Multigene Identification of a Giant Wild Strain of Ganoderma mutabile (ZHM1939) and Screening of Its Culture Substrates
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
- (1)
- The strain: The wild strain was collected from a rotten wood of coniferous forest on Wulao Mountain in Lincang City, Yunnan Province, China, in June 2022. Its basidiocarp is deposited in Fungarium Union of China (in Hexi University, Zhangye City, Gansu Province, China) with the code FHXU-ZHM1939. The pure culture of the strain is stored in the Mycology Laboratory of West Yunnan University, Lincang, China, coded ZHM1939.
- (2)
- The pure culture: The pure cultures of ZHM1939 were obtained using tissue isolation and purification techniques. Mycelia from the pure cultures were used for microscopic observation of morphology and for extraction of genomic DNA [24].
- (3)
- The PDA medium: The potato dextrose agar (PDA) medium was formulated with 200 g fresh peeled potatoes, 20 g dextrose and 20 g agar, plus tap water to 1000 mL. After being pasteurized at 121 °C (0.10–0.12 Mpa) for 30 min in an autoclave, PDA plates were made by aseptically pouring the medium into sterile Φ90 mm petri dishes (18 mL in each petri dish); PDA slants were made by pouring 10 mL into each of the 20 mL test tubes. The PDA plates and slants were used to culture and store the fungus, respectively.
- (4)
- The primer pairs: The primer pairs ITS1/ITS4 [25], TEF1-983/TEF1-1567 [24] and RPB2-6f/fRPB2-7cR [26] were synthesized by Suoqin Biotech Co., Ltd., Beijing, China, and were applied to amplify the internal transcribed spacer (ITS), alpha 1 translation elongation factor gene (TEF1α) and RNA polymerase II second largest subunit gene (RPB2), respectively, through polymerase chain reaction (PCR).
- (5)
- The culture substrates: The mycelium growth of ZHM1939 was measured on 15 original, 5 propagation and 6 cultivation inoculum substrates (Table 1) for screening the optimal substrates to prepare inocula to be employed at different stages of domestication and cultivation.
- (6)
- Gene sequences downloaded: The ITS, TEF1α and RPB2 gene sequences (Table 2) of 55 strains of related Ganoderma spp. were downloaded from the NCBI GenBank database and were used to construct a phylogenic tree for identification of the present ZHM1939 strain.
2.2. Methods
- (1)
- Pure culture preparation: Initial cultures of ZHM1938 were isolated under aseptic conditions by transferring appropriately small amounts of internal tissues from the basidiocarp onto potato dextrose agar (PDA) medium and incubated at 25 ± 2 °C in darkness for 5 days. The tip mycelium from the edges of initial culture colonies was inoculated onto new PDA plates and incubated at 25 ± 2 °C and darkness for 9 days. Pure cultures of fungus were obtained by three repeated transfers with the same methods. After the PDA surface was fully covered with the fungal mycelium, the stock pure culture of the strain was deposited in the Applied Biology Laboratory of West Yunnan University, with the code ZHM1939. The cultures were maintained at 4 °C for further study.
- (2)
- Morphological observations: These included macro-structural and microscopic observations.
- The macro-structural morphology. The basidiocarp and pure culture colonies of ZHM1939 were directly observed. For the basidiocarp, its total height (from the matrix to the pileus top), length and width diameters, and thickness of the pileus were measured; the shape and color changes of wild fresh and lab-dried samples were described and photographed. For the pure culture colony, its shape and color were observed and photographed; colonies of five replicates (in Φ90 mm petri dishes) were prepared and their diameters were measured with the crossing method [24] to obtain the mean value.
- The microscopic morphology. From the basidiocarp, an appropriately small amount of the tissue was picked up from its internal ventral surface with a sterilized needle, and the morphological characteristics of the skeleton hyphae and basidiospores were observed and 30 of each structure were measured under a microscope. The culture slant was activated at room temperature for 24 h, and an appropriate amount of mycelium was picked up from the slant, placed at the center of the PDA plates (in Φ90 mm petri dishes, 5 replicates), and incubated at 25 ± 2 °C in an incubator for 21 days. The morphology (shape, color etc.) of the colony was observed (photographed) at an appropriate time and the diameter of each colony (mm) was measured. At the same time, the morphological characteristics of the hyphae and spores (color, shape, septa, etc.) were observed under a microscope; the size of 50 spores and the diameter of the hyphae (μm) were measured.
- (3)
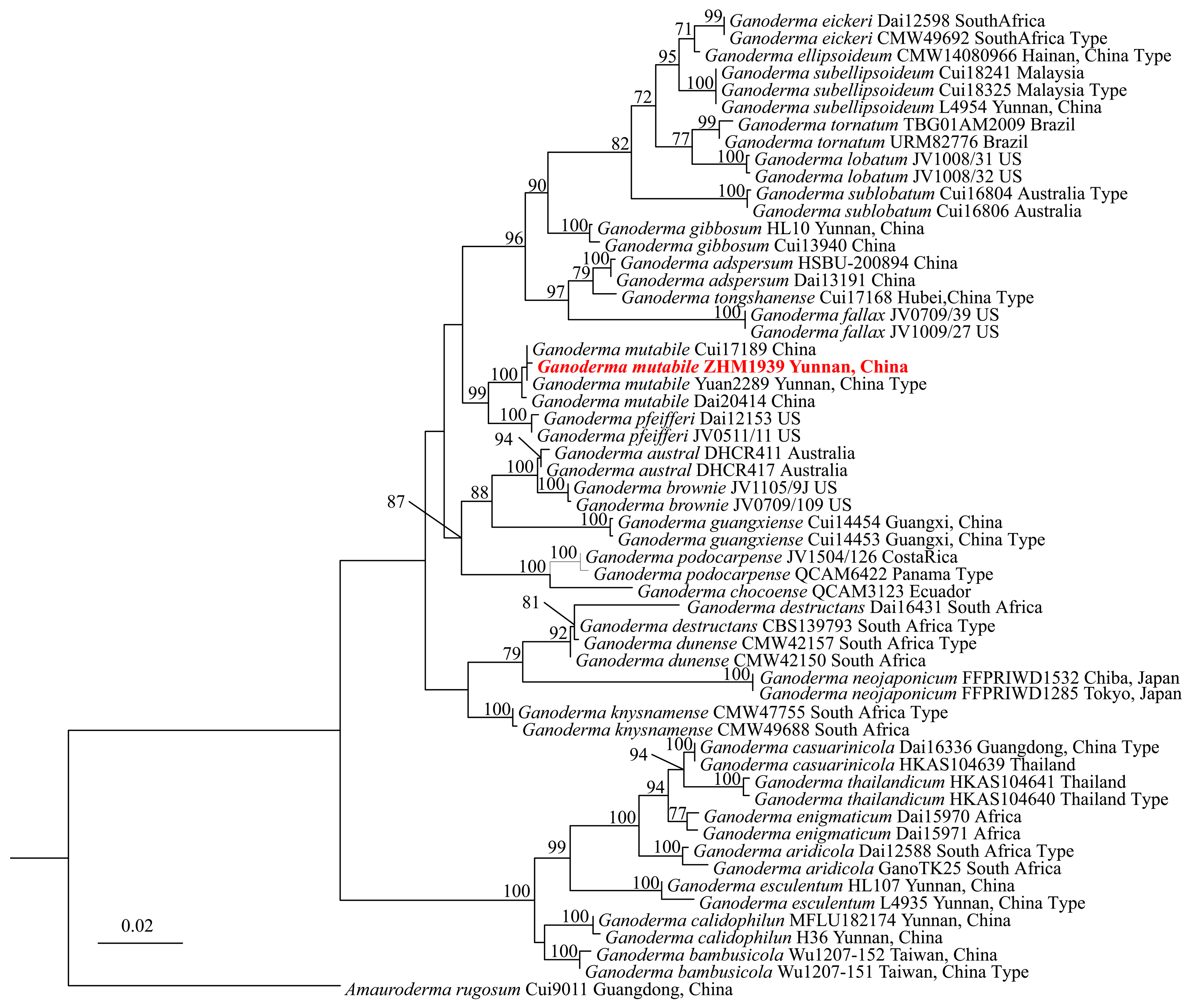
- Molecular identification: The genomic DNA was extracted from the mycelium of ZHM1939 with the cetearyltrimethylammonium bromide (CTAB) method (White et al., 2021) [27]. The ITS, TEF1α and RPB2 gene sequences were amplified from the DNA using the ITS1/ITS4, TEF1-983/TEF1-1567 and RPB2-6f/fRPB2-7cR primers. PCR amplification was performed following the method described by He et al. (2022) [24]. The PCR-amplified products were harvested and sequenced by Beijing Suoqin Biotech. Co. Ltd., China. The sequences were submitted to NCBI GenBank for registration of their accession numbers. Based on blast-n analyses of these sequences, ITS, TEF1α and RPB2 sequences of related Ganoderma spp. were downloaded from NCBI GenBank (Table 2). Using the concatenated sequences of these Ganoderma spp. as an in-group and that of Amauroderma rigosum Cui 9011 strain as a contrast out-group, a maximum likelihood phylogenetic tree was constructed by RAxML-HPC2 on XSEDE on CIPRES Science Gateway [28]. The phylogenetic tree was visualized via Fig Treeversion 1.4.0 and edited with Power Point 2023 software. Finally, the species name of ZHM1939 was determined according to the phylogenetic tree.
- (4)
- Preparation of substrates: The formulae of 15 original, 5 propagation and 6 cultivation inoculum substrates (Table 1) were designed and tested. Their preparation methods are briefly described as the follows.
- Original inoculum substrates. The materials of each substrate were fully mixed or dissolved in an appropriate amount of distilled water and then the volume of the medium was made to 1000 mL by adding distilled water. The solution of each medium was poured into 250 mL flasks and sterilized at 121 °C (0.10–0.12 Mpa) in an autoclave for 30 min. After cooling down to about 60 °C, the medium was poured into Φ90 mm petri dishes to make plates for use. The pH of substrate 7 was adjusted to pH6.5, and the pH of the other substrates was not adjusted. For substrates 1 to 7, the potatoes and/or carrots were peeled, washed and cut into about 1 cm3 cubes, and were boiled until being softened (but not rotten) and filtrated with double-layered gauze. The onion (in medium 13) was boiled with the same amount of water for 30 min and then filtered.
- Propagation inoculum substrates: These substrates were maintained at 65% of moisture and natural pH values. Each substrate was put into cylindrical glass bottles (Φ85 mm and height 118 mm); the bottles were filled fully with the substrate and sealed with moisture-proof paper. The bottled substrate was autoclaved at 121 °C (0.10–0.12 Mpa) for 90 min; after cooling down for 24 h, it was sterilized again at 121 °C (0.10–0.12 Mpa) for 60 min. The wheat grains of all the substrates were soaked in water for 24 h and then boiled until grains were softened; the grains were maintained in the pot for 5 min and the water was filtered from the wheat grains. The broad-leaf wood sawdust and pine needles (10–20 mm sections) were pre-soaked for 6 h and the red soil was sieved with a 200-mesh sieve before being used.
- Cultivation inoculum substrates: The substrates were maintained at about 65% of moisture and natural pH value. Each substrate was put into cylindrical polythene bags (Φ170 mm and height 330 mm); the bags were fully filled with the substrate and sealed with sticky paper. The substrate in the bags was autoclaved at 121 °C (0.14–0.14 Mpa) for 90 min; after cooling down for 24 h, it was sterilized again at 121 °C (0.10–0.12 Mpa) for 90 min. The wheat grains of all the substrates were soaked in water for 24 h and then boiled until 95% of the grains were softened; the grains were maintained in the pot for 5 min and the water was filtered from the wheat grains. The broad-leafed tree wood sawdust and pine needles (10–15 mm sections) were pre-soaked for 4–6 h and the red soil was sieved with a 200-mesh sieve before being used. The sawdust, cottonseed shells, corncobs, broad-leaf tree leaves, fungal bran, and broad bean shells were pre-soaked with tap water for 6–12 h; the cow-dung and red soil were crushed and sieved with a 200-mesh sieve; and the sugar was dissolved in an appropriate amount of tap water before it was were used.
- (5)
- Test of ZHM1939 growth on different substrates: The growth test methods of ZHM1939 mycelium on original, propagation and cultivation inoculum substrates were as the follows:
- Test of original inoculum substrates: Φ5 mm mycelial discs were prepared by punching from the edges of pure culture colony of ZHM1939. One mycelial disc was placed at the center of a PDA plate and five plates were inoculated. The plates were then incubated at 25 ± 2 °C in darkness in an incubator; the moisture was that within the petri dishes (RH > 90%). The new colony on each PDA plate was observed and its diameter was measured 7 days post inoculation. The colony growth rate was calculated (mm/day): r (mm/day) = (mean radius − inoculated mycelia radius)/7.
- Test of propagation inoculum substrates: Φ10 mm mycelial discs were prepared by punching from the edges of the original inoculum colony of ZHM1939. Three of the mycelial discs were placed separately on the surface of each propagation inoculum substrate bottle. Each substrate treatment was replicated in five bottles each. The bottles were sealed and incubated at 25 ± 2 °C, under darkness and relatively high humidity, inside the bottles. The distance of mycelium growth between 5–15 days after inoculation was measured and the mean growth distance (D) of the 5 replicates was used for calculating the mycelium growth rate (mm/day): r = D/10.
- Test on cultivation inoculum substrates: Twenty grams of the ZHN1939-infested wheat grains from the propagation inoculum substrate was seeded uniformly on the surface of the cultivation inoculum substrate in each bag, and five bags were replicated for each substrate. The bags were sealed and incubated at 25 ± 2 °C under darkness and 65% humidity inside the culture bags. The distance of mycelium growth between 5–15 days after inoculation was measured and the mean growth distance (D) of the 5 replicates was used for calculating the mycelium growth rate (mm/day): r = D/10.
3. Results and Analysis
3.1. Morphological Characterization of ZHM1939
3.2. Molecular Identification of ZHM1939
3.3. Growth of ZHM1939 on Original, Propagation and Cultivation Inoculum Substrates
- (1)
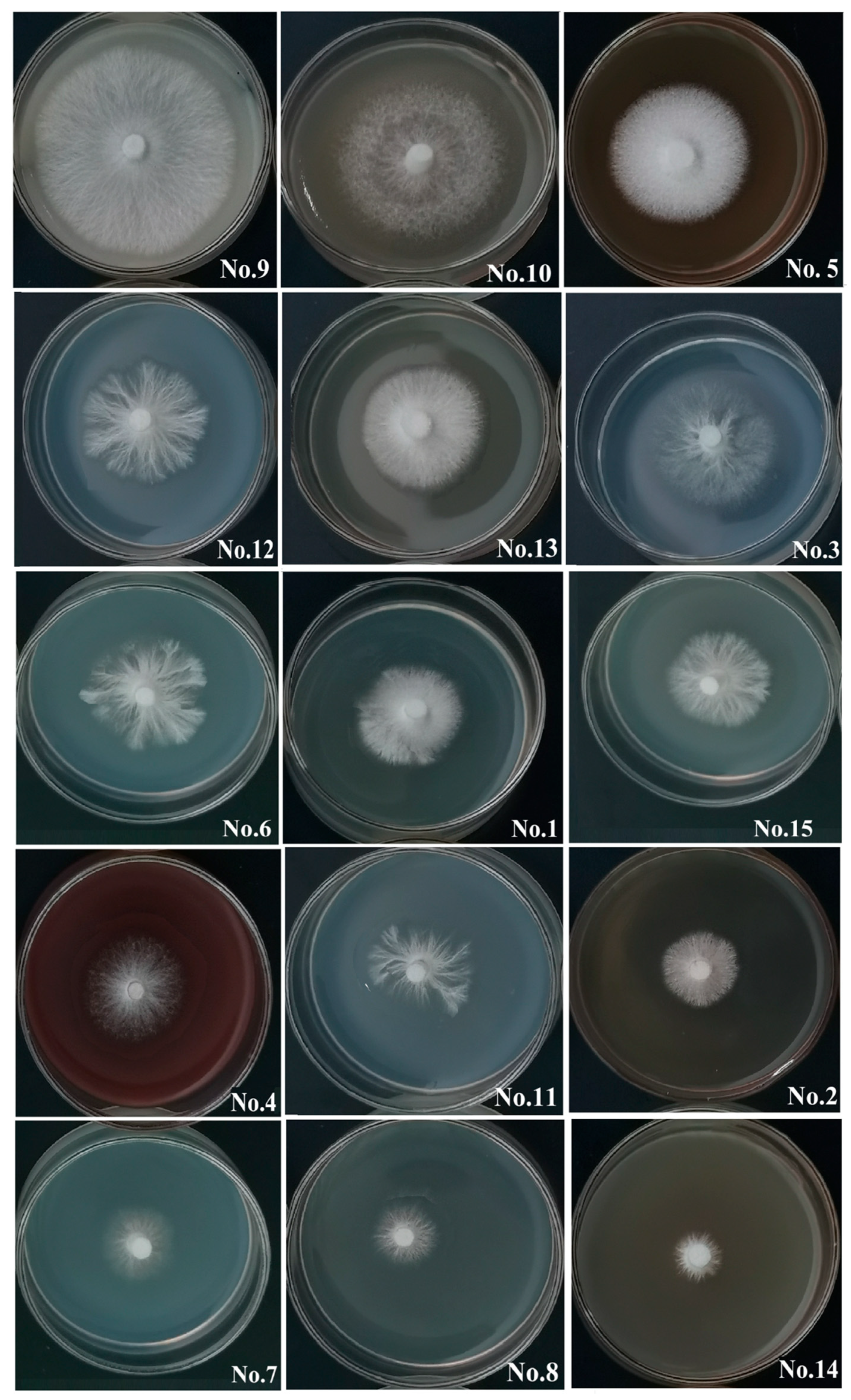
- Growth on original inoculum substrates: A number of 15 original inoculum substrates were tested and the results of ZHM1939 colony growth are presented in Table 3 and visually shown by Figure 3. ZHM1939 produced white colonies on all the substrates tested, but the colonies varied apparently in morphology and size, resulting in significantly different growth rates (p < 0.01) between those on various substrates. After culture at 25 °C on substrate 9 (corn meal 40 g, sucrose10 g, agar 20 g) for 7 days, the colony was the largest (Φ47.51 ± 2.42 mm) and its growth rate was calculated as 6.79 mm/day; the mycelium was the most condensed and robust (Figure 3). Contrarily, the colony (Φ13.32 ± 1.15 mm) formed on substrate 14 (apple juice 200 mL and agar 20 g) was the smallest, with the slowest growth rate (1.91 mm/day); the mycelium of the colony was relatively weak and scarce (Figure 3). Therefore, substrate 9 was the best for producing the original inoculum of ZHM1939.
- (2)
- Growth on propagation inoculum substrates: Mycelium growth in five different propagation inoculum substrates was observed and the results are presented in Table 4. These results indicate that the mycelium was able to grow to fill bottles of all the propagation inoculum substrates, but the time needed varied from 18.63 days (substrate 1) to 23.42 days (substrate 4). Accordingly, mycelial growth and growth rates were significantly different between different substrates (p < 0.01). The best one was cultivation substrate 1 (wheat grains 500 g, gypsum powder 6.5 g and calcium carbonate 2.0 g), in which the mycelium was most condensed and its growth rate was the highest (7.78 mm/dayay) among the five tested cultivation substrates. In cultivation substrate 2, the mycelium was also rather condensed and vigorous and grew fast with a growth rate of 6.65 mm/dayay. In substrate 4, however, the mycelium grew most slowly (growth rate = 4.02 mm/dayay) and much less densely. Consequently, substrate 1 was the best propagation inoculum substrate for culturing ZHM1939.
- (3)
- Growth on cultivation inoculum substrates: In Table 5, the results of the ZHM1939 mycelium growth in the 6 cultivation inoculum substrates tested are shown. Apparently, mycelium growth in cultivation substrate 2 was the fastest (7.64 mm/day) and most vigorous among all the tested substrates, whereas it was the slowest (4.43 mm/day) and was much less exuberant in cultivation substrate 6. As a result, the time needed for the mycelium to grow to the full cultivation bag was nearly 11 days less in substrate 2 (37.12 d) than in substrate 6 (48.03 days). Therefore, substrate 2 was the best cultivation substrate for growing ZHM1939.
4. Discussion
5. Conclusions
6. Patent
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xing, J.H. Studies on Species Diversity, Classification and Systematic Phylogeny of Ganoderma. Ph.D. Thesis, Beijing Forest University, Beijing, China, 2019. [Google Scholar]
- Zhao, X.P. Present status of studies on the medicinal fungi Ganoderma spp. and their cultivation. China Edible Fungi 2019, 38, 1–5. [Google Scholar]
- Cui, B.K.; Pan, X.H.; Pan, F.; Sun, Y.F.; Xing, J.H.; Dai, Y.C. Diversity and resources of Ganoderma spp. in China. Mycosystema 2023, 42, 170–178. [Google Scholar] [CrossRef]
- Mao, X.L. (Ed.) Macro-Fungi in China; Helan Science and Technology Press: Zhengzhou, China, 2000. [Google Scholar]
- Zhang, J.; Kang, J.C.; Wu, X.L. Progresses of molecular systematic studies on Ganoderma spp. Guizhou Sci. 2006, 24, 75–79. [Google Scholar]
- Zhou, Y.Q.; Yang, X.T.; Li, X.Q.; Mi, K.; Feng, H.Q.; Yang, Q.Y. Comparison and mechanism analysis of anti-tumor activity of ethanol extracts from Ganoderma spp. J. Shang Normal Univ. (Nat. Sci. Ed.) 2005, 2, 77–81. [Google Scholar]
- Wei, J.; Cao, Z.H.; Huang, C.Y. Current situation analysis, problems, and countermeasures of edible forest mushroom industry based on the big food view. J. Microbiol. 2025, 65, 1714–1725. [Google Scholar] [CrossRef]
- Li, J.J.; Hu, X.Q.; Zhang, X.F.; Liu, J.J.; Cao, L.S. Study on variation of main ingredients from spores and fruiting bodies of Ganoderma lucidum. Chin. Med. J. 2014, 39, 4246–4251. [Google Scholar] [CrossRef]
- Lu, Z.Q. Liquid deep fermentation technology of Ganoderma lucidum. Jiangshu Foods Ferment 2007, 3, 38–40. [Google Scholar]
- Oke, M.A.; Afolabi, F.J.; Oyeleke, O.O.; Kilani, T.A.; Adeosun, A.R.; Olanbiwoninu, A.A.; Adebayo, E.A. Ganoderma lucidum: Unutilized natural medicine and promising future solution to emerging diseases in Africa. Front. Pharmacol. 2022, 13, 952027. [Google Scholar] [CrossRef]
- Yu, H.Z.; Liu, Y.F.; Zhou, S.; Yan, M.Q.; Xue, L.K.; Tang, Q.J.; Zhang, J.S. Comparison of the polysaccharides from fruiting bodies, mycelia and spore powder of Ganoderma lingzhi. Mycosystema 2016, 35, 170–177. [Google Scholar] [CrossRef]
- Jong, S.C.; Birmingham, J.M. Medicinal benefits of the mushroom Ganoderma. Adv. Appl. Microb. 1992, 37, 101–134. [Google Scholar] [CrossRef]
- Luo, J.; Lin, Z.B. Advances in pharmacological effects of triterpenoids from Ganoderma lucidum. J. Med. 2002, 37, 574–578. [Google Scholar] [CrossRef]
- Shao, H.J.; Sun, J.; Liu, J.B.; Chen, R.Y.; Kang, J. Study on chemical constituents and pharmacological activities of triterpenes from Ganoderma lucidum. Mycol. Res. 2024, 22, 1–21. [Google Scholar]
- Lin, J.W.; Duan, Z.W.; Guan, S.Y.; Han, X.; Fan, W.L. Gene cloning, bio-informatic analysis and eukaryotic expression vector of FIP-gap gene from Ganoderma applanatum. J. Shenyang Agric. Univ. 2016, 47, 1–7. [Google Scholar]
- Li, Z.Q. Comparative study on medicinal efficacy of wild and cultivated Ganoderma applanatum. China Edib. Fungi 2002, 13, 16–18. [Google Scholar]
- Yang, Q.F. Study on Chemical Constituents of Ganoderma resinaceum. Ph.D. Thesis, Yunnan Nationalities University, Kunming, China, 2019. [Google Scholar]
- Wu, G.F.; Yan, H.D. Comparative analysis of total flavonoid content in wild macro-fungi. Chin. Agric. Sci. Bull. 2023, 29, 125–129. [Google Scholar]
- Huang, D.H.; Xiao, D.N.; Zhang, Z.Z. Research progress of wild edible fungi in China based on bibliometrics. Rural. Sci. Tech. 2025, 16, 71–78. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Pan, Q.Q.; Chen, J.X.; Ei, Y.Q.; Zhang, Y.; Ma, H.C.; Hu, Y.P.; Wu, J.R. Investigation of macro-fungal resources in Daweishan National Nature Reserve. J. Yunnan Agric. Univ. (Nat. Sci. Ed.) 2022, 37, 611–622. [Google Scholar]
- Zhang, Y.Z.; Zhou, H.M.; Bai, Y.Y.; Cai, H.M.; Gu, G.H.; Zhao, Y.T.; Tian, L.B.; Mao, M.J. Identification and domestication cultivation of wild type Ganoderma lingzhi from Lincang, Yunnan. J. Tropic. Crops 2020, 41, 2176–2182. [Google Scholar]
- Cao, Y.; Yuan, H.S. Ganoderma mutabile sp. nov. from southwestern China based on morphological and molecular data. Mycol. Progr. 2013, 12, 121–126. [Google Scholar] [CrossRef]
- Sui, Y.X.; Xu, W.L.; Li, C.D.; Guo, L. Screening a inoculum medium for culturing a wild strain of Lepista personata from grassland. Agric. Technol. 2023, 43, 14–17. [Google Scholar]
- He, J.; Han, X.; Luo, Z.L.; Tang, S.M.; Luo, H.M.; Niu, K.Y.; Su, X.; Li, S.H. Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microb. 2022, 13, 1035434. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Mycol. Stud. 2022, 101, 1–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zhou, H.M. (Ed.) Cultivation Techniques of Edible Fungi; China Agricultural University Press: Beijing, China, 2017. [Google Scholar]
- Steel, R.G.D.; Torrie, J.H. Duncan’s new multiple range test. In Principles and Procedures of Statistics; McGraw-Hill: New York, NY, USA, 1980; pp. 187–188. [Google Scholar]
- Mardones, M.; Carranza-Velázquez, J.; Mata-Hidalgo, M.; Amador-Fernández, X.; Urbina, H. Taxonomy and phylogeny of the genus Ganoderma (Polyporales, Basidiomycota) in Costa Rica. MycoKeys 2023, 100, 5–47. [Google Scholar] [CrossRef]
- Sun, Y.F.; Xing, J.H.; He, X.L.; Wu, D.M.; Song, C.G.; Liu, S.; Vlasák, J.; Gates, G.; Gibertoni, T.B.; Cui, B.K. Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 2022, 101, 287–415. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.D.; Ding, Y.; Wen, T.C.; Hapuarachchi, K.K.; Wei, W.D. Danoderma ovisporum sp. nov. (Polyporales, Polyporaceae) from Southwest China. Biod. Data J. 2022, 10, e80034. [Google Scholar] [CrossRef]
- Yuan, Y.; Bian, L.S.; Wu, Y.D.; Chen, J.J.; Wu, F.; Liu, H.G.; Zeng, G.Y.; Dai, Y.C. Species diversity of pathogenic wood-rotting fungi (Agaricomycetes, Basidiomycota) in China. Mycology 2023, 14, 204–226. [Google Scholar] [CrossRef]
- Xing, J.H.; Sun, Y.F.; Han, Y.L.; Cui, B.K.; Dai, B.C. Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys 2018, 34, 93–108. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukes, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef]
- Shen, P.; Chen, X.D. (Eds.) Microbiology, 8th ed.; Higher Education Press: Beijing, China, 2019; pp. 310–330. [Google Scholar]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Nat. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Lv, M.L.; Ying, G.H.; Si, J.P.; Cao, L.S.; Ye, R.H.; Li, L.L. The effects of cultivation medium on the appearance and yield of tree cultivated Ganoderma lucidum. China Edible Fungi 2008, 5, 22–24. [Google Scholar]
- Chen, S.; Liu, S.J.; Gao, Y.; Song, Z.K.; Ma, H.X. Biological characteristics and domestic cultivation of wild Ganoderma gibbosum. Mycosystema 2023, 42, 2218–2230. [Google Scholar] [CrossRef]
- Wannasawang, N.; Luangharn, T.; Thawthong, A.; Charoensup, R.; Jaidee, W.; Tongdeesoontorn, W.; Thongklang, N. Study of optimal conditions to grow Thai Ganoderma, fruiting test, proximate and their alpha glucosidase inhibitory activity. Life 2023, 13, 1887. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.M.; Peng, Z.Q.; He, J.; Tan, W.Z.; Wang, C.W.; Huang, F.Y.; Yu, L.F.; He, M.; Wu, D.; Zhou, Q.W.; et al. Ganoderma mutabile ZHM1939 and Its Mycelium Cultivation Method. Chinese Patent No. ZL2024 1 1285569.7, 17 April 2025. [Google Scholar]

| Substrate for | Number | Medium Components |
|---|---|---|
| Original inoculum | 1 | Potato 200 g, dextrose 20 g |
| 2 | Potato 200 g, sucrose 20 g | |
| 3 | Potato 200 g, maltose 20 g | |
| 4 | Potato 200 g, sucrose 20 g, peptone 10 g | |
| 5 | Potato 200 g, sucrose 20 g, Vitamin B1 1 mg mono potassium phosphate 3 g, magnesium sulfate 1.5 g | |
| 6 | Potato 200 g, carrot 80 g | |
| 7 | Sucrose 20 g, peptone 2 g, mono potassium phosphate 0.5 g, magnesium sulfate 0.5 g (pH 6.5) | |
| 8 | Sucrose 1 g, peptone 10 g | |
| 9 | Yeast powder 5 g, sucrose 20 g | |
| 10 | Sucrose 20 g, rice bran 50 g, monopotassium phosphate 0.3 g, mono potassium phosphate 0.2 g | |
| 11 | Sucrose 20 g, peptone 2 g, Vitamin B1 0.05 mg, monopotassium phosphate 0.46 g, mono potassium phosphate 0.5 g | |
| 12 | Sorghum powder 30 g | |
| 13 | Sucrose 50 g, onion 100 g, soy sauce 50 mL | |
| 14 | Apple juice 200 mL, agar 20 g (pH 6.4–6.8) | |
| 15 | Sucrose 20 g, peptone 2 g, yeast powder 2 g | |
| Propagation inoculum | 1 | Wheat grains 500 g, gypsum powder 6.5 g, calcium carbonate 2 g |
| 2 | Formula 1 + broad-leaf tree sawdust 25 g | |
| 3 | Formula 1 + red soil powder 25 g | |
| 4 | Formula 1 + wheat bran 25 g | |
| 5 | Formula 1 +, pine leaves 25 g | |
| Cultivation inoculum | 1 | Wood saw dust 75 g, rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g |
| 2 | Rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g, cottonseed hulls 75 g | |
| 3 | Rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g, corncob 75 g | |
| 4 | Rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g, corncob 60 g, mushroom bran 15 g | |
| 5 | Rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g, corncob 45 g, mushroom bran 15 g, broad bean skin 15 g | |
| 6 | Rice bran 12 g, tree leaves 5 g, corn meal 5 g, gypsum powder 1 g, sugar 1 g, red soil 1 g, corncob 30 g, mushroom bran 15 g, broad bean skin 15 g, dried cow dung 15 g |
| Species | Voucher/Strain | Origin | GenBank Accession Numbers | ||
|---|---|---|---|---|---|
| ITS | TEF 1α | RPB2 | |||
| G. adspersum | HSBU200894 | China | MG279154 | MG367542 | - |
| G. adspersum | Dai13191 | China | MG279153 | MG367541 | MG367492 |
| G. aridicola | Dai12588T | South Africa | KU572491 | KU572502 | - |
| G. aridicola | GanoTK25 | South Africa | JN105708 | - | - |
| G. australe | DHCR411 | Australia | MF436675 | MF436677 | - |
| G. australe | DHCR417 | Australia | MF436676 | MF436678 | - |
| G. bambusicola | Wu1207-151T | Taiwan, China | MN957781 | LC517941 | LC517944 |
| G. bambusicola | Wu1207-152 | Taiwan, China | MN957782 | LC517942 | LC517945 |
| G. brownii | JV0709/109 | US | KF605662 | MG367548 | MG367495 |
| G. brownii | JV1105/9J | US | MG279159 | MG367547 | MG367494 |
| G. calidophilun | MFLU182174 | Yunnan, China | MN398337 | - | - |
| G. calidophilun | H36 | Yunnan, China | MW750241 | MW838997 | MW839003 |
| G. casuarinicola | HKAS104639 | Thailand | MK817650 | MK871328 | MK840868 |
| G. casuarinicola | Dai16336 T | Cantong, China | MG279173 | MG367565 | MG367508 |
| G. chocoense | QCAM 3123 | Ecuador | MH890527 | - | - |
| G. destructans | CBS139793 T | South Africa | NR132919 | - | - |
| G. destructans | Dai16431 | South Africa | MG279177 | MG367569 | MG367512 |
| G. dunense | CMW42150 | SouthAfrica | MG020249 | MG020228 | - |
| G. dunense | CMW42157 T | South Africa | MG020255 | MG020227 | - |
| G. eickeri | CMW49692 T | South Africa | MH571690 | MH567287 | |
| G. eickeri | Dai 12598 | South Africa | MZ354965 | - | - |
| G. ellipsoideum | CMW14080966 | Hainan, Chin | MH106867 | MZ221652 | - |
| G. ellipsoideum | L4954 | Yunnan, China | ON994242 | OP508446 | - |
| G. enigmaticum | Dai15971 | Africa | KU572487 | KU572497 | MG367514 |
| G. enigmaticum | Dai15970 | Africa | KU572486 | KU572496 | MG367513 |
| G.esculentum | L4935T | Yunnan, China | MW750242 | MW838998 | MW839004 |
| G.esculentum | HL107 | Yunnan, China | ON994243 | OP508437 | OP508424 |
| G. fallax | JV1009/27 | US | KF605655 | - | - |
| G. fallax | JV0709/39 | US | KF605658 | - | - |
| G. gibbosum | HL10 | Yunnan, China | ON994245 | OP508434 | OP508421 |
| G. gibbosum | Cui13940 | China | MZ354972 | MZ221658 | MZ245404 |
| G. guangxiense | Cui14453T | Guangxi, China | MZ354939 | MZ221661 | MZ245407 |
| G. guangxiense | Cui14454 | Guangxi, China | MZ354941 | MZ221662 | MZ245408 |
| G. knysnamense | CMW47755T | South Africa | MH571681 | MH567261 | |
| G. knysnamense | CMW49688 | South Africa | MH571684 | MH567274 | |
| G. lobatum | JV1008/31 | US | KF605671 | MG367553 | MG367499 |
| G. lobatum | JV1008/32 | US | KF605670 | MG367554 | MG367500 |
| G. mutabile | Yuan 2289T | Yunnan, China | JN383977 | - | - |
| G. mutabile | Cui17189 | China | MZ354976 | MZ221679 | - |
| G. mutabile | Dai20414 | China | MZ354977 | MZ221680 | MZ345735 |
| G. mutabile | ZHM1939 | Yunnan, China | PV545123 | PV550967 | PV550966 |
| G. neojaponicum | FFPRI WD1532 | Chiba, Japan | MN957785 | - | - |
| G. neojaponicum | FFPRI WD1285 | Tokyo, Japan | MN957784 | - | - |
| G. pfeifferi | JV0511/11 | US | KF605660 | - | - |
| G. pfeifferi | Dai12153 | US | MG279164 | MG367559 | |
| G. podocarpense | QCAM 6422T | Panama | MF796661 | - | - |
| G. podocarpense | JV1504/126 | Costa Rica | MZ354942 | MZ221687 | MZ345737 |
| G. subellipsoideum | Cui18325T | Malaysia | - | MZ221702 | - |
| G. subellipsoideum | Cui18241 | Malaysia | - | MZ221701 | - |
| G. sublobatum | Cui16804T | Australia | MZ354973 | MZ221704 | MZ345747 |
| G. sublobatum | Cui16806 | Australia | MZ354974 | MZ221705 | - |
| G. thailandicum | HKAS104640 T | Thailand | MK848681 | MK875829 | MK875831 |
| G. thailandicum | HKAS104641 | Thailand | MK848682 | MK875830 | MK875832 |
| G. tongshanense | Cui17168T | Hubei, China | MZ354975 | MZ221706 | - |
| G. tornatum | TBG01AM2009 | Brazil | JQ514108 | - | - |
| G. tornatum | URM82776 | Brazil | JQ514110 | - | - |
| Amauroderma rugosum | Cui9011 | Kantong, China | KJ531664 | KU572504 | MG367506 |
| Substrate No. | Colony Diameter (mm) | Growth Rate (mm/day) | Growth Vigorousness |
|---|---|---|---|
| 9 | 47.51 ± 2.42 A | 3.04 ± 0.06 A | +++ |
| 10 | 39.92 ± 0.91 B | 2.49 ± 0.13 B | +++ |
| 5 | 31.67 ± 2.31 C | 1.90 ± 0.33 C | ++ |
| 12 | 31.27 ± 2.53 C | 1.90 ± 0.08 C | ++ |
| 13 | 27.76 ± 1.84 CD | 1.62 ± 0.08 CD | ++ |
| 3 | 27.58 ± 1.12 CD | 1.61 ± 0.16 CD | ++ |
| 6 | 26.67 ± 1.33 DE | 1.55 ± 0.19 DE | ++ |
| 1 | 25.65 ± 1.19 DE | 1.48 ± 0.17 DE | ++ |
| 15 | 23.09 ± 1.47 DEF | 1.29 ± 0.21 DEF | ++ |
| 4 | 22.33 ± 0.56 EF | 1.23 ± 0.08 EF | + |
| 11 | 22.08 ± 0.49 EF | 1.22 ± 0.07 EF | + |
| 2 | 21.92 ± 2.03 EF | 1.21 ± 0.29 EF | + |
| 7 | 20.65 ± 1.05 F | 1.12 ± 0.15 F | + |
| 8 | 18.58 ± 0.21 F | 0.97 ± 0.03 F | + |
| 14 | 13.32 ± 1.15 G | 0.60 ± 0.04 G | + |
| Substrate | Colony Full Time (d) | Colony Distance (mm) | Grow. Rate (mm/day) | Grow. Vigorousness |
|---|---|---|---|---|
| 1 | 18.63 ± 0.82 D | 77.82 ± 2.15 A | 7.78 ± 0.21 A | ++ |
| 2 | 21.02 ± 2.25 B | 66.50 ± 2.82 B | 6.65 ± 0.28 B | ++ |
| 5 | 20.38 ± 2.15 BC | 22.17 ± 1.26 D | 1.12 ± 0.13 D | ++ |
| 3 | 22.50 ± 1.36 AB | 44.83 ± 2.84 C | 4.28 ± 0.28 C | + |
| 4 | 23.42 ± 3.20 A | 40.21 ± 2.13 C | 4.00 ± 0.21 C | + |
| Substrate | Colony Full Time (d) | Colony Growth (mm) | Growth Rate (mm/day) | Growth Vigorousness |
|---|---|---|---|---|
| 2 | 37.12 ± 1.92 CD | 76.63 ± 2.08 A | 7.64 | ++ |
| 4 | 39.21 ± 2.04 C | 63.32 ± 1.49 B | 6.31 | ++ |
| 3 | 41.16 ± 2.25 BC | 61.71 ± 1.92 B | 6.16 | ++ |
| 1 | 43.96 ± 2.85 B | 54.47 ± 1.23 C | 5.61 | ++ |
| 5 | 46.87 ± 3.26 A | 45.38 ± 0.74 D | 4.54 | + |
| 6 | 48.03 ± 2.92 A | 44.36 ± 0.91 D | 4.43 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Bao, L.; Peng, Z.; Bai, Y.; Su, Q.; Yu, L.; Ma, C.; He, J.; Tan, W. Multigene Identification of a Giant Wild Strain of Ganoderma mutabile (ZHM1939) and Screening of Its Culture Substrates. Life 2025, 15, 1475. https://doi.org/10.3390/life15091475
Zhou H, Bao L, Peng Z, Bai Y, Su Q, Yu L, Ma C, He J, Tan W. Multigene Identification of a Giant Wild Strain of Ganoderma mutabile (ZHM1939) and Screening of Its Culture Substrates. Life. 2025; 15(9):1475. https://doi.org/10.3390/life15091475
Chicago/Turabian StyleZhou, Huiming, Longqian Bao, Zeqin Peng, Yuying Bai, Qiqian Su, Longfeng Yu, Chunlian Ma, Jun He, and Wanzhong Tan. 2025. "Multigene Identification of a Giant Wild Strain of Ganoderma mutabile (ZHM1939) and Screening of Its Culture Substrates" Life 15, no. 9: 1475. https://doi.org/10.3390/life15091475
APA StyleZhou, H., Bao, L., Peng, Z., Bai, Y., Su, Q., Yu, L., Ma, C., He, J., & Tan, W. (2025). Multigene Identification of a Giant Wild Strain of Ganoderma mutabile (ZHM1939) and Screening of Its Culture Substrates. Life, 15(9), 1475. https://doi.org/10.3390/life15091475






