Microarray Analysis of Differentially Expressed Genes in Peripheral Blood of Postpartum Women with Gestational Diabetes Mellitus and Type 2 Diabetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Extraction and Microarray Processing
2.3. Microarray Enrichment Analysis
2.4. Validation of Selected Genes Using RT-PCR
2.5. Statistical Analysis
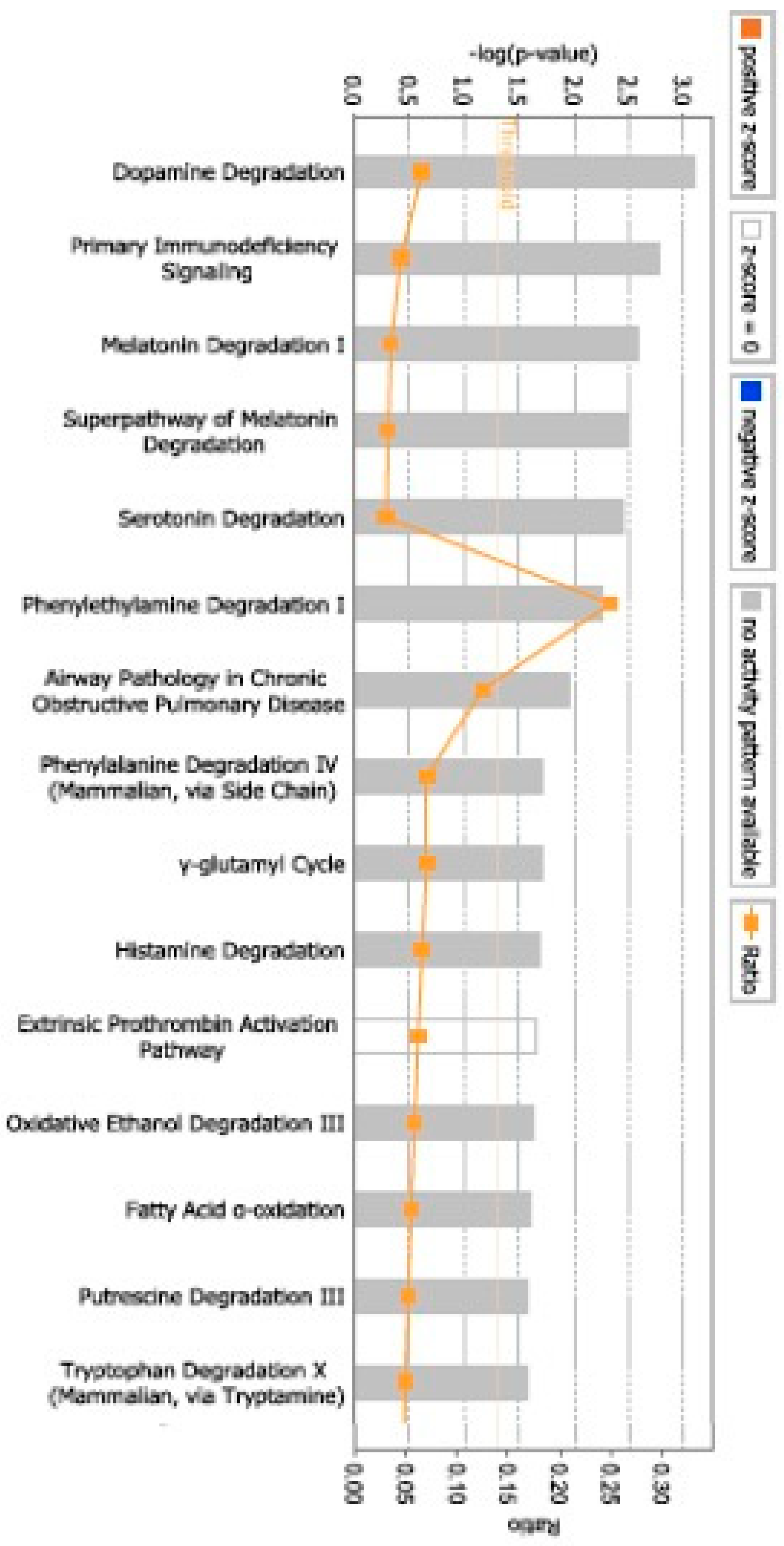
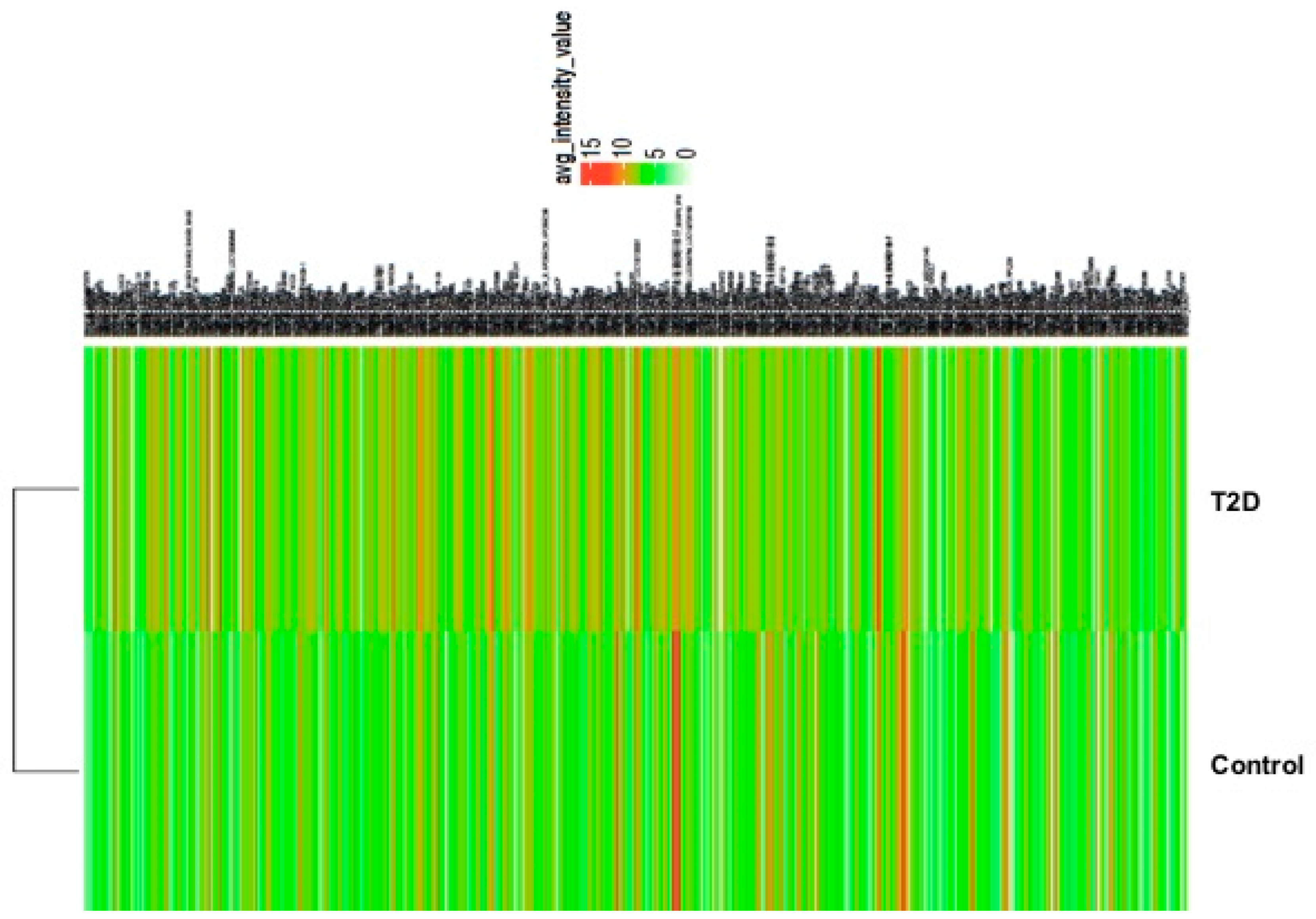
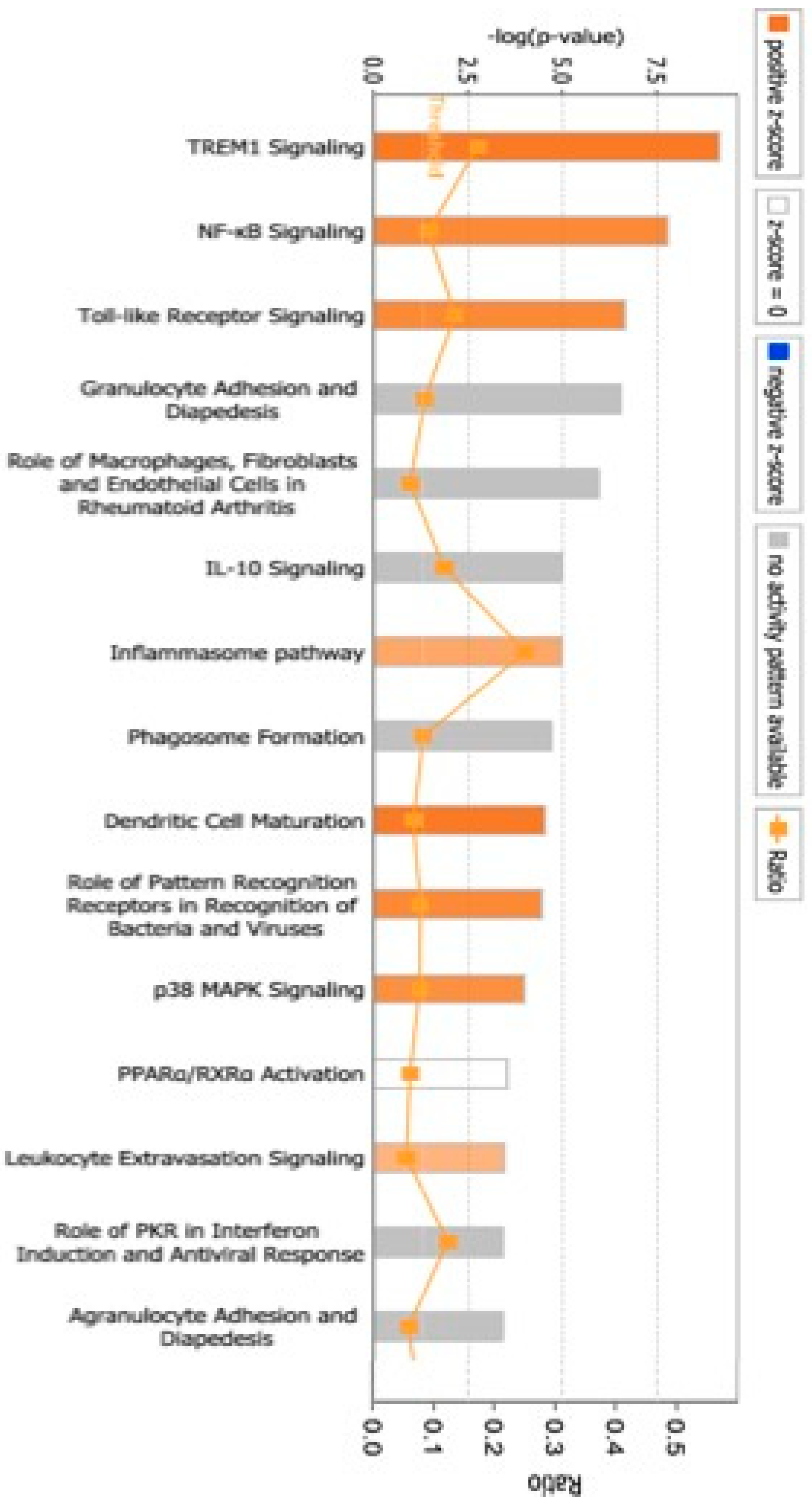
3. Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Młynarska, E.; Czarnik, W.; Dzieża, N.; Jędraszak, W.; Majchrowicz, G.; Prusinowski, F.; Stabrawa, M.; Rysz, J.; Franczyk, B. Type 2 diabetes mellitus: New pathogenetic mechanisms, treatment and the most important complications. Int. J. Mol. Sci. 2025, 26, 1094. [Google Scholar] [CrossRef]
- Immanuel, J.; Cheung, N.W.; Mohajeri, M.; Simmons, D.J.; Hague, W.M.; Teede, H.; Nolan, C.J.; Peek, M.J.; Flack, J.R.; McLean, M.; et al. Association between glycemia, glycemic variability, and pregnancy complications in early GDM. Diabetes Care 2025, 48, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, M.; Rahmati, M.; Azizi, F.; Ramezani Tehrani, F. Lactation duration and lifetime progression to metabolic syndrome in women according to their history of gestational diabetes: A prospective longitudinal community-based cohort study. J. Transl. Med. 2023, 21, 177. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, L.; Casas, J.P.; Hingorani, A.D.; Williams, D. Type 2 diabetes mellitus after gestational diabetes: A systematic review and meta-analysis. Lancet 2009, 373, 1773–1779. [Google Scholar] [CrossRef]
- Gunderson, E.P.; Chiang, V.; Pletcher, M.J.; Jacobs, D.R., Jr.; Quesenberry, C.P., Jr.; Sidney, S.; Lewis, C.E. History of gestational diabetes mellitus and future risk of atherosclerosis in mid-life: The coronary artery risk development in young adults study. J. Am. Heart Assoc. 2014, 3, e000490. [Google Scholar] [CrossRef]
- Rayanagoudar, G.; Hashi, A.A.; Zamora, J.; Khan, K.S.; Hitman, G.A.; Thangaratinam, S. Quantification of the type 2 diabetes risk in women with gestational diabetes: A systematic review and meta-analysis of 95,750 women. Diabetologia 2016, 59, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Baptiste-Roberts, K.; Barone, B.B.; Gary, T.L.; Golden, S.H.; Wilson, L.M.; Bass, E.B.; Nicholson, W.K. Risk factors for type 2 diabetes among women with gestational diabetes: A systematic review. Am. J. Med. 2009, 122, 207–214.e204. [Google Scholar] [CrossRef]
- Göbl, C.S.; Bozkurt, L.; Yarragudi, R.; Prikoszovich, T.; Tura, A.; Pacini, G.; Koppensteiner, R.; Kautzky-Willer, A. Biomarkers of endothelial dysfunction in relation to impaired carbohydrate metabolism following pregnancy with gestational diabetes mellitus. Cardiovasc. Diabetol. 2014, 13, 138. [Google Scholar] [CrossRef]
- Vrachnis, N.; Augoulea, A.; Iliodromiti, Z.; Lambrinoudaki, I.; Sifakis, S.; Creatsas, G. Previous gestational diabetes mellitus and markers of cardiovascular risk. Int. J. Endocrinol. 2012, 2012, 458610. [Google Scholar] [CrossRef]
- Van, J.A.; Luo, Y.; Danska, J.S.; Dai, F.; Alexeeff, S.E.; Gunderson, E.P.; Rost, H.; Wheeler, M.B. Postpartum defects in inflammatory response after gestational diabetes precede progression to type 2 diabetes: A nested case-control study within the SWIFT study. Metabolism 2023, 149, 155695. [Google Scholar] [CrossRef]
- Bassaw, B.; Fletcher, H.; Rattray, C.; McIntyre, G.; Sarkharkar, V.; Sankat, S.; Sirjusingh, A.; Chinnia, J. Screening for gestational diabetes mellitus: A Caribbean perspective. J. Obstet. Gynaecol. 2018, 38, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, T.A. Pancreatic B-cell defects in gestational diabetes: Implications for the pathogenesis and prevention of type 2 diabetes. J. Clin. Endocrinol. Metab. 2001, 86, 989–993. [Google Scholar] [CrossRef]
- Williams, M.A.; Qiu, C.; Dempsey, J.C.; Luthy, D.A. Familial aggregation of type 2 diabetes and chronic hypertension in women with gestational diabetes mellitus. J. Reprod. Med. 2003, 48, 955–962. [Google Scholar]
- Lee, C.; An, D.; Park, J. Hyperglycemic memory in metabolism and cancer. Horm. Mol. Biol. Clin. Investig. 2016, 26, 77–85. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, H.; Guan, W.; Kang, X.; Tai, X.; Shen, Y. Metabolic memory in mitochondrial oxidative damage triggers diabetic retinopathy. BMC Ophthalmol. 2018, 18, 258. [Google Scholar] [CrossRef]
- Keating, S.T.; Plutzky, J.; El-Osta, A. Epigenetic changes in diabetes and cardiovascular risk. Circ. Res. 2016, 118, 1706–1722. [Google Scholar] [CrossRef]
- Sultan, S.; Alzahrani, N.; Al-Sakkaf, K. The postpartum effect of maternal diabetes on the circulating levels of sirtuins and superoxide dismutase. FEBS Open Bio. 2018, 8, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Metzger, B.E.; Gabbe, S.G.; Persson, B.; Lowe, L.P.; Dyer, A.R.; Oats, J.J.; Buchanan, T.A. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy: Response to Weinert. Diabetes Care 2010, 33, e98. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Zhao, M.; Yao, Z.; Zhang, Y.; Ma, L.; Pang, W.; Ma, S.; Xu, Y.; Wei, L. Predictive value of machine learning for the progression of gestational diabetes mellitus to type 2 diabetes: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2025, 25, 18. [Google Scholar] [CrossRef]
- Wang, F.; Zhao, Y.; Xing, J.; Gong, L. Avicularin as a Promising Therapeutic Agent for Gestational Diabetes Mellitus: Modulation of Inflammatory, Oxidative Stress, and Apoptotic Pathways. In Doklady Biochemistry and Biophysics; Springer: Berlin/Heidelberg, Germany, 2025; pp. 206–220. [Google Scholar]
- Elguoshy, A.; Yamamoto, K.; Hirao, Y.; Yanagita, K.; Uchimoto, T.; Kamimura, T.; Takazawa, T.; Yamamoto, T. Machine Learning-Driven Glycoproteomic Profiling Identifies Novel Diabetes-Associated Glycosylation Biomarkers. medRxiv 2025. [Google Scholar] [CrossRef]
- Liu, H.; Yao, Q.; Wang, X.; Xie, H.; Yang, C.; Gao, H.; Xie, C. The research progress of crosstalk mechanism of autophagy and apoptosis in diabetic vascular endothelial injury. Biomed. Pharmacother. 2024, 170, 116072. [Google Scholar] [CrossRef]
- Sheng, N.; Zhang, Z.; Zheng, H.; Ma, C.; Li, M.; Wang, Z.; Wang, L.; Jiang, J.; Zhang, J. Scutellarin rescued mitochondrial damage through ameliorating mitochondrial glucose oxidation via the Pdk-Pdc axis. Adv. Sci. 2023, 10, 2303584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef]
- Yao, Y.; Shi, J.; Zhang, C.; Gao, W.; Huang, N.; Liu, Y.; Yan, W.; Han, Y.; Zhou, W.; Kong, L. Pyruvate dehydrogenase kinase 1 protects against neuronal injury and memory loss in mouse models of diabetes. Cell Death Dis. 2023, 14, 722. [Google Scholar] [CrossRef]
- Huang, P.; Sun, R.; Xu, C.; Jiang, Z.; Zuo, M.; Li, Y.; Liu, R.; Gong, P.; Han, Y.; Fang, J.; et al. Glucocorticoid activates STAT3 and NF-κB synergistically with inflammatory cytokines to enhance the anti-inflammatory factor TSG6 expression in mesenchymal stem/stromal cells. Cell Death Dis. 2024, 15, 70. [Google Scholar] [CrossRef]
- Dyer, D.P.; Salanga, C.L.; Johns, S.C.; Valdambrini, E.; Fuster, M.M.; Milner, C.M.; Day, A.J.; Handel, T.M. The anti-inflammatory protein TSG-6 regulates chemokine function by inhibiting chemokine/glycosaminoglycan interactions. J. Biol. Chem. 2016, 291, 12627–12640. [Google Scholar] [CrossRef]
- Sultan, S. Aberrant expression of proatherogenic cytokines and growth factors in human umbilical vein endothelial cells from newborns of type 2 diabetic women. SAGE Open Med. 2021, 9, 20503121211026832. [Google Scholar] [CrossRef]
- Sultan, S.A.; Liu, W.; Peng, Y.; Roberts, W.; Whitelaw, D.; Graham, A.M. The role of maternal gestational diabetes in inducing fetal endothelial dysfunction. J. Cell. Physiol. 2015, 230, 2695–2705. [Google Scholar] [CrossRef] [PubMed]
- Heitritter, S.M.; Solomon, C.G.; Mitchell, G.F.; Skali-Ounis, N.; Seely, E.W. Subclinical inflammation and vascular dysfunction in women with previous gestational diabetes mellitus. J. Clin. Endocrinol. Metab. 2005, 90, 3983–3988. [Google Scholar] [CrossRef]
- Ridker, P.M.; Hennekens, C.H.; Buring, J.E.; Rifai, N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N. Engl. J. Med. 2000, 342, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Festa, A.; D’Agostino, R., Jr.; Howard, G.; Mykkänen, L.; Tracy, R.P.; Haffner, S.M. Chronic subclinical inflammation as part of the insulin resistance syndrome: The Insulin Resistance Atherosclerosis Study (IRAS). Circulation 2000, 102, 42–47. [Google Scholar] [CrossRef]
- Bai, C.; Yang, W.; Qi, G.; Yang, L.; Wu, Q.; Peng, J.; Wang, N.; Liu, T. Causal association between matrix metalloproteinases and diabetic neuropathy: A two-sample Mendelian randomization study. Front. Endocrinol. 2025, 15, 1429121. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, C.; Li, Z.; Wang, X.; Shang, D.; Dong, W. Matrix metalloproteinase-triggered self-assembling peptides for biomedical applications. J. Mater. Chem. B. 2025, 13, 8298–8334. [Google Scholar] [CrossRef]
- Tanase, D.M.; Valasciuc, E.; Anton, I.B.; Gosav, E.M.; Dima, N.; Cucu, A.I.; Costea, C.F.; Floria, D.E.; Hurjui, L.L.; Tarniceriu, C.C.; et al. Matrix Metalloproteinases: Pathophysiologic Implications and Potential Therapeutic Targets in Cardiovascular Disease. Biomolecules 2025, 15, 598. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Su, L.; Loo, S.J.; Gao, Y.; Khin, E.; Kong, X.; Dalan, R.; Su, X.; Lee, K.O.; Ma, J.; et al. Matrix metallopeptidase 9 contributes to the beginning of plaque and is a potential biomarker for the early identification of atherosclerosis in asymptomatic patients with diabetes. Front. Endocrinol. 2024, 15, 1369369. [Google Scholar] [CrossRef]
- Silva, F.S.; Bortolin, R.H.; Araújo, D.N.; Marques, D.E.; Lima, J.P.M.; Rezende, A.A.; Vieira, W.H.; Silva, N.B.; Medeiros, K.C.; Ackermann, P.W.; et al. Exercise training ameliorates matrix metalloproteinases 2 and 9 messenger RNA expression and mitigates adverse left ventricular remodeling in streptozotocin-induced diabetic rats. Cardiovasc. Pathol. 2017, 29, 37–44. [Google Scholar] [CrossRef]
- Death, A.K.; Fisher, E.J.; McGrath, K.C.; Yue, D.K. High glucose alters matrix metalloproteinase expression in two key vascular cells: Potential impact on atherosclerosis in diabetes. Atherosclerosis 2003, 168, 263–269. [Google Scholar] [CrossRef]
- Pustovrh, C.; Jawerbaum, A.; Sinner, D.; Pesaresi, M.; Baier, M.; Micone, P.; Gimeno, M.; Gonzalez, E.T. Membrane-type matrix metalloproteinase-9 activity in placental tissue from patients with pre-existing and gestational diabetes mellitus. Reprod. Fertil. Dev. 2000, 12, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Dufner, A.; Pownall, S.; Mak, T.W. Caspase recruitment domain protein 6 is a microtubule-interacting protein that positively modulates NF-κB activation. Proc. Natl. Acad. Sci. USA 2006, 103, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Ho, E.; Quan, N.; Tsai, Y.H.; Lai, W.; Bray, T.M. Dietary zinc supplementation inhibits NFκB activation and protects against chemically induced diabetes in CD1 mice. Exp. Biol. Med. 2001, 226, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, Y.; Wu, G.; Kong, Y.; Zhang, Y.; Zha, X. Circ-CARD6 inhibits oxidative stress-induced apoptosis and autophagy in ARPE-19 cells via the miR-29b-3p/PRDX6/PI3K/Akt axis. Exp. Eye Res. 2024, 238, 109690. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Chan, P.S. Metabolic memory in diabetes–from in vitro oddity to in vivo problem: Role of Apoptosis. Brain Res. Bull. 2010, 81, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Baelde, H.J.; Eikmans, M.; Doran, P.P.; Lappin, D.W.; de Heer, E.; Bruijn, J.A. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am. J. Kidney Dis. 2004, 43, 636–650. [Google Scholar] [CrossRef]
- Ma, R.; Deng, X.L.; Aleteng, Q.Q.G.; Li, L.; Zhu, J. Genome-Wide Transcriptome Analysis in Type 2 Diabetes Patients Treated by Sitagliptin. Diabetes Metab. Syndr. Obes. 2022, 15, 1761–1770. [Google Scholar] [CrossRef]



| Primers | Sequences |
|---|---|
| PDK3 | Forward: 5′-GAGCAATCCCAGCAGTGAAC-3′ Reverse: 5′-ATAACTGTGATGCCACGCTC-3′ |
| TNFAIP6 | Forward: 5′-GCTGGATGGATGGCTAAGGG-3′ Reverse: 5′-CCTTTGCGTGTGGGTTGTAG-3′ |
| MMP9 | Forward: 5′-GGTGATTGACGACGCCTTTG-3′ Reverse: 5′-GGACCACAACTCGTCATCGT-3′ |
| CARD6 | Forward: 5′-CGAGAGTACTCCCTCAGAGAT-3′ Reverse: 5′-GCCCCCATAGATTGAGGAGG-3′ |
| β-actin | Forward: 5′-AGCGGGAAATCGTGCGTGAC-3′ Reverse: 5′-CGGACTCGTCATACTCCTGCT-3′ |
| Mothers’ Status | Controls (n = 3) | pGDM (n = 4) | T2D (n = 3) | P (pGDM vs. Controls/T2D vs. Controls) |
|---|---|---|---|---|
| Age (years) | 28.6 ± 0.9 | 30 ± 1 | 31.7 ± 1.6 | 0.29/0.2 |
| BMI | 28.8 ± 3 | 30 ± 1.3 | 31.3 ± 3.9 | 0.37/0.32 |
| HbA1c (%) RPG (mM) 2h OGTT | - 4 ± 0.3 - | 6.1 ± 0.1 5.3 ± 0.9 9.9 ± 1 | 6.3 ± 0.5 5.5 ± 0.2 - | - 0.28/0.01 * - |
| Gene Symbol | Fold Change | p-Value | Gene Name |
|---|---|---|---|
| MGAM2 | 4.7 | 0.0343 | maltase-glucoamylase 2 (putative) |
| LOC105372578 | 4.36 | 7.38 × 10−7 | uncharacterized LOC105372578 |
| TRGV5 | 4.24 | 0.0414 | T-cell receptor gamma variable 5 |
| LOC100507639 | 3.94 | 0.0147 | uncharacterized LOC100507639 |
| TRDJ1 | 3.86 | 0.0019 | T-cell receptor delta joining 1 |
| TRDJ4 | 3.57 | 0.0095 | T-cell receptor delta joining 4 |
| CST7 | 3.55 | 8.22 × 10−5 | cystatin F (leukocystatin) |
| CD177 | 3.54 | 0.0129 | CD177 molecule |
| LOC102723373 | 3.35 | 0.0051 | uncharacterized LOC102723373 |
| HCG26 | 3.12 | 0.0326 | HLA complex group 26 (non-protein-coding) |
| LINC01061 | 2.96 | 0.0353 | long intergenic non-protein-coding RNA 1061 |
| RNU6-59P | 2.94 | 0.0004 | RNA, U6 small nuclear 59, pseudogene |
| KIAA1324 | 2.86 | 0.0025 | KIAA1324 |
| ANPEP | 2.75 | 0.0059 | alanyl (membrane) aminopeptidase |
| TRDJ2 | 2.71 | 0.0493 | T-cell receptor delta joining 2 |
| RNU5B-1 | 2.64 | 0.0031 | RNA, U5B small nuclear 1 |
| SULT1B1 | 2.6 | 0.0237 | sulfotransferase family 1B member 1 |
| CEP170P1 | 2.57 | 0.0024 | centrosomal protein 170 kDa pseudogene 1 |
| TNFAIP6 | 2.51 | 0.0088 | tumor necrosis factor, alpha-induced protein 6 |
| BACH1-IT2 | 2.46 | 0.002 | BACH1 intronic transcript 2 |
| MIR548K | 2.45 | 0.0159 | microRNA 548k |
| PSMD5-AS1 | 2.41 | 0.0203 | PSMD5 antisense RNA 1 (head-to-head) |
| DYSF | 2.38 | 0.025 | dysferlin |
| LOC105379818 | 2.37 | 0.0199 | uncharacterized LOC105379818 |
| F5 | 2.36 | 0.0333 | coagulation factor V (proaccelerin, labile factor) |
| CRISPLD2 | 2.35 | 4.47 × 10−5 | cysteine-rich secretory protein LCCL domain containing 2 |
| C3orf62 | 2.33 | 0.0123 | chromosome 3 open reading frame 62 |
| DDX12P; DDX11 | 2.3 | 0.0015 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 12, pseudogene; DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| CCHCR1 | 2.24 | 0.0014 | coiled-coil alpha-helical rod protein 1 |
| TRDC | 2.23 | 0.0432 | T-cell receptor delta constant |
| ALDH2 | 2.21 | 0.0432 | aldehyde dehydrogenase 2 family (mitochondrial) |
| SPATA1 | 2.18 | 0.0386 | spermatogenesis associated 1 |
| CARD6 | 2.17 | 0.044 | caspase recruitment domain family, member 6 |
| CCDC144CP | 2.17 | 0.0111 | coiled-coil domain containing 144C, pseudogene |
| BMS1P4 | 2.16 | 0.0027 | BMS1 ribosome biogenesis factor pseudogene 4 |
| FAM120A | 2.15 | 0.0068 | family with sequence similarity 120A |
| SDHAP2; LINC00969 | 2.14 | 0.0361 | succinate dehydrogenase complex subunit A, flavoprotein pseudogene 2; long intergenic non-protein-coding RNA 969 |
| MIR1299 | 2.11 | 0.0403 | microRNA 1299 |
| PDCD6 | 2.08 | 0.0014 | programmed cell death 6 |
| PDK3 | 2.06 | 0.0028 | pyruvate dehydrogenase kinase, isozyme 3 |
| RAB3D | 2.06 | 0.0037 | RAB3D, member of RAS oncogene family |
| FAM111B | 2.02 | 0.0195 | family with sequence similarity 111, member B |
| IGLL5; IGLV1-36 | 2 | 0.0024 | immunoglobulin lambda-like polypeptide 5; immunoglobulin lambda variable 1-36 |
| XGY2; XG | 2 | 0.0009 | Xg pseudogene, Y-linked 2; Xg blood group |
| CYP2J2 | 2 | 0.0017 | cytochrome P450, family 2, subfamily J, polypeptide 2 |
| SNORD14A | −2.01 | 0.0154 | small nucleolar RNA, C/D box 14A |
| MIR4279 | −2.13 | 0.046 | microRNA 4279 |
| MIR3137 | −2.17 | 0.0059 | microRNA 3137 |
| HBD | −2.18 | 0.0494 | hemoglobin, delta |
| MIR514A1 | −2.18 | 0.0399 | microRNA 514a-1 |
| IGHD | −2.2 | 0.016 | immunoglobulin heavy constant delta |
| MMP8 | −2.23 | 0.0204 | matrix metallopeptidase 8 |
| LOC101928794 | −2.36 | 0.0249 | uncharacterized LOC101928794 |
| SNORD1B | −2.48 | 0.0488 | small nucleolar RNA, C/D box 1B |
| Gene List | Enrichment Score * | Cluster |
|---|---|---|
| IGHD, TRDC, IGLL5, HBD | 2.4 | GO:0042571 immunoglobulin complex, circulating |
| TNFAIP6, F5, CRISPLD2, CST7, CD177, IGHD, TREML5P, MMP8, KIAA1324, ANPEP | 0.9 | GO:0005615 extracellular space, glycosylation site: N-linked (GlcNAc), glycoprotein |
| F5, MMP8, ANPEP, PDCD6 | 0.8 | GO:0006508 proteolysis |
| Network Functions | Enrichment Score * | Gene in Network |
|---|---|---|
| Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking | 17 | ANPEP, CYP2J2, CD177, IGHD, TNFAIP6, DYSF, IGLL1, IGLL5 |
| Cell Cycle, Gene Expression, Cellular Development | 17 | mir-506, PDCD6, PDK3 |
| Cancer, Cell Cycle, Cellular Assembly and Organization | 2 | ANPEP, mir-506, mir-548, CD177, CST7, PDK3 |
| Cell-To-Cell Signaling and Interaction, Cancer, Gastrointestinal Disease | 2 | DYSF, F5, CD177, ANPEP, CYP2J2, PDCD6 IGHD, TNFAIP6 ALDH2, C3orf62, mir-506 |
| Hereditary Disorder, Neurological Disease, Organismal Injury and Abnormalities | 2 | TNFAIP6, ANPEP, F5, HBD, MMP8, ALDH2, PDK3 |
| Gene Symbol | Fold Change | p-Value | Gene Name |
|---|---|---|---|
| TMTC1 | 6.75 | 0.0129 | transmembrane and tetratricopeptide repeat containing 1 |
| TRDJ4 | 5.25 | 0.0049 | T-cell receptor delta joining 4 |
| CLEC12A | 4.74 | 0.0225 | C-type lectin domain family 12, member A |
| DYSF | 4.65 | 0.0013 | dysferlin |
| MT1L; MT1M | 4.4 | 0.0012 | metallothionein 1L (gene/pseudogene) |
| MGAM2 | 4.38 | 0.0455 | maltase-glucoamylase 2 (putative) |
| CLEC12B | 4.22 | 0.0174 | C-type lectin domain family 12, member B |
| ANPEP | 4.13 | 0.0008 | alanyl (membrane) aminopeptidase |
| ALPL | 4.01 | 0.0018 | alkaline phosphatase, liver/bone/kidney |
| LOC102724231 | 3.95 | 0.0088 | uncharacterized LOC102724231 |
| FCGR1A | 3.93 | 0.0289 | Fc fragment of IgG, high-affinity Ia, receptor (CD64) |
| CST7 | 3.82 | 0.0004 | cystatin F (leukocystatin) |
| IL3RA | 3.58 | 0.0007 | interleukin 3 receptor, alpha (low-affinity) |
| BMX | 3.25 | 0.0027 | BMX non-receptor tyrosine kinase |
| ADGRG3 | 3.24 | 0.0174 | adhesion G-protein-coupled receptor G3 |
| ICAM1 | 3.23 | 0.0072 | intercellular adhesion molecule 1 |
| MIR3939 | 3.19 | 0.0242 | microRNA 3939 |
| TLR5 | 3.03 | 0.0319 | Toll-like receptor 5 |
| HAUS4 | 3 | 0.0004 | HAUS augmin-like complex subunit 4 |
| CASP5 | 2.99 | 0.0063 | caspase 5 |
| PIK3AP1 | 2.95 | 0.0056 | phosphoinositide-3-kinase adaptor protein 1 |
| KREMEN1 | 2.95 | 0.0104 | kringle containing transmembrane protein 1 |
| HCG26 | 2.94 | 0.0259 | HLA complex group 26 (non-protein-coding) |
| CARD17 | 2.91 | 0.0109 | caspase recruitment domain family, member 17 |
| TECPR2 | 2.89 | 0.0094 | tectonin beta-propeller repeat containing 2 |
| SSH1 | 2.83 | 0.0033 | slingshot protein phosphatase 1 |
| IL3RA | 2.82 | 0.0005 | interleukin 3 receptor, alpha (low-affinity) |
| TREML2 | 2.82 | 0.0069 | triggering receptor expressed on myeloid cell-like 2 |
| NQO2 | 2.8 | 0.0244 | NAD(P)H dehydrogenase, quinone 2 |
| IL1B | 2.65 | 0.0209 | interleukin 1 beta |
| CEP19 | 2.63 | 0.0002 | centrosomal protein 19kDa |
| ADGRE1 | 2.63 | 0.002 | adhesion G-protein-coupled receptor E1 |
| SEMA4A | 2.63 | 0.0178 | sema domain, immunoglobulin domain (Ig) |
| MEFV | 2.62 | 0.0012 | Mediterranean fever |
| SLC25A44 | 2.62 | 0.013 | solute carrier family 25, member 44 |
| LINC00173 | 2.61 | 0.001 | long intergenic non-protein-coding RNA 173 |
| BCL6 | 2.61 | 0.0068 | B-cell CLL/lymphoma 6 |
| SLC26A8 | 2.61 | 0.0072 | solute carrier family 26 (anion exchanger), member 8 |
| ALDH2 | 2.61 | 0.009 | aldehyde dehydrogenase 2 family (mitochondrial) |
| ORAI2 | 2.59 | 0.0163 | ORAI calcium-release-activated calcium modulator 2 |
| NR6A1 | 2.58 | 0.0013 | nuclear receptor subfamily 6, group A, member 1 |
| CHSY1 | 2.58 | 0.0224 | chondroitin sulfate synthase 1 |
| CARD6 | 2.57 | 0.0057 | caspase recruitment domain family, member 6 |
| LINC01272 | 2.03 | 0.0062 | long intergenic non-protein-coding RNA 1272 |
| SLC6A6 | 2.03 | 0.0096 | solute carrier family 6 (neurotransmitter transporter) |
| MMP9 | 2.03 | 0.0129 | matrix metallopeptidase 9 |
| MIR133A1; MIR133A1HG | −2.01 | 0.0193 | microRNA 133a-1; MIR133A1 host gene (non-protein-coding) |
| SNORD9 | −2.05 | 0.0025 | small nucleolar RNA, C/D box 9 |
| HK1 | −2.07 | 0.0181 | hexokinase 1 |
| TRAJ28 | −2.07 | 0.0419 | T-cell receptor alpha joining 28 |
| CUL4A | −2.08 | 0.0021 | cullin 4A |
| GYPA | −4.36 | 0.0114 | glycophorin A |
| MMP8 | −4.72 | 0.0214 | matrix metallopeptidase 8 |
| AHSP | −5.3 | 0.0366 | alpha hemoglobin stabilizing protein |
| IFIT1B | −6.9 | 0.0017 | interferon-induced protein with tetratricopeptide repeats 1B |
| Gene List | Enrichment Score * | GO ID/Cluster |
|---|---|---|
| ADGRE1, FFAR2, HCK, MSRB1, TLR5, TLR8, LILRB2, IFIT2, NLRC4, MYD88, TNFSF13B, MEFV, FCGR1A, LILRA4, BCL6, FCGR2A, CLEC4D, EIF2AK2, APOBEC3A_B, TBKBP1, AKIRIN2, SEMA4A | 3.7 | GO:0045087 innate immune response |
| CGB1, IER3, OR1A1, NDST1, MMP9, TREML5P, MMP8, GYPA, SIRPB2, CXCR2, ANPEP, TLR5, FCRL6, TLR8, CANT1, SMPDL3A, ITPRIP, HPSE, LTB4R, LILRA4, CSF3R, SEMA3C, SERPINA1, CLEC4D | 3.2 | GO:0005886 plasma membrane, glycosylation site: N-linked (GlcNAc), glycoprotein |
| CASP5, TNFRSF1A, IRAK3, NLRC4, MYD88, MEFV, CARD17, NLRP12, CARD6, CASP1 BCL3, BCL6 | 3.1 | IPR011029 death-like domains |
| Network Functions | Enrichment Score * | Genes in Network |
|---|---|---|
| Inflammatory Disease, Inflammatory Response, Cellular Movement | 39 | MMP8, MMP9, ICAM1, IL1B, IL3RA, IMPDH1, MGAM, mir-21, mir-368 |
| Infectious Diseases, Inflammatory Response, Organismal Injury and Abnormalities | 35 | BASP1, BMX, BPGM, CACNA1E, CANT1, CARD6, CCR1, CD274, CPD, CSF3R, CSTA, DGAT2, DYSF, ELL, FCAR |
| Cellular Movement, Hematological System | 31 | C5AR2, CCR1, CXCR2, DEFA1, MMP9, PLAUR, SEMA4A, SERPINA1, TIMP2, TNFRSF1A |
| Cell Death and Survival, Cancer | 29 | BCL6, CD274, FCGR3B, CARD6, ICAM1, IL1B, IL1RN, MYD88, NFIL3, NFKBIA, SIGLEC9, TNFRSF1A, TNFSF13B, MMP9 |
| Connective Tissue Disorders, Immunological Disease | 23 | MMP25, MMP8, MMP9, NFKBIA, SAMD9L, TIMP2, TLE3, TLR8, TNFRSF1A, TNFSF13B, TRIM21, ZFP36, ZNF281 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sultan, S. Microarray Analysis of Differentially Expressed Genes in Peripheral Blood of Postpartum Women with Gestational Diabetes Mellitus and Type 2 Diabetes. Life 2025, 15, 1270. https://doi.org/10.3390/life15081270
Sultan S. Microarray Analysis of Differentially Expressed Genes in Peripheral Blood of Postpartum Women with Gestational Diabetes Mellitus and Type 2 Diabetes. Life. 2025; 15(8):1270. https://doi.org/10.3390/life15081270
Chicago/Turabian StyleSultan, Samar. 2025. "Microarray Analysis of Differentially Expressed Genes in Peripheral Blood of Postpartum Women with Gestational Diabetes Mellitus and Type 2 Diabetes" Life 15, no. 8: 1270. https://doi.org/10.3390/life15081270
APA StyleSultan, S. (2025). Microarray Analysis of Differentially Expressed Genes in Peripheral Blood of Postpartum Women with Gestational Diabetes Mellitus and Type 2 Diabetes. Life, 15(8), 1270. https://doi.org/10.3390/life15081270







