Prophage Activation: An In Silico Platform for Identifying Prophage Regulatory Elements to Inform Phage Engineering Against Drug-Resistant Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Pipeline Analysis of Prophage Activation
3. Results
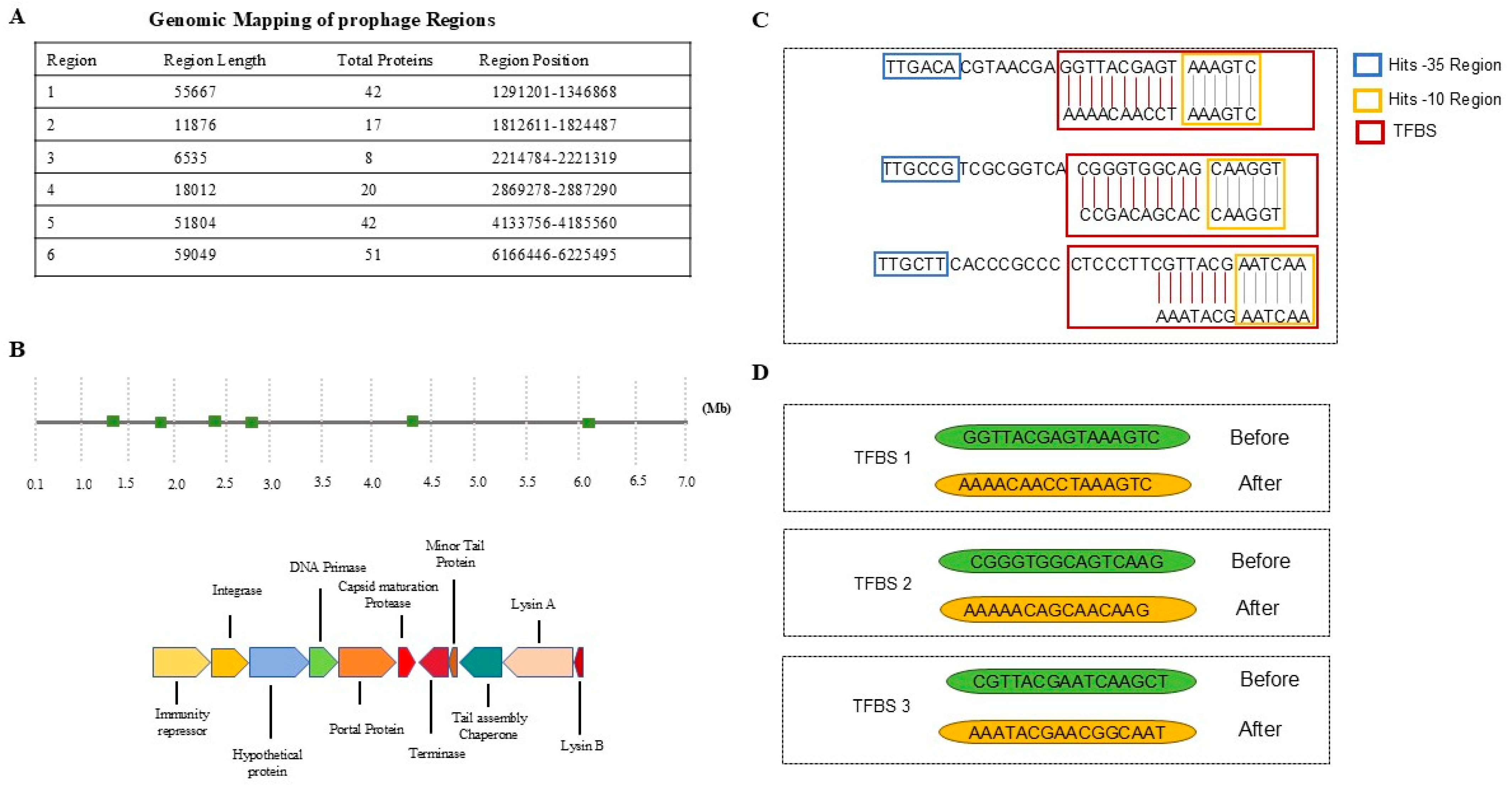
3.1. The Prophage Activation Platform: Proof of Concept
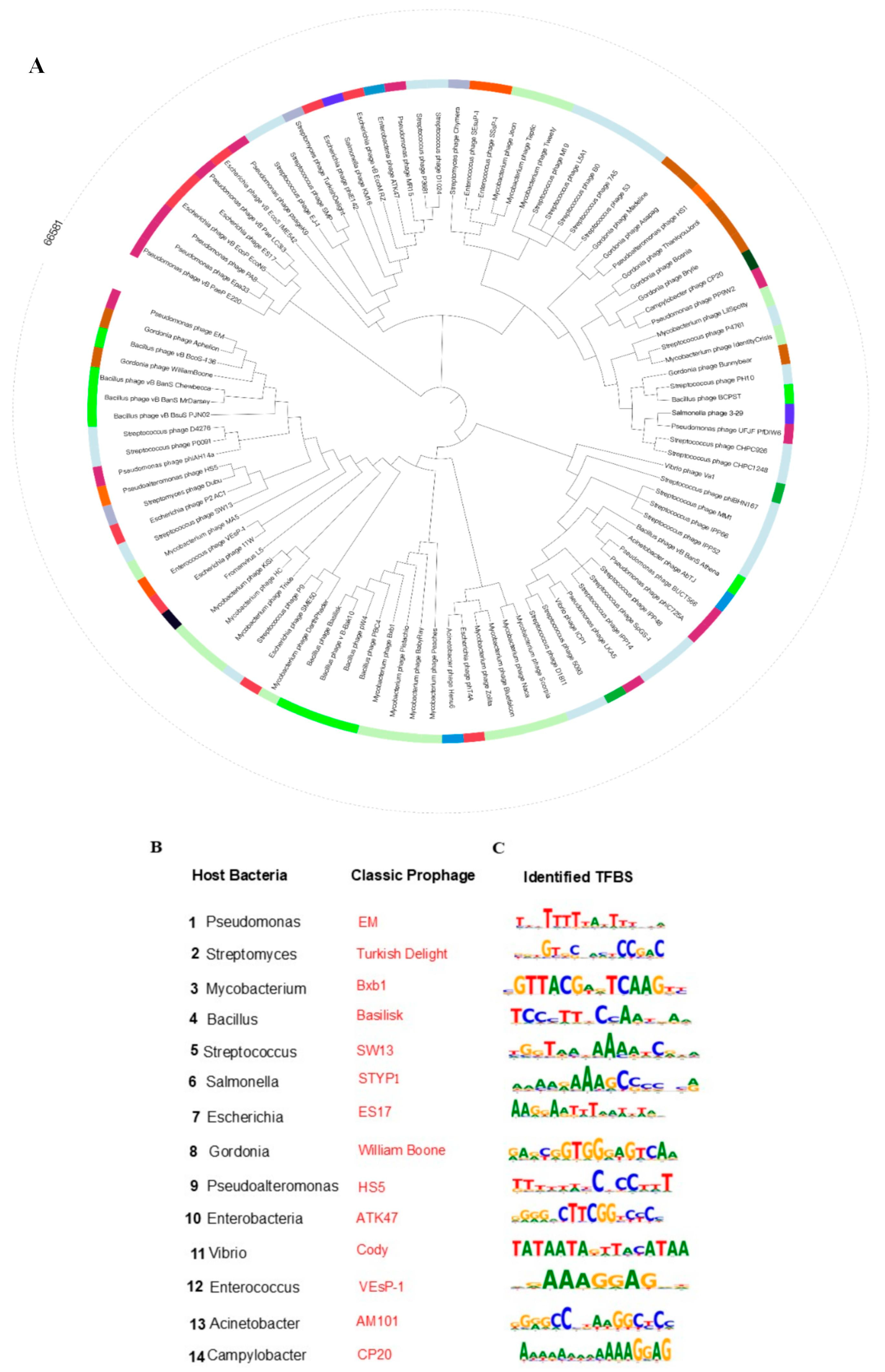
3.2. A Universal Platform for Prophage Characterization and Modulation in Diverse Bacterial Lineages
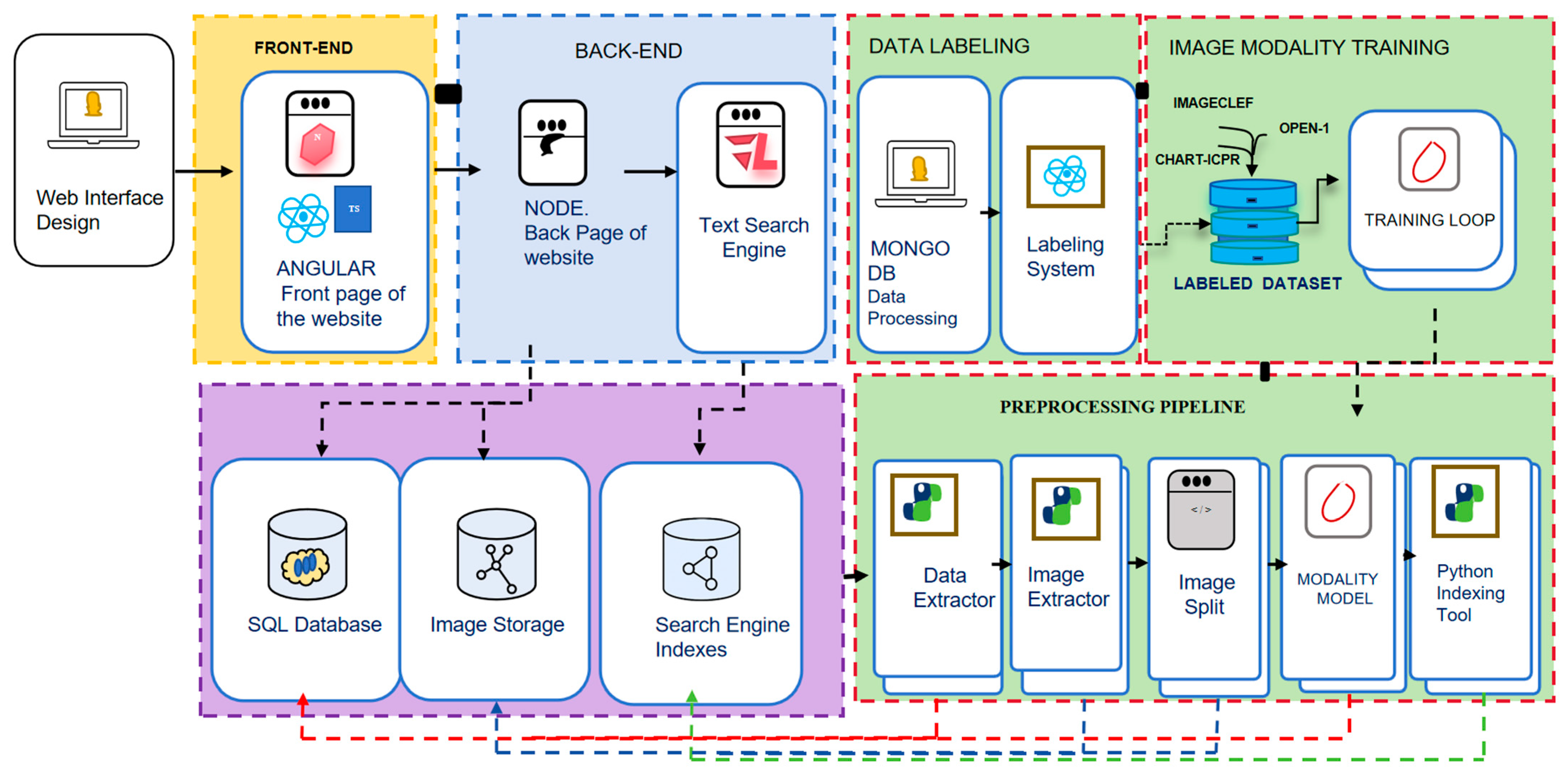
3.3. A Systematic, Open, and User-Friendly Phage Research Tool
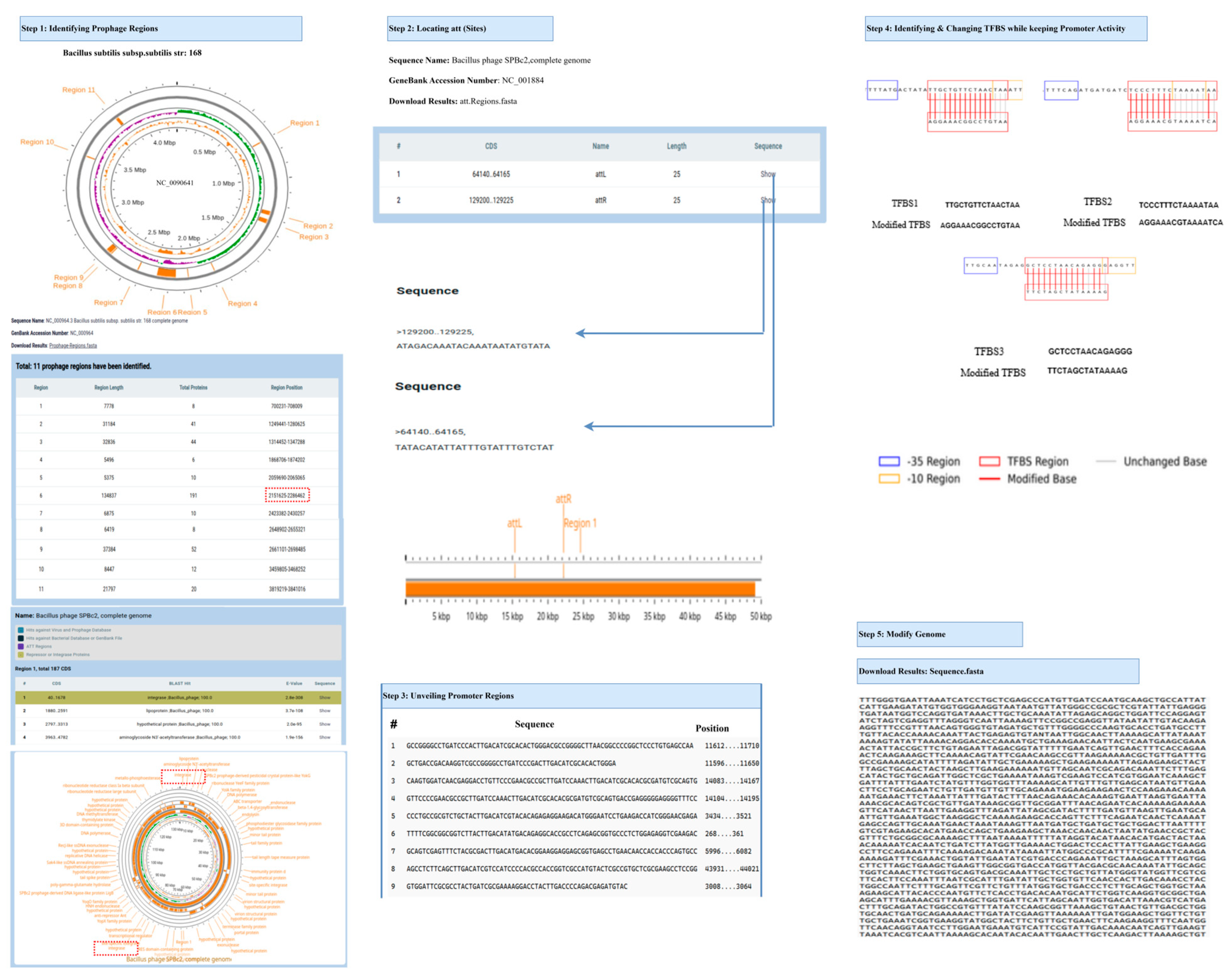
3.4. Comprehensive Workflow of the Prophage Activation Platform Across Diverse Bacterial Hosts
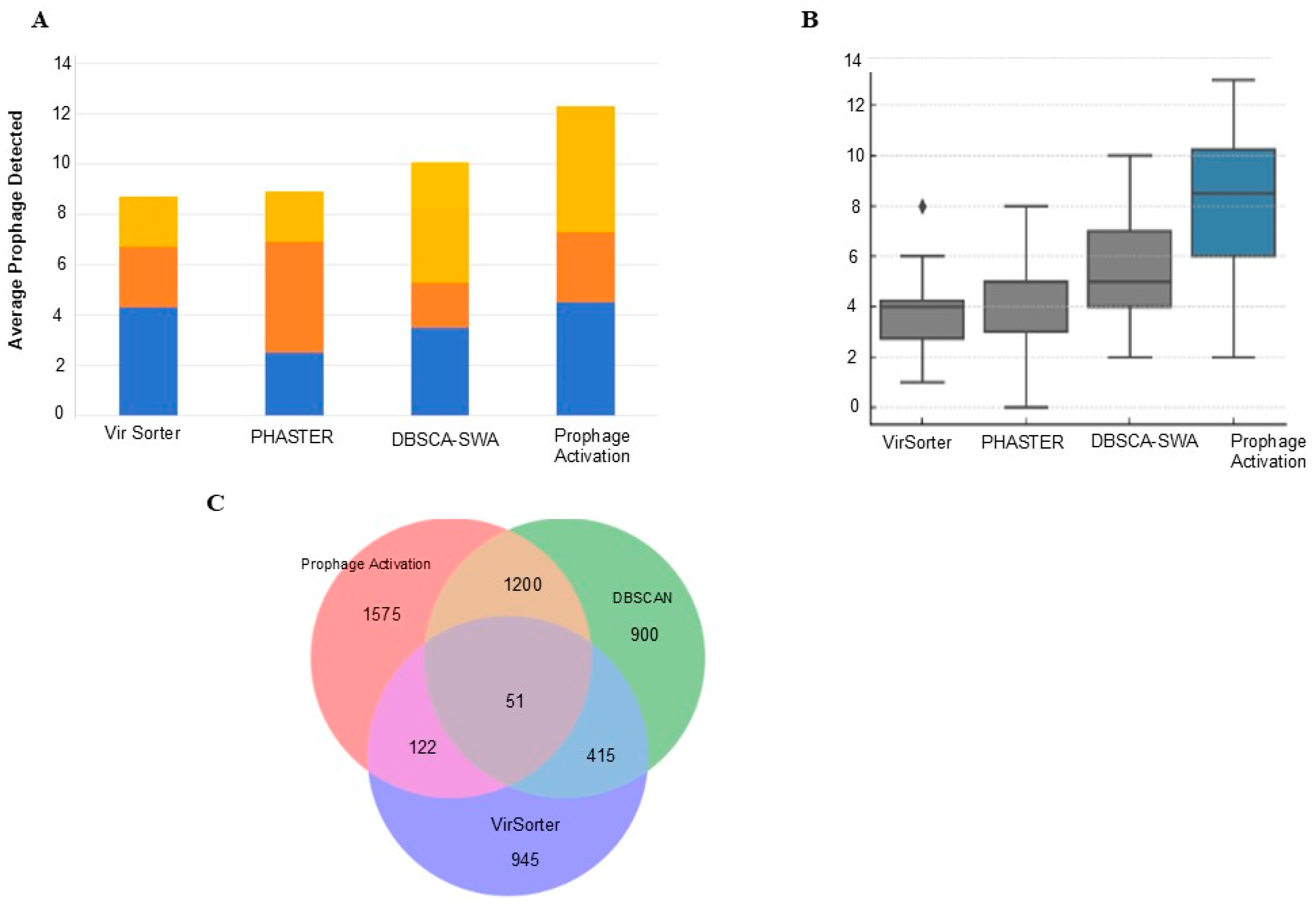
3.5. Comparative Performance Assessment of Prophage Prediction Tools
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| AMR | Antimicrobial Resistance |
| MCC | Matthews Correlation Coefficient |
| NCBI | National Center for Biotechnology Institute |
| ORF | Open Reading Frame |
| PWM | Position Weight Matrix |
| RF | Random Forest |
| SIE | Super Infection Exclusion |
| TFBS | Transcription Factor Binding Site |
References
- Morrison, L.; Zembower, T.R. Antimicrobial Resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, W.E.; Yang, Q. Clinical Perspective of Antimicrobial Resistance in Bacteria. Infect. Drug Resist. 2022, 15, 735–746. [Google Scholar] [CrossRef]
- Ugwu, M.C.; Shariff, M.; Nnajide, C.M.; Beri, K.; Okezie, U.M.; Iroha, I.R.; Esimone, C.O. Phenotypic and Molecular Characterization of β-Lactamases among Enterobacterial Uropathogens in Southeastern Nigeria. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 5843904. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Miedzybrodzki, R.; Borysowski, J.; Weber-Dabrowska, B.; Lobocka, M.; Fortuna, W.; Letkiewicz, S.; Zimecki, M.; Filby, G. Bacteriophage therapy for the treatment of infections. Curr. Opin. Investig. Drugs 2009, 10, 766–774. [Google Scholar]
- Duckworth, D.H.; Gulig, P.A. Bacteriophages: Potential treatment for bacterial infections. BioDrugs 2002, 16, 57–62. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Davidson, A.R. When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness. J. Microbiol. 2014, 52, 235–242. [Google Scholar] [CrossRef]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef]
- Bertani, G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef]
- Amanian, M.; Demetrick, S.; Gana, J.; Tam, T. Sub-inhibitory treatment of gentamicin in Escherichia coli decreases T7 bacteriophage infectivity and cell lysis. UJEMI 2019, 5, 1–11. [Google Scholar]
- Pires, D.P.; Melo, L.D.; Azeredo, J. Understanding the complex phage-host interactions in biofilm communities. Annu. Rev. Virol. 2021, 8, 73–94. [Google Scholar] [CrossRef]
- Sui, B.; Han, L.; Ren, H.; Liu, W.; Zhang, C. A novel polyvalent bacteriophage vB_EcoM_swi3 infects pathogenic Escherichia coli and Salmonella enteritidis. Front. Microbiol. 2021, 12, 649673. [Google Scholar] [CrossRef]
- Onsea, J.; Soentjens, P.; Djebara, S.; Merabishvili, M.; Depypere, M.; Spriet, I.; De Munter, P.; Debaveye, Y.; Nijs, S.; Vanderschot, P. Bacteriophage application for difficult-to-treat musculoskeletal infections: Development of a standardized multidisciplinary treatment protocol. Viruses 2019, 11, 891. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity-and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.-X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef]
- Gan, R.; Zhou, F.; Si, Y.; Yang, H.; Chen, C.; Ren, C.; Wu, J.; Zhang, F. DBSCAN-SWA: An integrated tool for rapid prophage detection and annotation. Front. Genet. 2022, 13, 885048. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Gussow, A.B.; Benler, S.; Wolf, Y.I.; Koonin, E.V. Seeker: Alignment-free identification of bacteriophage genomes by deep learning. Nucleic Acids Res. 2020, 48, e121. [Google Scholar] [CrossRef]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef]
- Knaus, R.; Bujard, H. Principles governing the activity of E. coli promoters. In Nucleic Acids and Molecular Biology 4; Springer: Berlin/Heidelberg, Germany, 1990; pp. 110–122. [Google Scholar]
- Record, M.T., Jr. Escherichia coli RNA polymerase Eσ70, promoters, and the kinetics of the steps of transcription initiation. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Ed.; ASM Press: Washington, DC, USA, 1996; Volume 7, pp. 792–821. [Google Scholar]
- Kieft, K.; Anantharaman, K. Deciphering active prophages from metagenomes. Msystems 2022, 7, e00084-22. [Google Scholar] [CrossRef]
- Russell, D.A.; Hatfull, G.F. PhagesDB: The actinobacteriophage database. Bioinformatics 2017, 33, 784–786. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Santos-Zavaleta, A.; Salgado, H.; Gama-Castro, S.; Sánchez-Pérez, M.; Gómez-Romero, L.; Ledezma-Tejeida, D.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Muñiz-Rascado, L.J.; Peña-Loredo, P. RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019, 47, D212–D220. [Google Scholar] [CrossRef]
- García-Calderón, C.B.; Casadesús, J.; Ramos-Morales, F. Rcs and PhoPQ regulatory overlap in the control of Salmonella enterica virulence. J. Bacteriol. 2007, 189, 6635–6644. [Google Scholar] [CrossRef]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.R.; Hatfull, G.F. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 2005, 123, 861–873. [Google Scholar] [CrossRef]
- Ghosh, P.; Bibb, L.A.; Hatfull, G.F. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc. Natl. Acad. Sci. USA 2008, 105, 3238–3243. [Google Scholar] [CrossRef]
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2004. [Google Scholar]
- Holik, M.; Horvath, T.; Oujezsky, V.; Munster, P.; Tomasov, A.; Valach, S. Mongodb database as storage for gpon frames. Sensors 2020, 20, 6208. [Google Scholar] [CrossRef]
- Chiaretta, S. Front-End Development with ASP. NET Core, Angular, and Bootstrap; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Bangare, S.; Gupta, S.; Dalal, M.; Inamdar, A. Using Node. Js to build high speed and scalable backend database server. Int. J. Res. Advent. Technol. 2016, 4, 19. [Google Scholar]
- Cincovic, J.; Delcev, S.; Draskovic, D. Architecture of web applications based on Angular Framework: A Case Study. Methodology 2019, 7, 254–259. [Google Scholar]
- Chauhan, A. A review on various aspects of mongodb databases. Int. J. Eng. Res. Technol. (IJERT) 2019, 8, 90–92. [Google Scholar]
- Croce, L.d.; Salinas, J.A. Flexibilidad en Bases de Datos NoSQL Sobre Ambientes Web Mining. Master’s Thesis, Universidad Nacional de La Plata, La Plata, Argentina, 2016. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Varoquaux, G.; Buitinck, L.; Louppe, G.; Grisel, O.; Pedregosa, F.; Mueller, A. Scikit-learn: Machine learning without learning the machinery. GetMobile Mob. Comput. Commun. 2015, 19, 29–33. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musrrat, S.; Han, Z.; Wang, K.; Huang, Y.; Xiang, Y.; Liu, S.; Yin, W. Prophage Activation: An In Silico Platform for Identifying Prophage Regulatory Elements to Inform Phage Engineering Against Drug-Resistant Bacteria. Life 2025, 15, 1417. https://doi.org/10.3390/life15091417
Musrrat S, Han Z, Wang K, Huang Y, Xiang Y, Liu S, Yin W. Prophage Activation: An In Silico Platform for Identifying Prophage Regulatory Elements to Inform Phage Engineering Against Drug-Resistant Bacteria. Life. 2025; 15(9):1417. https://doi.org/10.3390/life15091417
Chicago/Turabian StyleMusrrat, Saher, Zequan Han, Kai Wang, Yunhai Huang, Yanhui Xiang, Sen Liu, and Wen Yin. 2025. "Prophage Activation: An In Silico Platform for Identifying Prophage Regulatory Elements to Inform Phage Engineering Against Drug-Resistant Bacteria" Life 15, no. 9: 1417. https://doi.org/10.3390/life15091417
APA StyleMusrrat, S., Han, Z., Wang, K., Huang, Y., Xiang, Y., Liu, S., & Yin, W. (2025). Prophage Activation: An In Silico Platform for Identifying Prophage Regulatory Elements to Inform Phage Engineering Against Drug-Resistant Bacteria. Life, 15(9), 1417. https://doi.org/10.3390/life15091417






