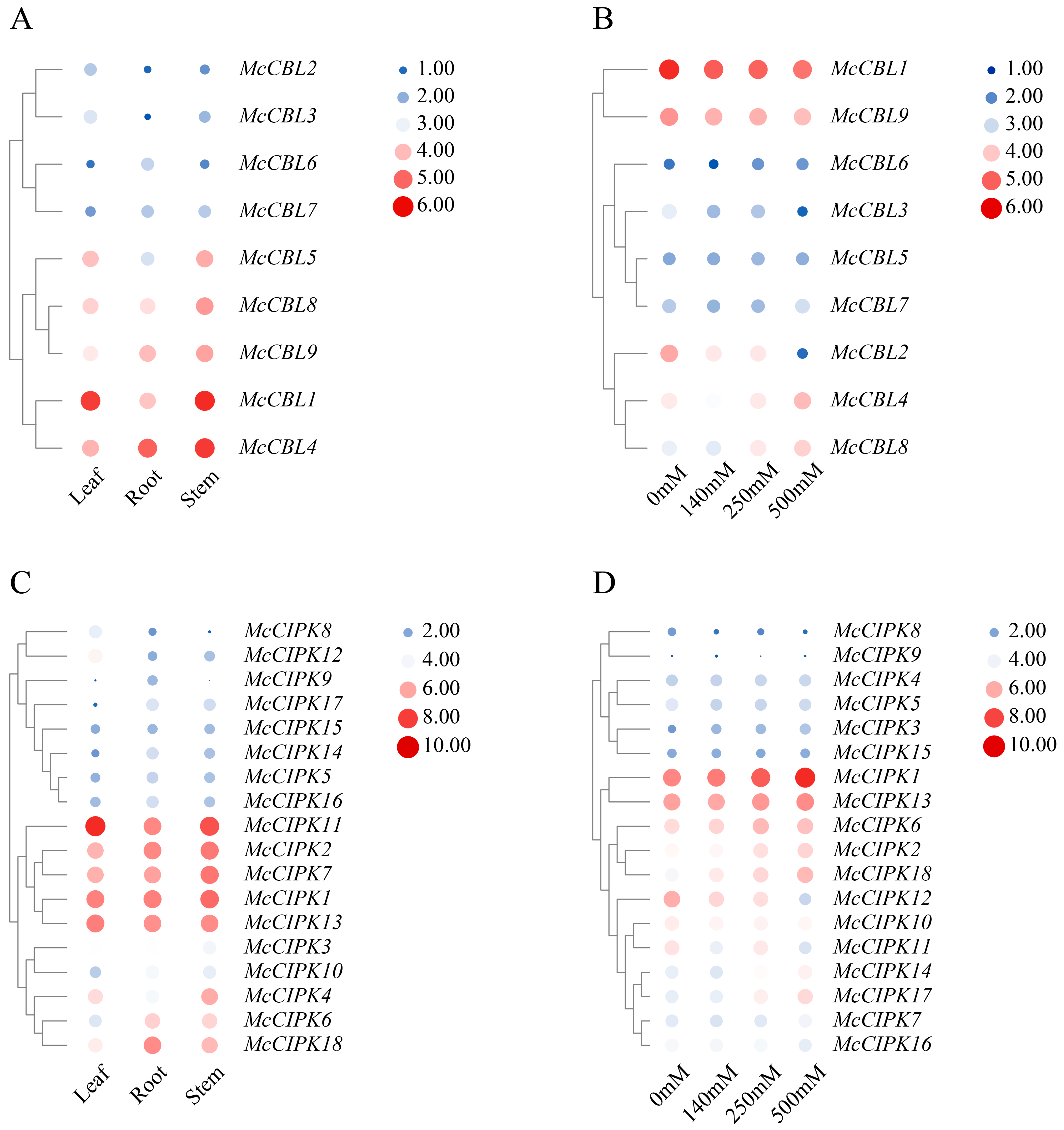
2.8. Gene Expression Profile Analysis
3. Results
3.1. Identification and Phylogenetic Analysis of CBL-CIPK Gene Family Members in 24 Plant Species
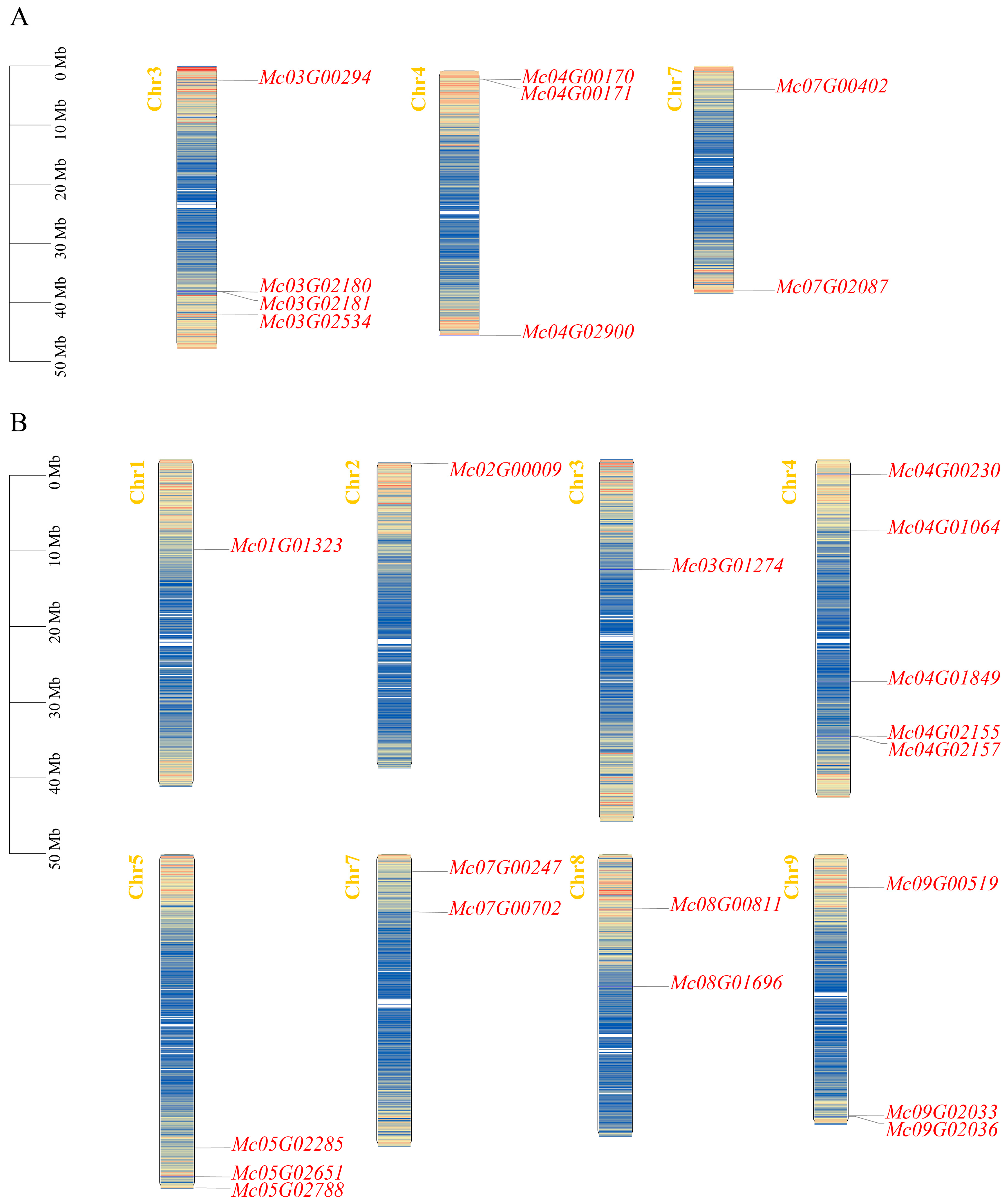
3.2. Chromosomal Localization of Ice Plant CBL-CIPK Gene Family Members
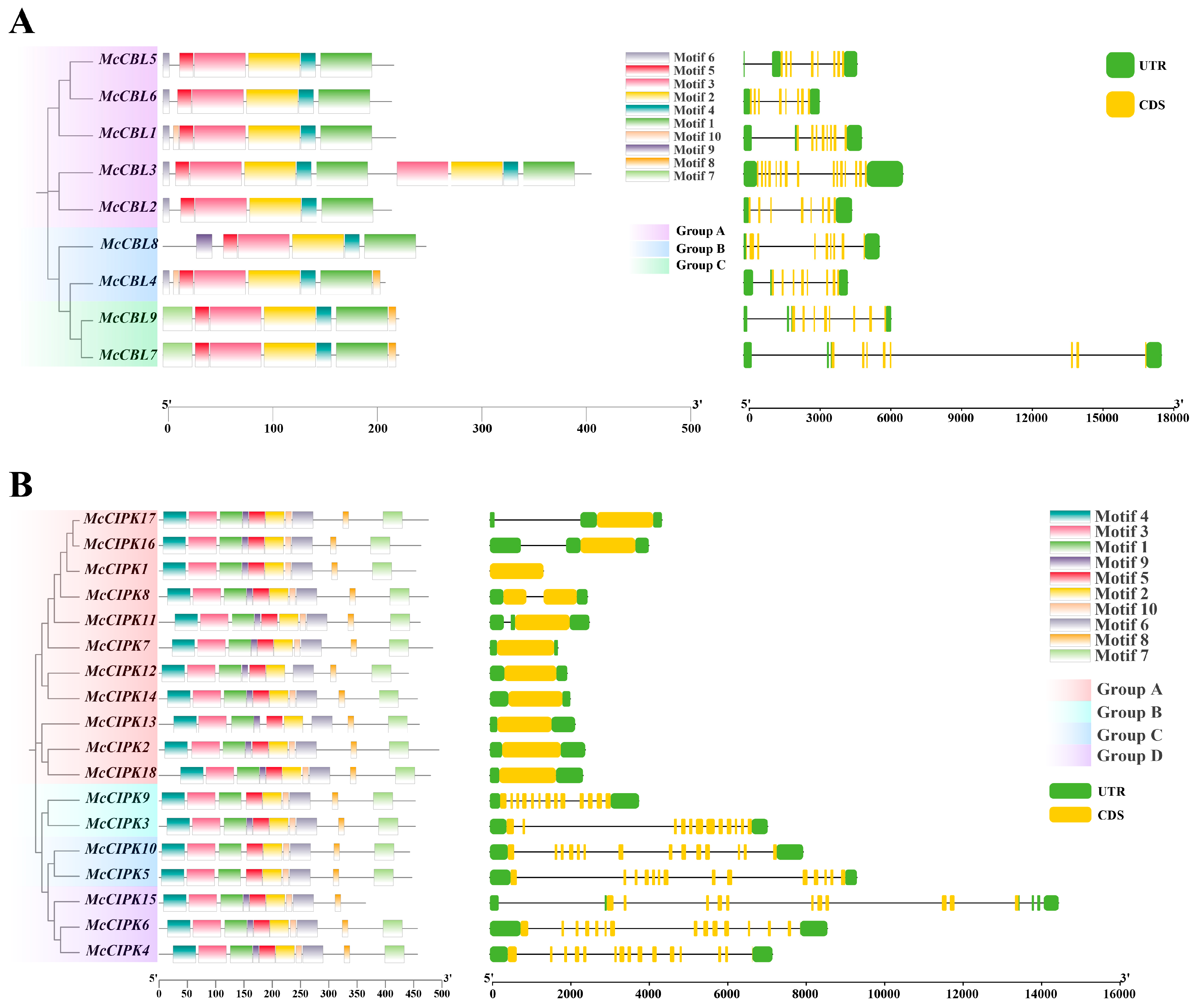
3.3. Analysis of Subfamilies, Conserved Motifs, and Gene Structure of Ice Plant CBL-CIPK Genes
3.4. Physicochemical Properties and Subcellular Localization of Ice Plant CBL-CIPK Proteins
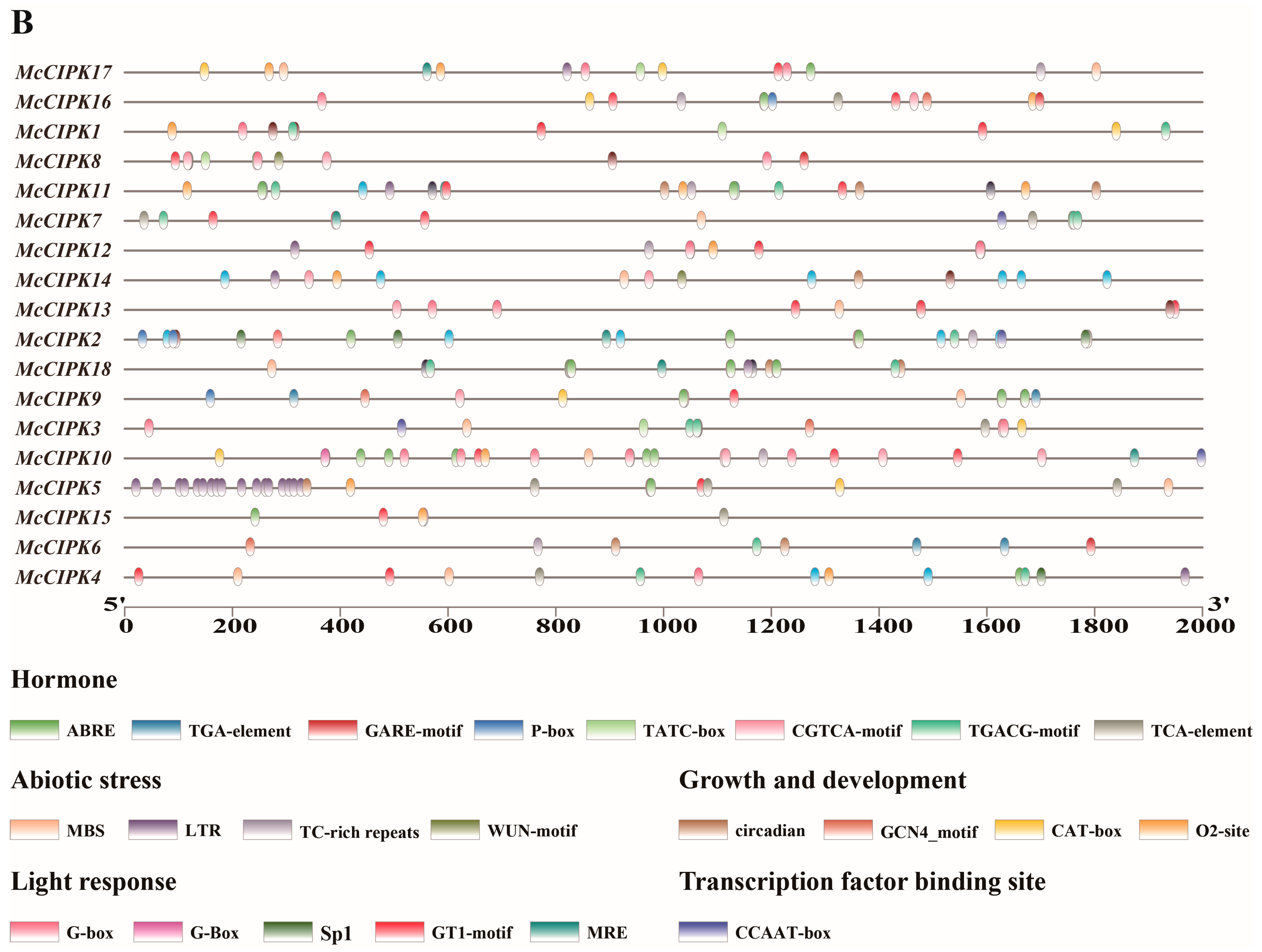
3.5. Analysis of Cis-Acting Regulatory Elements in Ice Plant CBL-CIPK Gene Family Members
3.6. Intraspecies Collinearity Analysis of Ice Plant CBL-CIPK Gene Family Members
3.7. Expression Profile Analysis of Ice Plant CBL-CIPK Genes
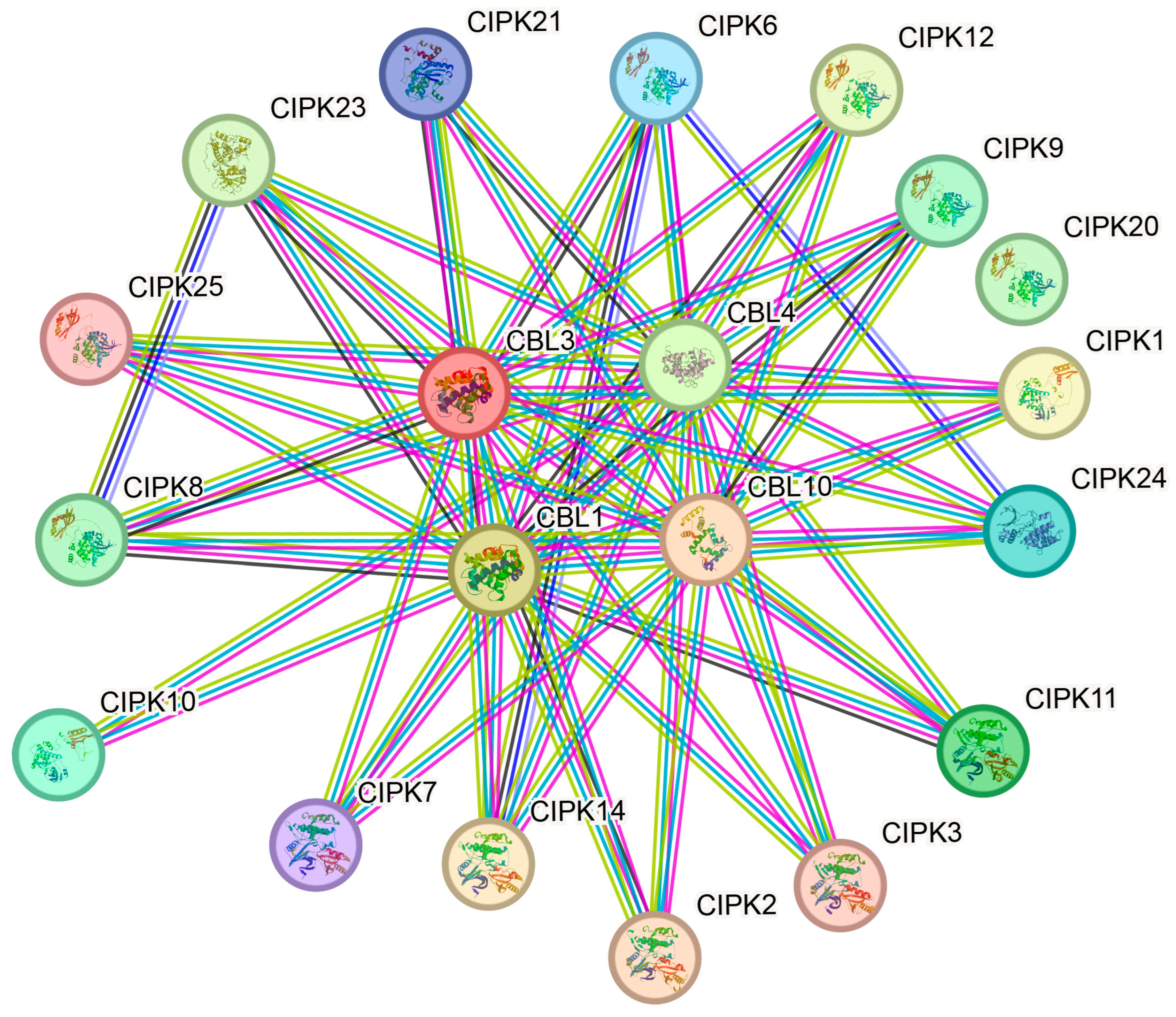
3.8. Construction of 3D Models and Protein Interaction Network Analysis for Ice Plant CBL-CIPK Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sato, R.; Kondo, Y.; Agarie, S. The first released available genome of the common ice plant (Mesembryanthemum crystallinum L.) extended the research region on salt tolerance, C3-CAM photosynthetic conversion, and halophilism. F1000Research 2023, 12, 448. [Google Scholar] [CrossRef]
- Shen, S.; Li, N.; Wang, Y.; Zhou, R.; Sun, P.; Lin, H.; Chen, W.; Yu, T.; Liu, Z.; Wang, Z.; et al. High-quality ice plant reference genome analysis provides insights into genome evolution and allows exploration of genes involved in the transition from C3 to CAM pathways. Plant Biotechnol. J. 2022, 20, 2107–2122. [Google Scholar] [CrossRef]
- Chiang, C.P.; Yim, W.C.; Sun, Y.H.; Ohnishi, M.; Mimura, T.; Cushman, J.C.; Yen, H.E. Identification of Ice Plant (Mesembryanthemum crystallinum L.) MicroRNAs Using RNA-Seq and Their Putative Roles in High Salinity Responses in Seedlings. Front. Plant Sci. 2016, 7, 1143. [Google Scholar] [CrossRef] [PubMed]
- Gamage Kaushalya Madhavi, B.; Mun Choi, G.; Entaz Bahar, M.; Eun Moon, B.; Eun Kim, N.; Lee, H.-W.; Tae Kim, H. Assessment of different salt concentrations on the growth and phytochemical change of the ice plants. J. King Saud Univ.-Sci. 2022, 34, 102168. [Google Scholar] [CrossRef]
- Lee, D.; Kim, S.J.; Choi, Y.J.; Rho, Y.H.; Kang, T.S.; Kim, Y.G.; Kang, K.S. The Glucose-Lowering Effect of Mesembryanthemum crystallinum and D-Pinitol: Studies on Insulin Secretion in INS-1 Cells and the Reduction of Blood Glucose in Diabetic Rats. Nutrients 2025, 17, 193. [Google Scholar] [CrossRef]
- Kang, Y.W.; Joo, N.M. Optimization of Nutrient-Rich Ice Plant (Mesembryanthemum crystallinum L.) Paste Fresh Noodle Pasta Using Response Surface Methodology. Foods 2023, 12, 2482. [Google Scholar] [CrossRef]
- Kim, H.L.; Jung, Y.; Kim, H.I.; Sung, N.Y.; Kim, M.J.; Han, I.J.; Kim, G.; Nho, E.Y.; Park, S.Y.; Han, Y.; et al. Antidiabetic Effect of Fermented Mesembryanthemum crystallinum L. in db/db Mice Involves Regulation of PI3K-Akt Pathway. Curr. Issues Mol. Biol. 2023, 45, 6415–6431. [Google Scholar] [CrossRef]
- Hong, H.T.K.; Trang, P.T.H.; Ho, T.-T.; Dang, J.; Sato, R.; Yoshida, K.; Silaguntsuti, P.; Agarie, S. Reproductive growth characteristics of Mesembryanthemum crystallinum L. in High-Salinity stress conditions. Sci. Hortic. 2024, 331, 113172. [Google Scholar] [CrossRef]
- Sene, J.H.B.; Faye, E.; Tine, A.K. Curbing the Salinization of Arable Land and Agronomically Restoring Salt-affected Soils, a food security challenge: Assessment and prospects, the case of Senegal, West Africa. Mosc. Univ. Soil Sci. Bull. 2023, 78, 461–466. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Suzuki, T.; Nishikawa, K.; Agarie, S.; Ishiguro, S.; Higashiyama, T. RNA-seq analysis of the response of the halophyte, Mesembryanthemum crystallinum (ice plant) to high salinity. PLoS ONE 2015, 10, e0118339. [Google Scholar] [CrossRef]
- Park, K.S.; Kim, S.K.; Cho, Y.-Y.; Cha, M.K.; Jung, D.H.; Son, J.E. A coupled model of photosynthesis and stomatal conductance for the ice plant (Mesembryanthemum crystallinum L.), a facultative CAM plant. Hortic. Environ. Biotechnol. 2016, 57, 259–265. [Google Scholar] [CrossRef]
- Fan, S.; Yang, S.; Shi, K.; Yang, L.; An, M.; Wang, F.; Qi, Y.; Feng, M.; Wang, M.; Geng, P.; et al. Genome-wide identification of the LRX gene family in Cucurbitaceae and expression analysis under salt and drought stress in cucumber. Veg. Res. 2024, 4, e026. [Google Scholar] [CrossRef]
- Peng, Y.; Zhu, H.; Wang, Y.; Kang, J.; Hu, L.; Li, L.; Zhu, K.; Yan, J.; Bu, X.; Wang, X.; et al. Revisiting the role of light signaling in plant responses to salt stress. Hortic. Res. 2025, 12, uhae262. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, S.; Feng, X.; Li, N.; Song, X. Two lncRNAs of Chinese cabbage confer Arabidopsis with heat and drought tolerance. Veg. Res. 2024, 4, e029. [Google Scholar] [CrossRef]
- Zhang, X.X.; Ren, X.L.; Qi, X.T.; Yang, Z.M.; Feng, X.L.; Zhang, T.; Wang, H.J.; Liang, P.; Jiang, Q.Y.; Yang, W.J.; et al. Evolution of the CBL and CIPK gene families in Medicago: Genome-wide characterization, pervasive duplication, and expression pattern under salt and drought stress. BMC Plant Biol. 2022, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, K.; Gong, Y.; Yang, Y.; Yue, Y. Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis). Genes 2022, 13, 795. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.J.; Wang, C.; Li, K.; Luan, S. The CBL-CIPK Calcium Signaling Network: Unified Paradigm from 20 Years of Discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhou, B.; Song, Z.; Tan, Z.; Liu, R.; Luo, Y.; Guo, Z.; Lu, S. A calmodulin-like protein PvCML9 negatively regulates salt tolerance. Plant Physiol. Biochem. 2024, 210, 108642. [Google Scholar] [CrossRef]
- Arthikala, M.-K.; Blanco, L.; Alvarado-Affantranger, X.; Márquez-Guzmán, J.; Lara, M.; Nanjareddy, K. Identification of CBL and CIPK Gene Families and Functional Characterization of PvCIPK7 as an Essential Regulator of Root Nodule Development and Nitrogen Fixation in Phaseolus vulgaris. J. Plant Biol. 2023, 66, 535–549. [Google Scholar] [CrossRef]
- Huang, L.; Li, Z.; Fu, Q.; Liang, C.; Liu, Z.; Liu, Q.; Pu, G.; Li, J. Genome-Wide Identification of CBL-CIPK Gene Family in Honeysuckle (Lonicera japonica Thunb.) and Their Regulated Expression Under Salt Stress. Front. Genet. 2021, 12, 751040. [Google Scholar] [CrossRef]
- Xiaolin, Z.; Baoqiang, W.; Xian, W.; Xiaohong, W. Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa. BMC Genom. 2022, 23, 447. [Google Scholar] [CrossRef]
- Bihani, S.C.; Tarushi; Srivastava, A.K. Decoding the calcium signal: Structural insights into CBL-CIPK pathway in plants. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2025, 1869, 130819. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef]
- Ma, X.; Li, Q.-H.; Yu, Y.-N.; Qiao, Y.-M.; Haq, S.U.; Gong, Z.-H. The CBL–CIPK Pathway in Plant Response to Stress Signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef]
- Mao, J.; Mo, Z.; Yuan, G.; Xiang, H.; Visser, R.G.F.; Bai, Y.; Liu, H.; Wang, Q.; van der Linden, C.G. The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaptation. Plant Cell Environ. 2023, 46, 3012–3022. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, S.; He, K.; Yu, T.; Fu, J.; Gao, S.; Li, H. Exploring salt tolerance mechanisms using machine learning for transcriptomic insights: Case study in Spartina alterniflora. Hortic. Res. 2024, 11, uhae082. [Google Scholar] [CrossRef] [PubMed]
- Gharat, S.A.; Parmar, S.; Tambat, S.; Vasudevan, M.; Shaw, B.P. Transcriptome Analysis of the Response to NaCl in Suaeda maritima Provides an Insight into Salt Tolerance Mechanisms in Halophytes. PLoS ONE 2016, 11, e0163485. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Liu, Z.; Yu, T.; Zhou, R.; Yang, Q.; Cao, R.; Nie, F.; Ma, X.; Bai, Y.; Song, X. Flowering genes identification, network analysis, and database construction for 837 plants. Hortic. Res. 2024, 11, uhae013. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Z.; Chen, H.; Li, N.; Yu, T.; Zhou, R.; Nie, F.; Guo, D.; Ma, X.; Song, X. PHGD: An integrative and user-friendly database for plant hormone-related genes. iMeta 2024, 3, e164. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, C.; He, J.; Li, C.; Fu, Y.; Zhou, Y.; Cao, R.; Liu, H.; Song, X. plantGIR: A genomic database of plants. Hortic. Res. 2024, 11, uhae342. [Google Scholar] [CrossRef]
- Shen, L.; Yang, S.; Xia, X.; Nie, W.; Yang, X. Genome-wide identification of Kip-related protein (KRP) gene family members in eggplant and the function of SmKRP3 under salt stress. Veg. Res. 2024, 4, e013. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Fong, D.; Gao, C.; Churas, C.; Pillich, R.; Lenkiewicz, J.; Pratt, D.; Pico, A.R.; Hanspers, K.; Xin, Y.; et al. Cytoscape Web: Bringing network biology to the browser. Nucleic Acids Res. 2025, 53, W203–W212. [Google Scholar] [CrossRef]
- Cushman, J.C.; Tillett, R.L.; Wood, J.A.; Branco, J.M.; Schlauch, K.A. Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). J. Exp. Bot. 2008, 59, 1875–1894. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Wang, S.T.; Mei, Q.; Sun, T.; Hu, J.T.; Xiao, G.S.; Chen, H.; Xuan, Y.H. The role of CBL–CIPK signaling in plant responses to biotic and abiotic stresses. Plant Mol. Biol. 2024, 114, 53. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C.; Uğurlar, F.; Adamakis, I.-D.S. Molecular Mechanisms of CBL-CIPK Signaling Pathway in Plant Abiotic Stress Tolerance and Hormone Crosstalk. Int. J. Mol. Sci. 2024, 25, 5043. [Google Scholar] [CrossRef] [PubMed]
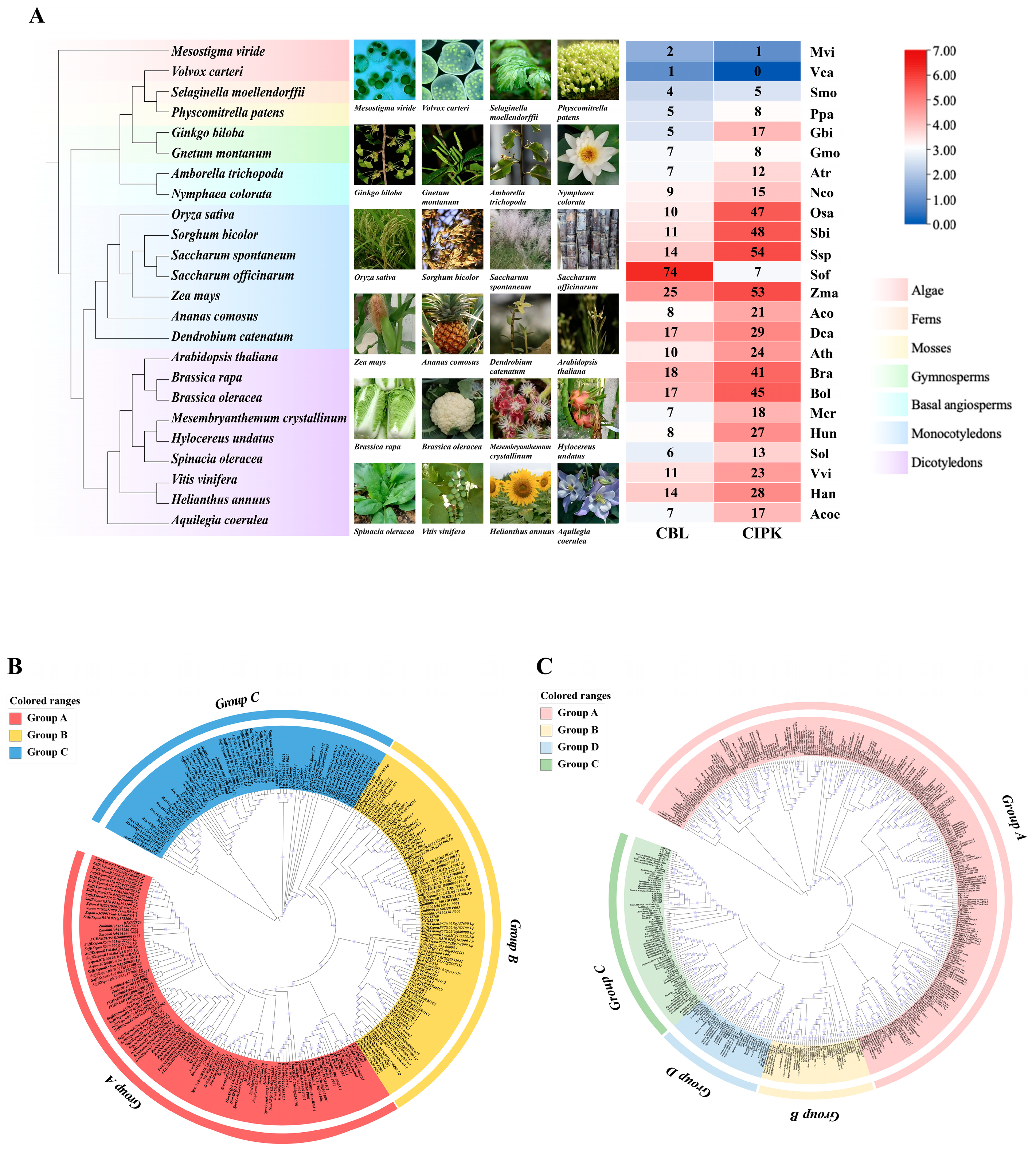

| Classification | Species | CBL | CIPK | Photosynthetic Type |
|---|---|---|---|---|
| Algae | Mesostigma viride | 2 | 1 | C3 |
| Volvox carteri | 1 | 0 | C3 | |
| Ferns | Selaginella moellendorffii | 4 | 5 | C3 |
| Mosses | Physcomitrella patens | 5 | 8 | C3 |
| Gymnosperms | Ginkgo biloba | 5 | 17 | C3 |
| Gnetum montanum | 7 | 8 | C3 | |
| Basal angiosperms | Amborella trichopoda | 7 | 12 | C3 |
| Nymphaea colorata | 9 | 15 | C3 | |
| Monocotyledons | Oryza sativa | 10 | 47 | C3 |
| Sorghum bicolor | 11 | 48 | C4 | |
| Saccharum spontaneum | 14 | 54 | C4 | |
| Saccharum officinarum | 74 | 7 | C4 | |
| Zea mays | 25 | 53 | C4 | |
| Ananas comosus | 8 | 21 | CAM | |
| Dendrobium catenatum | 17 | 29 | Facultative CAM plants | |
| Dicotyledons | Arabidopsis thaliana | 10 | 24 | C3 |
| Brassica rapa | 18 | 41 | C3 | |
| Brassica oleracea | 17 | 45 | C3 | |
| Mesembryanthemum crystallinum | 7 | 18 | Facultative CAM plants | |
| Hylocereus undatus | 8 | 27 | CAM | |
| Spinacia oleracea | 6 | 13 | C3 | |
| Vitis vinifera | 11 | 23 | C3 | |
| Helianthus annuus | 14 | 28 | C3 | |
| Aquilegia coerulea | 7 | 17 | CAM |
| Gene | Num of Amino Acids | Molecular Weight | Isoelectric Point | Instability Index | Aliphatic Index | Hydrophilicity Coefficient | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| McCBL1 | 223 | 25,396.97 | 4.71 | 39.57 | 97.4 | −0.18 | nucl |
| McCBL2 | 219 | 25,112.75 | 4.76 | 32.49 | 93.01 | −0.206 | cyto |
| McCBL3 | 410 | 47,339.71 | 4.64 | 40.35 | 93.17 | −0.262 | cyto |
| McCBL4 | 213 | 24,471.69 | 4.81 | 41.34 | 84.18 | −0.236 | chlo |
| McCBL5 | 221 | 25,620.26 | 4.71 | 52.21 | 89.5 | −0.342 | cyto |
| McCBL6 | 219 | 25,434.09 | 5.24 | 29.47 | 86.3 | −0.43 | cyto |
| McCBL7 | 226 | 25,880.42 | 4.8 | 35.93 | 89.34 | −0.261 | cyto |
| McCBL8 | 252 | 28,688.76 | 4.76 | 37.43 | 98.33 | −0.03 | chlo |
| McCBL9 | 226 | 26,371.12 | 5 | 49.16 | 92.3 | −0.25 | cyto |
| McCIPK1 | 454 | 51,318.55 | 8.9 | 30.37 | 93.59 | −0.341 | cyto |
| McCIPK2 | 495 | 55,330.92 | 6.51 | 42.6 | 83.15 | −0.331 | chlo |
| McCIPK3 | 453 | 50,507.81 | 7.58 | 37.74 | 95.92 | −0.264 | cyto |
| McCIPK4 | 457 | 51,572.96 | 7.66 | 36.76 | 86.19 | −0.365 | chlo |
| McCIPK5 | 447 | 50,779.19 | 6.05 | 40.81 | 93.04 | −0.259 | plas |
| McCIPK6 | 457 | 51,285.25 | 9.14 | 38.82 | 88.51 | −0.31 | mito |
| McCIPK7 | 484 | 53,608.84 | 7.97 | 32.36 | 89.24 | −0.216 | chlo |
| McCIPK8 | 476 | 53,910.13 | 9.1 | 40.96 | 80.08 | −0.405 | chlo |
| McCIPK9 | 453 | 51,118.89 | 6.36 | 32.3 | 93.66 | −0.267 | cyto |
| McCIPK10 | 443 | 50,296.10 | 8.54 | 38.11 | 92.6 | −0.171 | chlo |
| McCIPK11 | 462 | 52,190.72 | 8.6 | 38.95 | 83.96 | −0.393 | plas |
| McCIPK12 | 441 | 50,028.82 | 7.94 | 40.58 | 85.49 | −0.293 | cyto |
| McCIPK13 | 460 | 50,734.46 | 9.16 | 43.44 | 88.11 | −0.225 | chlo |
| McCIPK14 | 457 | 51,885.06 | 9.14 | 27.89 | 82.71 | −0.314 | cyto |
| McCIPK15 | 365 | 41,340.50 | 6.48 | 33.67 | 84.38 | −0.343 | chlo |
| McCIPK16 | 463 | 53,024.01 | 8.64 | 36.53 | 86.35 | −0.505 | cyto |
| McCIPK17 | 476 | 53,892.50 | 8.95 | 35.41 | 86.62 | −0.312 | cyto |
| McCIPK18 | 480 | 54,329.23 | 8.52 | 36.82 | 82.83 | −0.463 | cyto |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Li, N.; Sun, H.; Xu, T.; He, J.; Zhang, C.; Meng, Z.; Zhang, X.; Zhou, R.; Zhang, Y.; et al. Genome-Wide Identification of the CBL-CIPK Gene Family in the Ice Plant and Functional Analysis of Salt Stress Tolerance. Life 2025, 15, 1476. https://doi.org/10.3390/life15091476
Wang C, Li N, Sun H, Xu T, He J, Zhang C, Meng Z, Zhang X, Zhou R, Zhang Y, et al. Genome-Wide Identification of the CBL-CIPK Gene Family in the Ice Plant and Functional Analysis of Salt Stress Tolerance. Life. 2025; 15(9):1476. https://doi.org/10.3390/life15091476
Chicago/Turabian StyleWang, Can, Nan Li, Haifeng Sun, Tianyue Xu, Jinghua He, Chenhao Zhang, Zipeng Meng, Xinyao Zhang, Rong Zhou, Yingchao Zhang, and et al. 2025. "Genome-Wide Identification of the CBL-CIPK Gene Family in the Ice Plant and Functional Analysis of Salt Stress Tolerance" Life 15, no. 9: 1476. https://doi.org/10.3390/life15091476
APA StyleWang, C., Li, N., Sun, H., Xu, T., He, J., Zhang, C., Meng, Z., Zhang, X., Zhou, R., Zhang, Y., & Song, X. (2025). Genome-Wide Identification of the CBL-CIPK Gene Family in the Ice Plant and Functional Analysis of Salt Stress Tolerance. Life, 15(9), 1476. https://doi.org/10.3390/life15091476







