Prediction of Breast Cancer Response to Neoadjuvant Therapy with Machine Learning: A Clinical, MRI-Qualitative, and Radiomics Approach
Abstract
1. Introduction
2. Materials and Methods
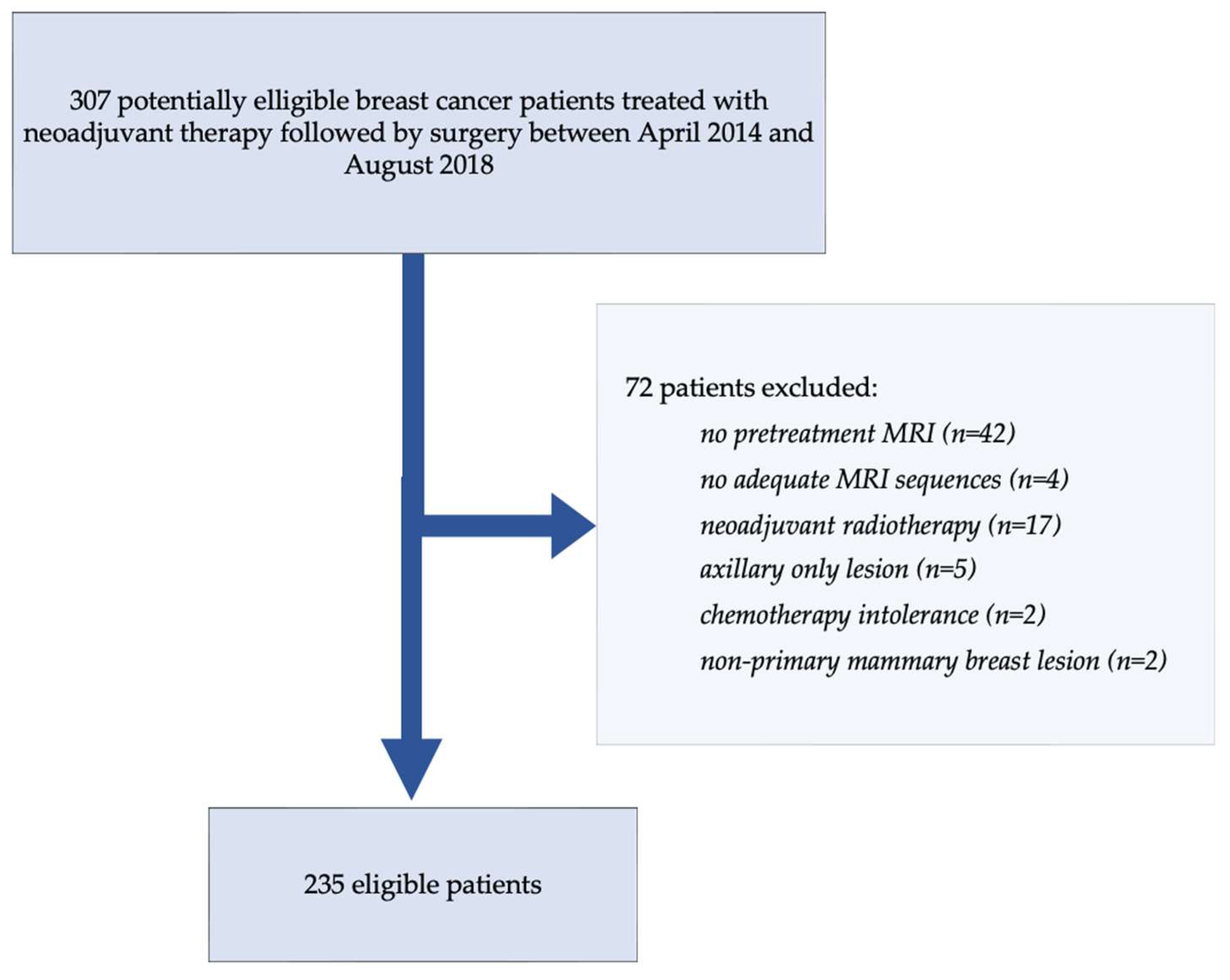
2.1. Cohort Description
2.2. Data Sources and Feature Extraction
2.2.1. Clinical Features
- Age at diagnosis (in years, as a continuous variable);
- Menopausal status (pre- or postmenopausal);
- Tumor histological type (e.g., invasive ductal carcinoma, lobular carcinoma, mucinous carcinoma);
- Histological grade (Scarff–Bloom–Richardson classification, grades 1–3);
- TNM clinical staging: including T stage (tumor size and extension) and N stage (lymph node involvement), based on initial imaging and clinical exam prior to NAST;
- Hormonal receptor expression: estrogen receptor (ER) and progesterone receptor (PR), determined by immunohistochemistry;
- HER2 status: assessed via immunohistochemistry and in situ hybridization;
- Ki-67 proliferation index;
- Molecular subtype: categorized as HER2-positive, triple-negative (ER-, PR-, HER2-), or hormone receptor-positive/HER2-negative;
- Presence of germline mutations: BRCA1, BRCA2, TP53, or others, when genetic testing had been performed;
- Neoadjuvant treatment regimen: including details on chemotherapy (anthracyclines, taxanes), targeted therapy (trastuzumab, pertuzumab).
2.2.2. Radiological Features
- Mass lesions: categorized based on shape (round, oval, irregular), margins (circumscribed, irregular, spiculated), and internal enhancement pattern (homogeneous, heterogeneous, rim enhancement);
- Non-mass enhancement (NME): characterized by distribution (focal, linear, segmental, regional, multiple regions, diffuse) and internal enhancement (homogeneous, heterogeneous, clumped, clustered ring);
- Parietal invasion: defined as direct contact or disruption of the anterior pectoral fascia, chest wall muscles;
- Tumor size: measured along the greatest dimension on T2-weighted axial images and early post-contrast T1-weighted axial images;
- Signal intensity on T2-weighted images: visually graded as hypointense, isointense, or hyperintense compared to surrounding normal fibroglandular breast tissue;
- Edema evaluation: the presence and type of associated edema on fat-saturated T2-weighted sequences were assessed and categorized as follows: absent, peritumoral edema (localized signal hyperintensity in subcutaneous fat or stroma adjacent to the lesion), pre-pectoral edema (signal hyperintensity in the space anterior to the pectoral muscle), and diffuse subcutaneous edema (extensive skin and fat stranding involving at least one quadrant).
2.2.3. Radiomic Features
- Histogram normalization: to rescale intensity values to a common dynamic range across patients and devices, reducing bias from contrast injection timing or scanner calibration.
- Voxel size resampling: all volumes were resampled to isotropic voxels of 1.0 mm × 1.0 mm × 1.0 mm using linear interpolation to ensure spatial consistency and allow accurate shape and texture analysis.
- Intensity discretization: grey-level values were quantized using a fixed bin number (64 bins), with relative discretization strategy (Lloyd–Max algorithm) applied separately for each sequence, allowing consistent texture calculation while accounting for intra-tumoral heterogeneity.
- First-order histogram features (n = 19): describing global intensity distribution (e.g., mean, standard deviation, skewness, kurtosis, percentiles);
- Shape-based features (n = 17): quantifying geometric properties of the tumor volume (e.g., volume, surface area, sphericity, compactness, elongation);
- Texture features (n = 72): computed from five matrices—GLCM (Gray-Level Co-occurrence Matrix), GLRLM (Run-Length Matrix), GLSZM (Size Zone Matrix), NGTDM (Neighborhood Grey-Tone Difference Matrix), and GLDM (Dependence Matrix)—to characterize intra-tumoral heterogeneity and spatial relationships.
2.3. Tumor Segmentation Protocol
2.4. MRI Acquisition and Heterogeneity
2.5. Data Analysis
2.5.1. Train-Test Split
2.5.2. Data Preprocessing
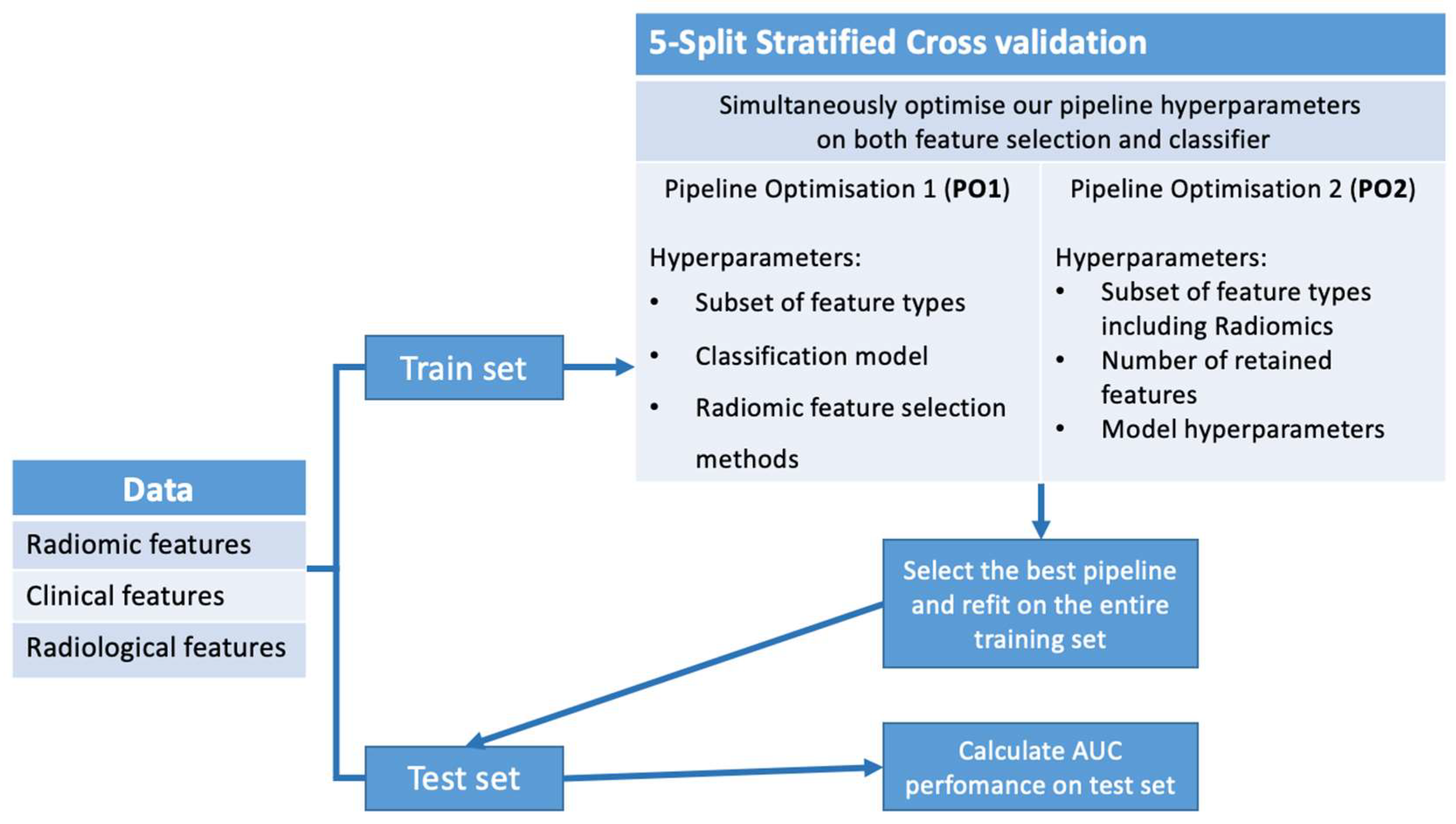
2.5.3. Pipeline Optimization
Pipeline Optimization 1 (PO1)
- Feature subsets: all combinations of Radiomics, Radiological, and Clinical features;
- Classification models: Logistic Regression, Support Vector Machines (SVM), Random Forest, Bagging Classifier, and K-Nearest Neighbors classifiers;
- Feature selection methods: Fisher score, ANOVA selection, MRMR (Maximum Relevance Minimum Redundancy selection), reliefF scoring test for supervised methods, and PCA decomposition for unsupervised selection.
Pipeline Optimization 2 (PO2)
- ■
- Classifier hyperparameters:
- ○
- For Random Forest models: maximum tree depth, minimum samples per split, and per leaf;
- ○
- For SVM models: kernel type (linear or RBF) and regularization parameter.
- ■
- Selection hyperparameters: feature retention count and feature subset selection (including Radiological features).
3. Results
3.1. Results of PO1
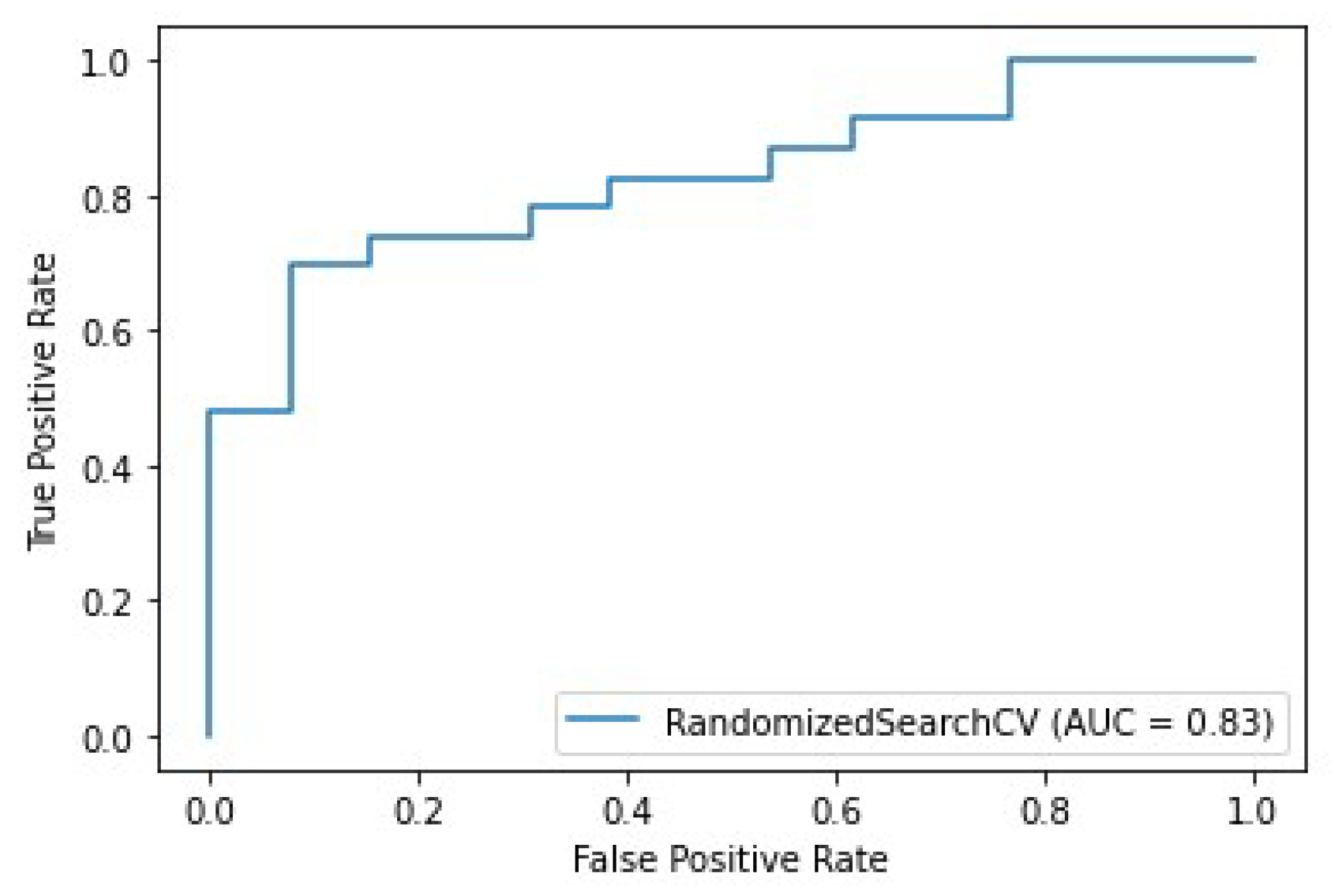
3.2. Prediction of pCR
3.3. Prediction of pCR in Different Breast Cancer Subgroups
3.4. MRI Response Performance
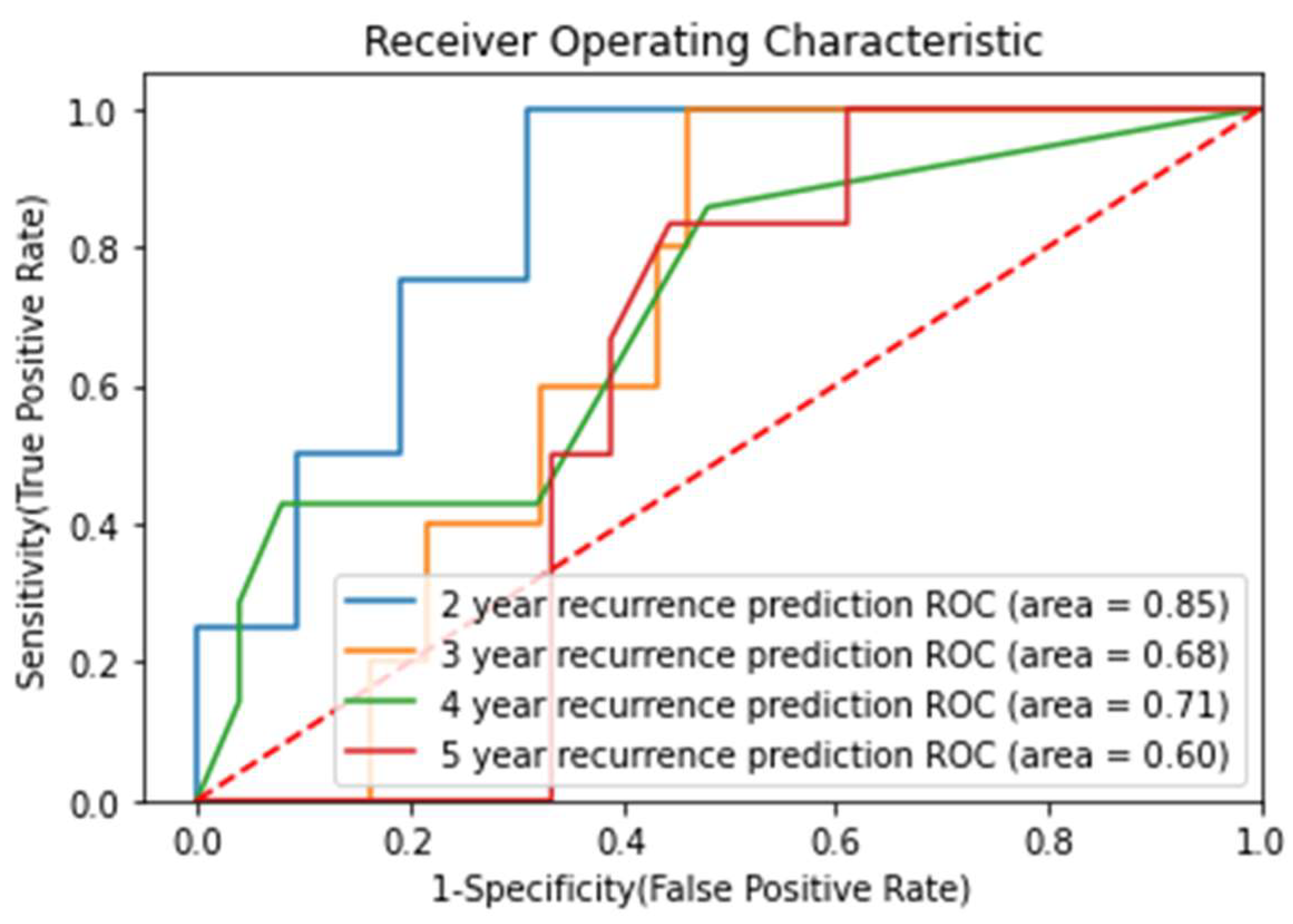
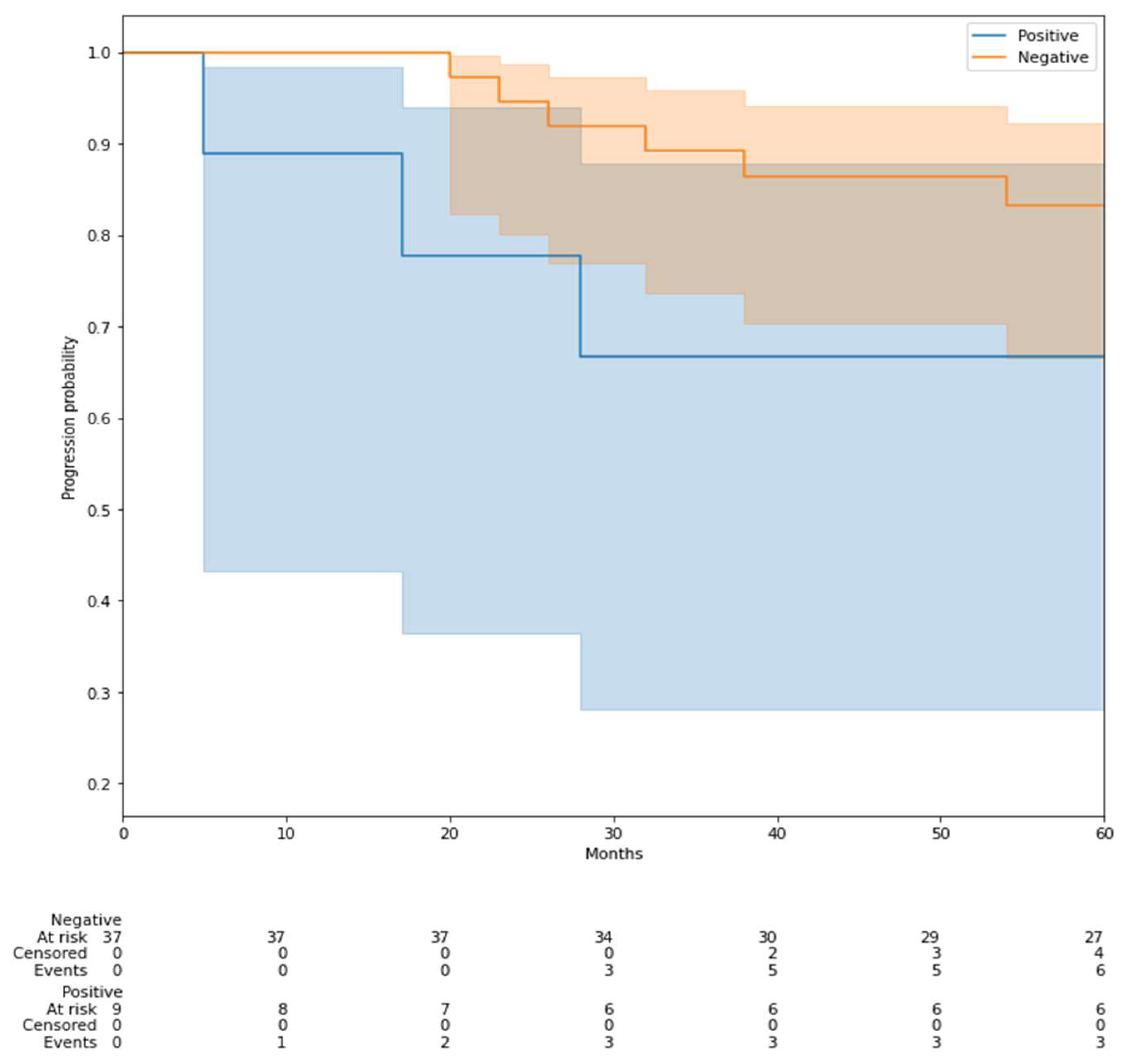
3.5. Prediction of Recurrence-Free Survival (RFS)
3.6. Additional Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area Under the Receiver Operating Characteristic Curve |
| BI-RADS | Breast Imaging Reporting and Data System |
| DCE | Dynamic Contrast-Enhanced |
| ER | Estrogen receptor |
| GLDM | Gray Level Dependence Matrix |
| GLCM | Gray Level Co-occurrence Matrix |
| GLRLM | Gray Level Run Length Matrix |
| GLSZM | Gray Level Size Zone Matrix |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| IBSI | Image Biomarker Standardization Initiative |
| IHC | Immunohistochemistry |
| KNN | K-Nearest Neighbors |
| MRIMagnetic | Resonance Imaging |
| MRMR | Maximum Relevance Minimum Redundancy |
| NME | Non-mass enhancement |
| PCA | Principal Component Analysis |
| PR | Progesterone Receptor |
| pCR | Pathological complete response |
| PO1 | Pipeline Optimization 1 |
| PO2 | Pipeline Optimization 2 |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic |
| RBF | Radial Basis Function |
| ROI | Region of Interest |
| SBR | Scarff–Bloom–Richardson (grading system) |
| SVM | Support Vector Machine |
| T1w | T1-weighted |
| T2w | T2-weighted |
| TNM | Tumor Node Metastasis (staging system) |
| VOI | Volume of interest |
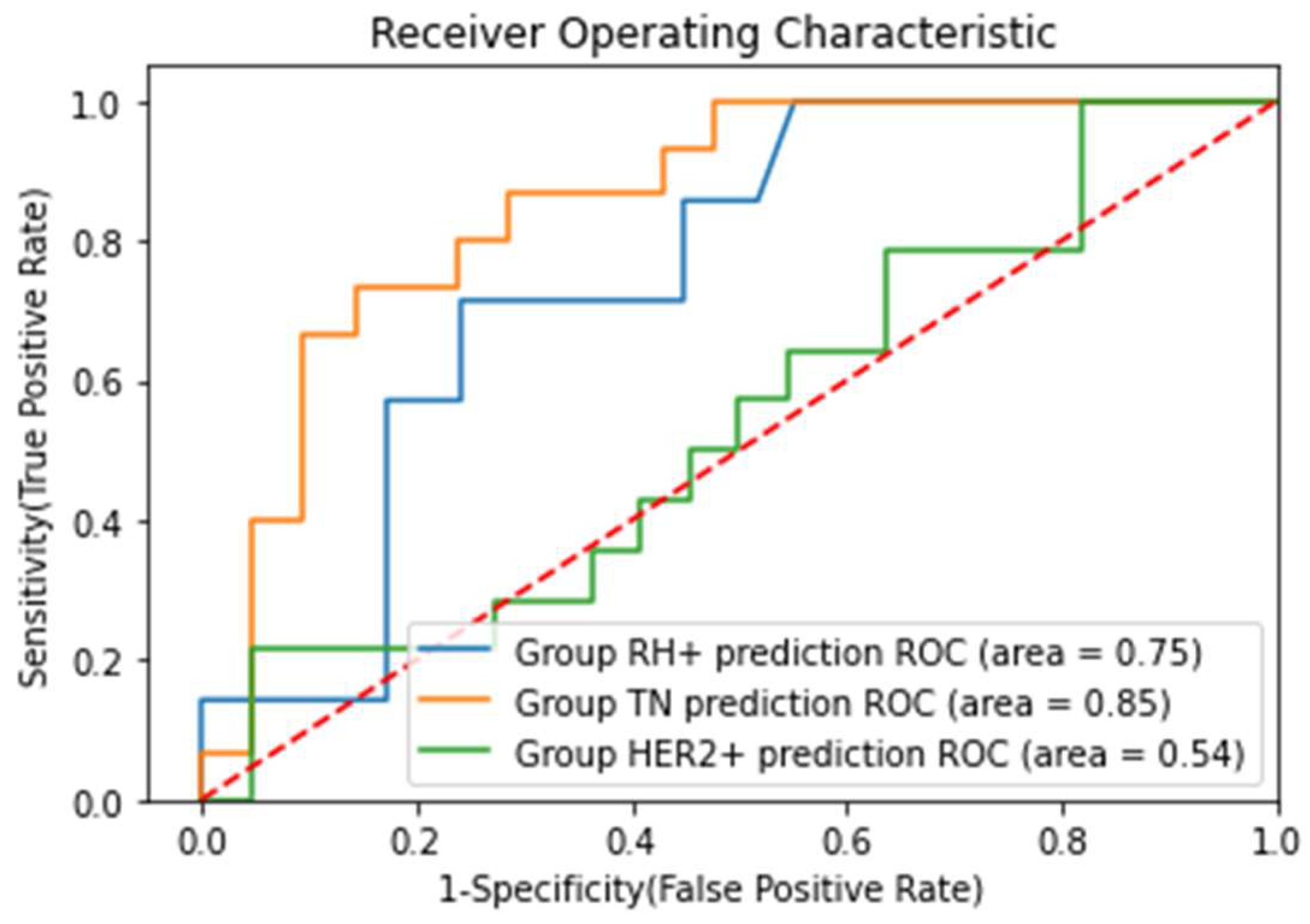
Appendix A. Prediction of Molecular Subtypes of Breast Cancer
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gianni, L.; Pienkowski, T.; Im, Y.-H.; Roman, L.; Tseng, L.-M.; Liu, M.-C.; Lluch, A.; Staroslawska, E.; de la Haba-Rodriguez, J.; Im, S.-A.; et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012, 13, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Mieog, J.S.D.; van der Hage, J.A.; van de Velde, C.J.H. Neoadjuvant chemotherapy for operable breast cancer. Br. J. Surg. 2007, 94, 1189–1200. [Google Scholar] [CrossRef]
- Symmans, W.F.; Peintinger, F.; Hatzis, C.; Rajan, R.; Kuerer, H.; Valero, V.; Assad, L.; Poniecka, A.; Hennessy, B.; Green, M.; et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 2007, 25, 4414–4422. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Lee, S.-J.; Ohtani, S.; Im, Y.-H.; Lee, E.-S.; Yokota, I.; Kuroi, K.; Im, S.-A.; Park, B.-W.; Kim, S.-B.; et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Montemurro, F.; Nuzzolese, I.; Ponzone, R. Neoadjuvant or Adjuvant Chemotherapy in Early Breast Cancer? Expert Opin. Pharmacother. 2020, 21, 1071–1082. [Google Scholar] [CrossRef]
- Heil, J.; Kuerer, H.M.; Pfob, A.; Rauch, G.; Sinn, H.P.; Golatta, M.; Liefers, G.J.; Vrancken Peeters, M.J. Eliminating the breast cancer surgery paradigm after neoadjuvant systemic therapy: Current evidence and future challenges. Ann. Oncol. 2020, 31, 61–71. [Google Scholar] [CrossRef]
- Garland, M.L.; Vather, R.; Bunkley, N.; Pearse, M.; Bissett, I.P. Clinical tumour size and nodal status predict pathologic complete response following neoadjuvant chemoradiotherapy for rectal cancer. Int. J. Color. Dis. 2014, 29, 301–307. [Google Scholar] [CrossRef]
- Hylton, N.M.; Blume, J.D.; Bernreuter, W.K.; Pisano, E.D.; Rosen, M.A.; Morris, E.A.; Weatherall, P.T.; Lehman, C.D.; Newstead, G.M.; Polin, S.; et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—Results from ACRIN 6657/I-SPY TRIAL. Radiology 2012, 263, 663–672. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, H.H.; Ahn, J.H.; Kim, S.-B.; Jung, K.H.; Gong, G.; Son, B.H.; Ahn, S.H. Comparison of mammography, sonography, MRI and clinical examination in patients with locally advanced or inflammatory breast cancer who underwent neoadjuvant chemotherapy. Br. J. Radiol. 2011, 84, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, X.; Li, L.; Shi, J.; Zeng, X.; Huang, Y.; Chen, H.; Jiang, F.; Yin, T.; Nickel, D.; et al. Early prediction of pathologic complete response of breast cancer after neoadjuvant chemotherapy using longitudinal ultrafast dynamic contrast-enhanced MRI. Diagn. Interv. Imaging 2023, 104, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Rella, R.; Bufi, E.; Belli, P.; Petta, F.; Serra, T.; Masiello, V.; Scrofani, A.R.; Barone, R.; Orlandi, A.; Valentini, V.; et al. Association between background parenchymal enhancement and tumor response in patients with breast cancer receiving neoadjuvant chemotherapy. Diagn. Interv. Imaging 2020, 101, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Stancanello, J.; El Naqa, I. Beyond imaging: The promise of radiomics. Phys. Med. 2017, 38, 122–139. [Google Scholar] [CrossRef]
- Song, J.; Yin, Y.; Wang, H.; Chang, Z.; Liu, Z.; Cui, L. A review of original articles published in the emerging field of radiomics. Eur. J. Radiol. 2020, 127, 108991. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2016, 2, 1636–1642. [Google Scholar] [CrossRef]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging 2020, 20, 33. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning Methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef]
- Granzier, R.W.Y.; van Nijnatten, T.J.A.; Woodruff, H.C.; Smidt, M.L.; Lobbes, M.B.I. Exploring Breast Cancer Response Prediction to Neoadjuvant Systemic Therapy Using MRI-Based Radiomics: A Systematic Review. Eur. J. Radiol. 2019, 121, 108736. [Google Scholar] [CrossRef]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and Peritumoral Radiomics for the Pretreatment Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy Based on Breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Invest. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Cain, E.H.; Saha, A.; Harowicz, M.R.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Multivariate Machine Learning Models for Prediction of Pathologic Response to Neoadjuvant Therapy in Breast Cancer Using MRI Features: A Study Using an Independent Validation Set. Breast Cancer Res. Treat. 2019, 173, 455–463. [Google Scholar] [CrossRef]
- Doran, S.J.; Kumar, S.; Orton, M.; d’Arcy, J.; Kwaks, F.; O’Flynn, E.; Ahmed, Z.; Downey, K.; Dowsett, M.; Turner, N.; et al. “Real-World” Radiomics from Multi-Vendor MRI: An Original Retrospective Study on the Prediction of Nodal Status and Disease Survival in Breast Cancer, as an Exemplar to Promote Discussion of the Wider Issues. Cancer Imaging 2021, 21, 37. [Google Scholar] [CrossRef]
- Morris, E.A.; Comstock, C.E.; Lee, C.H. ACR BI-RADS® Magnetic Resonance Imaging. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Da-ano, R.; Masson, I.; Lucia, F.; Doré, M.; Robin, P.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Reinhold, C.; Castelli, J.; et al. Performance Comparison of Modified ComBat for Harmonization of Radiomic Features for Multicenter Studies. Sci. Rep. 2020, 10, 10248. [Google Scholar] [CrossRef]
- Orlhac, F.; Lecler, A.; Savatovski, J.; Goya-Outi, J.; Nioche, C.; Charbonneau, F.; Ayache, N.; Frouin, F.; Duron, L.; Buvat, I. How Can We Combat Multicenter Variability in MR Radiomics? Validation of a Correction Procedure. Eur. Radiol. 2021, 31, 2272–2280. [Google Scholar] [CrossRef]
- Saint Martin, M.-J.; Orlhac, F.; Akl, P.; Khalid, F.; Nioche, C.; Buvat, I.; Malhaire, C.; Frouin, F. A Radiomics Pipeline Dedicated to Breast MRI: Validation on a Multi-Scanner Phantom Study. MAGMA 2021, 34, 355–366. [Google Scholar] [CrossRef]
- Bitencourt, A.G.; Gibbs, P.; Saccarelli, C.R.; Daimiel, I.; Gullo, R.L.; Fox, M.J.; Thakur, S.; Pinker, K.; Morris, E.A.; Morrow, M.; et al. MRI-Based Machine Learning Radiomics Can Predict HER2 Expression Level and Pathologic Response after Neoadjuvant Therapy in HER2 Overexpressing Breast Cancer. EBioMedicine 2020, 61, 103042. [Google Scholar] [CrossRef]
- Chen, J.H.; Feig, B.; Agrawal, G.; Yu, H.; Carpenter, P.M.; Mehta, R.S.; Nalcioglu, O.; Su, M.Y. MRI Evaluation of Pathologically Complete Response and Residual Tumors in Breast Cancer after Neoadjuvant Chemotherapy. Cancer 2008, 112, 17–26. [Google Scholar] [CrossRef]
- Eun, N.L.; Kang, D.; Son, E.J.; Park, J.S.; Youk, J.H.; Kim, J.-A.; Gweon, H.M. Texture Analysis with 3.0-T MRI for Association of Response to Neoadjuvant Chemotherapy in Breast Cancer. Radiology 2020, 294, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Dave, R.V.; Millican-Slater, R.; Dodwell, D.; Horgan, K.; Sharma, N. Neoadjuvant Chemotherapy with MRI Monitoring for Breast Cancer. Br. J. Surg. 2017, 104, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Parikh, J.; Selmi, M.; Charles-Edwards, G.; Glendenning, J.; Ganeshan, B.; Verma, H.; Mansi, J.; Harries, M.; Tutt, A.; Goh, V. Changes in Primary Breast Cancer Heterogeneity May Augment Midtreatment MR Imaging Assessment of Response to Neoadjuvant Chemotherapy. Radiology 2014, 272, 100–112. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer Based on Radiomics of T2-Weighted Fat-Suppression and Diffusion-Weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef]
- Zhao, B.; Tan, Y.; Tsai, W.-Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of Radiomics for Deciphering Tumor Phenotype with Imaging. Sci. Rep. 2016, 6, 23428. [Google Scholar] [CrossRef]
- Papadimitroulas, P.; Brocki, L.; Christopher Chung, N.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial Intelligence: Deep Learning in Oncological Radiomics and Challenges of Interpretability and Data Harmonization. Phys. Medica 2021, 83, 108–121. [Google Scholar] [CrossRef]
- Gibała, S.; Obuchowicz, R.; Lasek, J.; Piórkowski, A.; Nurzynska, K. Textural Analysis Supports Prostate MR Diagnosis in PIRADS Protocol. Appl. Sci. 2023, 13, 9871. [Google Scholar] [CrossRef]
- Obuchowicz, R.; Nurzynska, K.; Pierzchala, M.; Piorkowski, A.; Strzelecki, M. Texture Analysis for the Bone Age Assessment from MRI Images of Adolescent Wrists in Boys. J. Clin. Med. 2023, 12, 2762. [Google Scholar] [CrossRef]



| Characteristics | Value |
|---|---|
| Age at diagnosis (years, mean ± SD) | 46.47 ± 11.56 |
| Histologic subtype | |
| 208 (88.5%) |
| 14 (6%) |
| 2 (0.9%) |
| 2 (0.9%) |
| 2 (0.9%) |
| 5 (2.8%) |
| Histological grade | |
| 9 (3.8%) |
| 90 (38.2%) |
| 130 (55.3%) |
| 6 (2.7%) |
| Molecular subtype | |
| 91 (38.8%) |
| 72 (30.6%) |
| 72 (30.6%) |
| Genetic mutation | |
| 214 (91%) |
| 9 (3.7%) |
| 8 (3.5%) |
| 4 (1.8%) |
| T staging | |
| 4 (1.8%) |
| 138 (58.7%) |
| 71 (30.2%) |
| 5 (2.1%) |
| 4 (1.8%) |
| 13 (5.4%) |
| Lymph node invasion | 135 (57%) |
| MRI Models | Constructor | MRI Field (Tesla) | Exams Number |
|---|---|---|---|
| OptimaMR450w | GE | 1.5 | 61 a |
| Aera | Siemens | 1.5 | 53 |
| DiscoveryMR750w | GE | 3 | 36 b |
| Avanto | Siemens | 1.5 | 18 |
| OptimaMR360 | GE | 1.5 | 13 |
| Spectra | Siemens | 3 | 11 |
| SignaHDxt | GE | 1.5 | 11 |
| Ingenia | Philips | 3 | 7 |
| PanoramaHFO | Philips | 1 | 5 |
| Essenza | Siemens | 1.5 | 4 |
| Amira | Siemens | 1.5 | 3 |
| Achieva | Siemens | 1.5 | 3 |
| Titan | Canon | 1.5 | 2 |
| Skyra | Siemens | 3 | 2 |
| Signa Explorer | GE | 1.5 | 2 |
| Signa Excite | GE | 1.5 | 1 |
| Signa Architect | GE | 3 | 1 |
| Undetermined | 1.5 | 2 |
| Subset of Feature | AUC (mean +/− SD) |
|---|---|
| Clinical | 0.603 ± 0.042 |
| Radiologic | 0.583 ± 0.038 |
| Radiologic + Clinical | 0.646 ± 0.044 |
| Radiomic | 0.681 ± 0.049 |
| Radiomic + Clinical | 0.668 ± 0.046 |
| Radiomic + Radiologic | 0.664 ± 0.051 |
| Radiomic + Clinical + Radiologic | 0.650 ± 0.045 |
| Technique of Feature Selection | Mean F1 Score | Mean AUC Score |
|---|---|---|
| Fischer | 0.771 | 0.669 |
| MRMR | 0.782 | 0.662 |
| ANOVA | 0.777 | 0.655 |
| TSCR | 0.771 | 0.643 |
| ReliefF | 0.770 | 0.640 |
| PCA | 0.764 | 0.601 |
| Radiomic Features | Importance |
|---|---|
| T2-weighted: original first-order 90th percentile | 0.101 |
| T2- weighted: original first-order variance | 0.081 |
| T2-weighted: original first-order 10th percentile | 0.080 |
| T2-weighted: Original Grey Level Size Zone Matrix Grey Level Variance | 0.074 |
| T2-weighted: original first-order minimum | 0.065 |
| Radiologic features | Importance |
| Axial diameter on T1-weighted enhanced | 0.088 |
| Peri-tumoral oedema | 0.026 |
| Irregular margin | 0.013 |
| Clinical features | Importance |
| Age at diagnosis | 0.049 |
| No hormonal receptor expression Histologic grade | 0.036 0.011 |
| Endpoint (Years) | Number of Recurrence | Number of Samples | Recurrence Percentage |
|---|---|---|---|
| 2 | 4 | 46 | 8.7 |
| 3 | 5 | 42 | 11.9 |
| 4 | 7 | 32 | 21.8 |
| 5 | 6 | 24 | 25 |
| Study (First Author, Year) | Cohort Size | Input Data | ML Method | Reported AUC | External Validation | Main Limitations |
|---|---|---|---|---|---|---|
| Liu et al., 2019 [24] | 364 | Multiparametric MRI (radiomics) | SVM | 0.79 | Yes | No integration of clinical outcomes |
| Braman et al., 2017 [22] | 117 | Intratumoral and peritumoral MRI features | Logistic Regression | 0.74 | No | Small cohort, radiomics only |
| Bitencourt et al., 2020 [32] | 93 | MRI-based features | Machine learning | 0.84 (Accuracy) | No | No radiomics or clinical variable comparison |
| Cain et al., 2019 [25] | 288 | MRI radiomics | SVM, Random Forest | 0.73 | Yes | Limited integration of clinical features |
| Current Study (Hajri et al.) | 235 | Clinical, radiologic, radiomic MRI | Random Forest, SVM | 0.72 | No | No external validation, limited DWI availability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hajri, R.; Aboudaram, C.; Lassau, N.; Assi, T.; Antoun, L.; Ribeiro, J.M.; Lacroix-Triki, M.; Ammari, S.; Balleyguier, C. Prediction of Breast Cancer Response to Neoadjuvant Therapy with Machine Learning: A Clinical, MRI-Qualitative, and Radiomics Approach. Life 2025, 15, 1165. https://doi.org/10.3390/life15081165
Hajri R, Aboudaram C, Lassau N, Assi T, Antoun L, Ribeiro JM, Lacroix-Triki M, Ammari S, Balleyguier C. Prediction of Breast Cancer Response to Neoadjuvant Therapy with Machine Learning: A Clinical, MRI-Qualitative, and Radiomics Approach. Life. 2025; 15(8):1165. https://doi.org/10.3390/life15081165
Chicago/Turabian StyleHajri, Rami, Charles Aboudaram, Nathalie Lassau, Tarek Assi, Leony Antoun, Joana Mourato Ribeiro, Magali Lacroix-Triki, Samy Ammari, and Corinne Balleyguier. 2025. "Prediction of Breast Cancer Response to Neoadjuvant Therapy with Machine Learning: A Clinical, MRI-Qualitative, and Radiomics Approach" Life 15, no. 8: 1165. https://doi.org/10.3390/life15081165
APA StyleHajri, R., Aboudaram, C., Lassau, N., Assi, T., Antoun, L., Ribeiro, J. M., Lacroix-Triki, M., Ammari, S., & Balleyguier, C. (2025). Prediction of Breast Cancer Response to Neoadjuvant Therapy with Machine Learning: A Clinical, MRI-Qualitative, and Radiomics Approach. Life, 15(8), 1165. https://doi.org/10.3390/life15081165









