CYTO-SV-ML: A Machine Learning Tool for Cytogenetic Structural Variant Analysis in Somatic Cell Type Using Genome Sequences
Abstract
1. Introduction
2. Materials and Methods
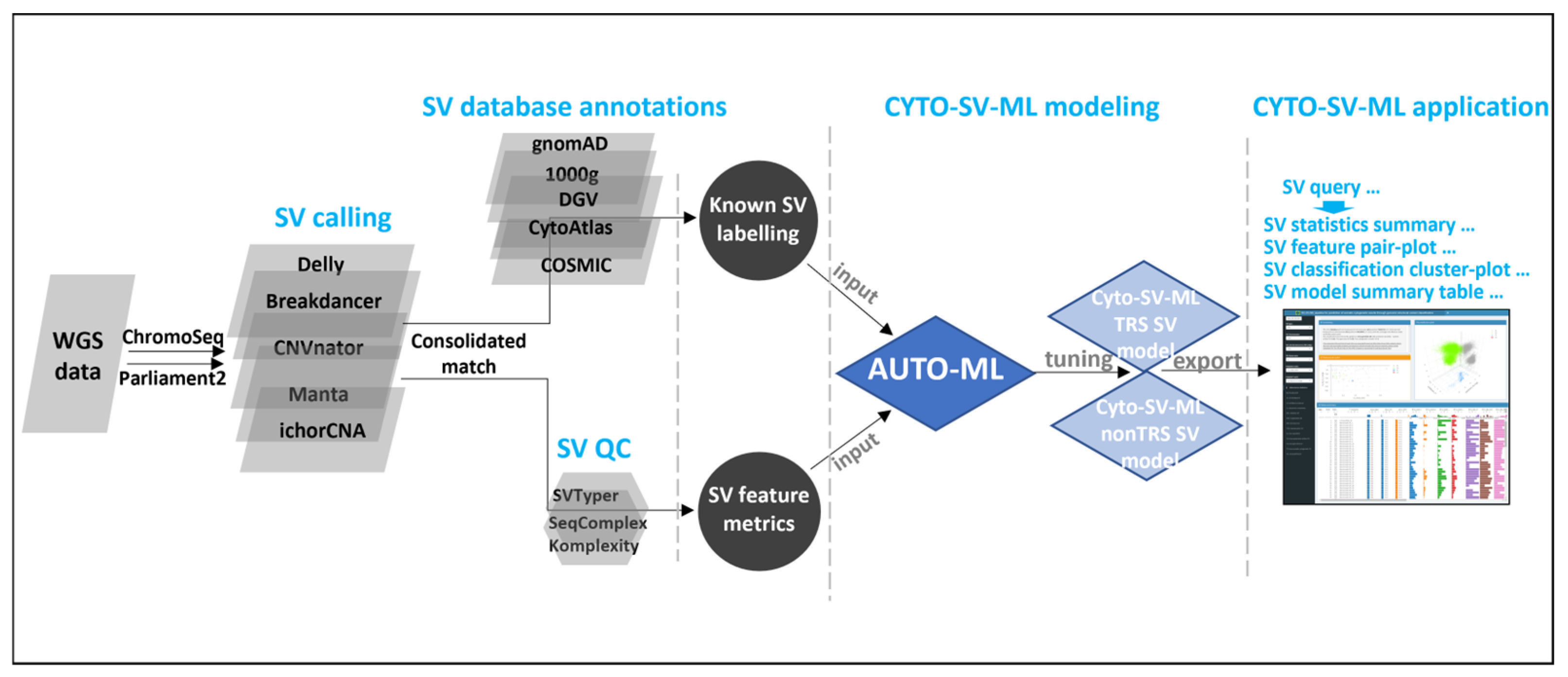
2.1. Implementation
2.1.1. WGS SV Preprocessing
2.1.2. Known SV Labeling
2.1.3. SV Classification Modeling
2.1.4. CYTO-SV-ML Interface Application
2.2. Biological Cohort, DNA Extraction
2.3. Whole Genome Sequencing
3. Results
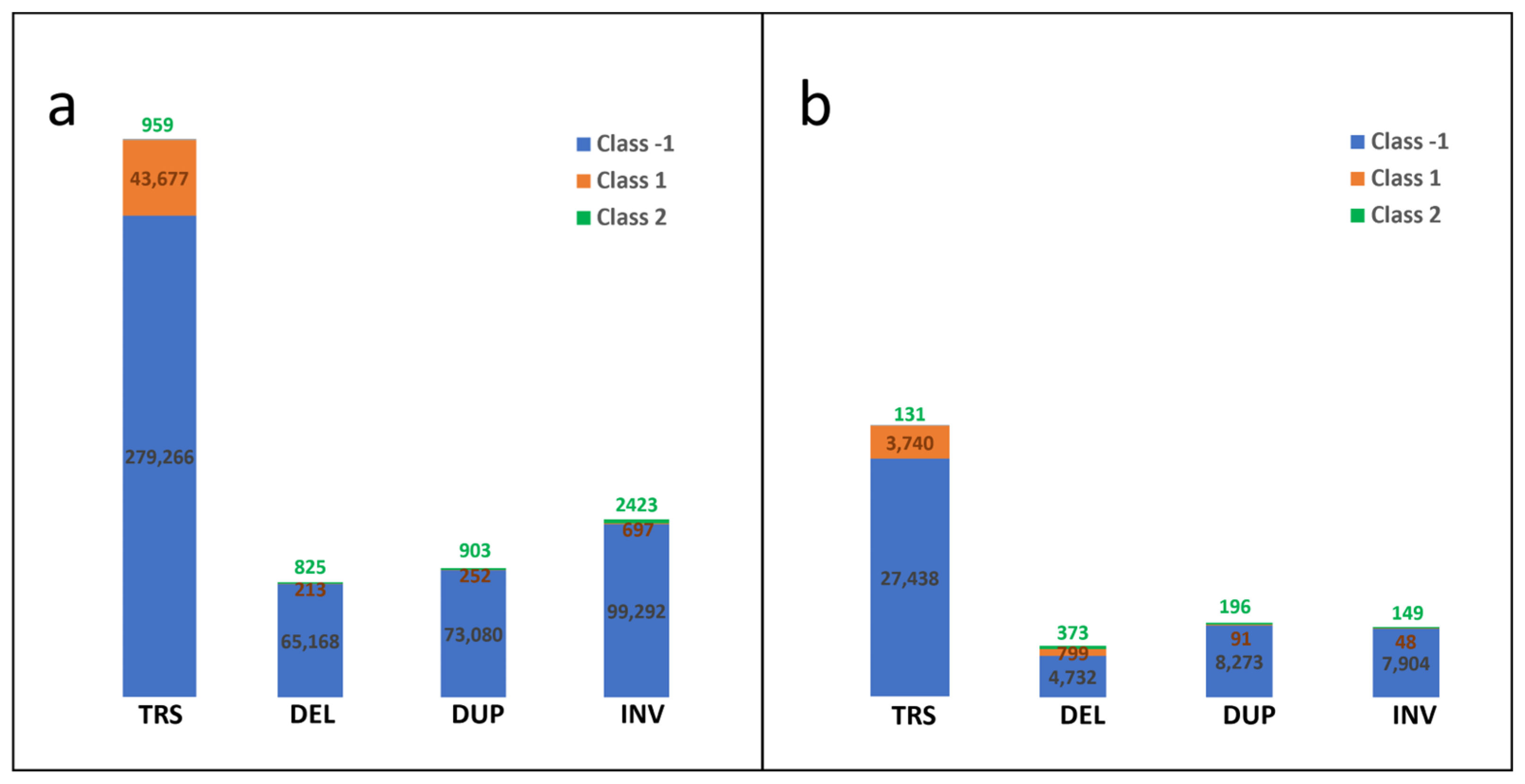
3.1. Preparation of Known SV Data of Cytogenetic Somatic SVs
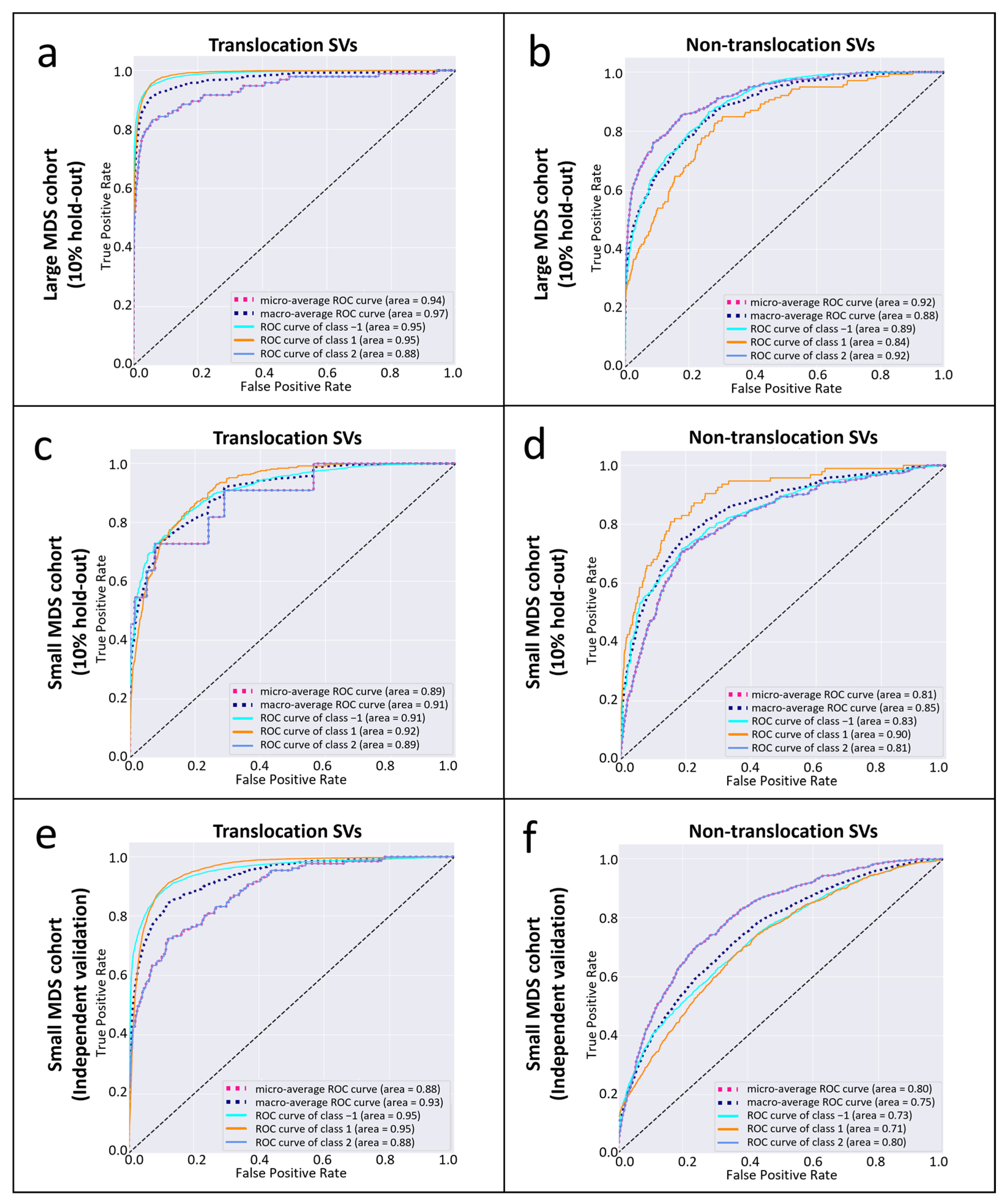
3.2. The Performance of CYTO-SV-ML Pipeline
3.3. Confirmation of SVs from Cytogenetic Records
3.4. The Overview of CYTO-SV-ML Application
3.5. The Runtime of CYTO-SV-ML Pipeline
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WGS | whole genome sequencing |
| SV | structural variation |
| CNV | copy number variation |
| MDS | myelodysplastic syndromes |
| ML | machine learning |
| AUC | area under the curve |
| FISH | fluorescence in situ hybridization |
| CMA | chromosomal microarray |
| COSMIC | Catalogue of Somatic Mutations in Cancer |
| gnomAD | Genome Aggregation Database |
| TRS | translocation |
| AF | allele frequency |
| PBC | peripheral blood cell |
| SHAP | Shapley Additive Explanations |
References
- Akkari, Y.M.N.; Baughn, L.B.; Dubuc, A.M.; Smith, A.C.; Mallo, M.; Dal Cin, P.; Diez Campelo, M.; Gallego, M.S.; Granada Font, I.; Haase, D.T.; et al. Guiding the global evolution of cytogenetic testing for hematologic malignancies. Blood 2022, 139, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Akkari, Y.; Baughn, L.B.; Kim, A.; Karaca, E.; Raca, G.; Shao, L.; Mikhail, F.M.; ACMG Laboratory Quality Assurance Committee. Section E6.1-6.6 of the American College of Medical Genetics and Genomics (ACMG) Technical Laboratory Standards: Cytogenomic studies of acquired chromosomal abnormalities in neoplastic blood, bone marrow, and lymph nodes. Genet. Med. 2024, 26, 101054. [Google Scholar] [CrossRef] [PubMed]
- Schwabkey, Z.I.; Al Ali, N.; Chan, O.; Sallman, D.A.; Padron, E.; Kuykendall, A.T.; Talati, C.; Sweet, K.; Lancet, J.E.; Komrokji, R.S. Fluorescence in Situ Hybridization (FISH) Utility for Risk Score Assessment in Patients With MDS With Normal Metaphase Karyotype. Clin. Lymphoma Myeloma Leuk. 2021, 21, e52–e56. [Google Scholar] [CrossRef]
- O’Malley, D.P.; Giudice, C.; Chang, A.S.; Chang, D.; Barry, T.S.; Hibbard, M.K.; Chen, R.; Chen, S.T. Comparison of array comparative genomic hybridization (aCGH) to FISH and cytogenetics in prognostic evaluation of chronic lymphocytic leukemia. Int. J. Lab. Hematol. 2011, 33, 238–244. [Google Scholar] [CrossRef]
- Mikhail, F.M.; Heerema, N.A.; Rao, K.W.; Burnside, R.D.; Cherry, A.M.; Cooley, L.D. Section E6.1-6.4 of the ACMG technical standards and guidelines: Chromosome studies of neoplastic blood and bone marrow-acquired chromosomal abnormalities. Genet. Med. 2016, 18, 635–642. [Google Scholar] [CrossRef]
- Alkan, C.; Coe, B.P.; Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Duncavage, E.J.; Schroeder, M.C.; O’Laughlin, M.; Wilson, R.; MacMillan, S.; Bohannon, A.; Kruchowski, S.; Garza, J.; Du, F.; Hughes, A.E.O.; et al. Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers. N. Engl. J. Med. 2021, 384, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Mack, E.K.M.; Marquardt, A.; Langer, D.; Ross, P.; Ultsch, A.; Kiehl, M.G.; Mack, H.I.D.; Haferlach, T.; Neubauer, A.; Brendel, C. Comprehensive genetic diagnosis of acute myeloid leukemia by next-generation sequencing. Haematologica 2019, 104, 277–287. [Google Scholar] [CrossRef]
- Mareschal, S.; Palau, A.; Lindberg, J.; Ruminy, P.; Nilsson, C.; Bengtzen, S.; Engvall, M.; Eriksson, A.; Neddermeyer, A.; Marchand, V.; et al. Challenging conventional karyotyping by next-generation karyotyping in 281 intensively treated patients with AML. Blood Adv. 2021, 5, 1003–1016. [Google Scholar] [CrossRef]
- Kayser, S.; Hills, R.K.; Langova, R.; Kramer, M.; Guijarro, F.; Sustkova, Z.; Estey, E.H.; Shaw, C.M.; Racil, Z.; Mayer, J.; et al. Characteristics and outcome of patients with acute myeloid leukaemia and t(8;16)(p11;p13): Results from an International Collaborative Study. Br. J. Haematol. 2021, 192, 832–842. [Google Scholar] [CrossRef]
- Uguen, K.; Jubin, C.; Duffourd, Y.; Bardel, C.; Malan, V.; Dupont, J.M.; El Khattabi, L.; Chatron, N.; Vitobello, A.; Rollat-Farnier, P.A.; et al. Genome sequencing in cytogenetics: Comparison of short-read and linked-read approaches for germline structural variant detection and characterization. Mol. Genet. Genom. Med. 2020, 8, e1114. [Google Scholar] [CrossRef] [PubMed]
- Lindstrand, A.; Eisfeldt, J.; Pettersson, M.; Carvalho, C.M.B.; Kvarnung, M.; Grigelioniene, G.; Anderlid, B.M.; Bjerin, O.; Gustavsson, P.; Hammarsjo, A.; et al. From cytogenetics to cytogenomics: Whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Med. 2019, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Asadi Fakhr, Z.; Mehrzad, V.; Izaditabar, A.; Salehi, M. Evaluation of the utility of peripheral blood vs bone marrow in karyotype and fluorescence in situ hybridization for myelodysplastic syndrome diagnosis. J. Clin. Lab. Anal. 2018, 32, e22586. [Google Scholar] [CrossRef]
- Coleman, J.F.; Theil, K.S.; Tubbs, R.R.; Cook, J.R. Diagnostic yield of bone marrow and peripheral blood FISH panel testing in clinically suspected myelodysplastic syndromes and/or acute myeloid leukemia: A prospective analysis of 433 cases. Am. J. Clin. Pathol. 2011, 135, 915–920. [Google Scholar] [CrossRef]
- Huret, J.L.; Ahmad, M.; Arsaban, M.; Bernheim, A.; Cigna, J.; Desangles, F.; Guignard, J.C.; Jacquemot-Perbal, M.C.; Labarussias, M.; Leberre, V.; et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013, 41, D920–D924. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Popic, V.; Rohlicek, C.; Cunial, F.; Hajirasouliha, I.; Meleshko, D.; Garimella, K.; Maheshwari, A. Cue: A deep-learning framework for structural variant discovery and genotyping. Nat. Methods 2023, 20, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, S.; Audano, P.A.; Meng, D.; Flores, J.I.; Kosters, W.; Yang, X.; Jia, P.; Marschall, T.; Beck, C.R.; et al. SVision: A deep learning approach to resolve complex structural variants. Nat. Methods 2022, 19, 1230–1233. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stutz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Fan, X.; Abbott, T.E.; Larson, D.; Chen, K. BreakDancer: Identification of Genomic Structural Variation from Paired-End Read Mapping. Curr. Protoc. Bioinform. 2014, 45, 15.6.1–15.6.11. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Kallberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.R.; Ziman, R.; Yuen, R.K.; Feuk, L.; Scherer, S.W. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014, 42, D986–D992. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, M.J.P.; Sanders, A.D.; Zhao, X.; Malhotra, A.; Porubsky, D.; Rausch, T.; Gardner, E.J.; Rodriguez, O.L.; Guo, L.; Collins, R.L.; et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019, 10, 1784. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Zhang, T.; Auer, P.; Dong, J.; Cutler, C.; Dezern, A.E.; Gadalla, S.M.; Deeg, H.J.; Nazha, A.; Carlson, K.S.; Spellman, S.; et al. Whole-genome sequencing identifies novel predictors for hematopoietic cell transplant outcomes for patients with myelodysplastic syndrome: A CIBMTR study. J. Hematol. Oncol. 2023, 16, 37. [Google Scholar] [CrossRef]
- Wang, W.; Auer, P.; Zhang, T.; Spellman, S.; Carlson, K.S.; Nazha, A.; Bolon, Y.T.; Saber, W. Impact of Epigenomic Hypermethylation at TP53 on Allogeneic Hematopoietic Cell Transplantation Outcomes for Myelodysplastic Syndromes. Transplant. Cell Ther. 2021, 27, 659.e1–659.e6. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; De Haro, L.P.; Wray, J.; Hromas, R. Mechanisms of leukemia translocations. Curr. Opin. Hematol. 2008, 15, 338–345. [Google Scholar] [CrossRef]
- Kosugi, S.; Momozawa, Y.; Liu, X.; Terao, C.; Kubo, M.; Kamatani, Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019, 20, 117. [Google Scholar] [CrossRef]
- Haferlach, T.; Hutter, S.; Meggendorfer, M. Genome Sequencing in Myeloid Cancers. N. Engl. J. Med. 2021, 384, e106. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Auer, P.; Spellman, S.R.; Dong, J.; Saber, W.; Bolon, Y.-T. CYTO-SV-ML: A Machine Learning Tool for Cytogenetic Structural Variant Analysis in Somatic Cell Type Using Genome Sequences. Life 2025, 15, 929. https://doi.org/10.3390/life15060929
Zhang T, Auer P, Spellman SR, Dong J, Saber W, Bolon Y-T. CYTO-SV-ML: A Machine Learning Tool for Cytogenetic Structural Variant Analysis in Somatic Cell Type Using Genome Sequences. Life. 2025; 15(6):929. https://doi.org/10.3390/life15060929
Chicago/Turabian StyleZhang, Tao, Paul Auer, Stephen R. Spellman, Jing Dong, Wael Saber, and Yung-Tsi Bolon. 2025. "CYTO-SV-ML: A Machine Learning Tool for Cytogenetic Structural Variant Analysis in Somatic Cell Type Using Genome Sequences" Life 15, no. 6: 929. https://doi.org/10.3390/life15060929
APA StyleZhang, T., Auer, P., Spellman, S. R., Dong, J., Saber, W., & Bolon, Y.-T. (2025). CYTO-SV-ML: A Machine Learning Tool for Cytogenetic Structural Variant Analysis in Somatic Cell Type Using Genome Sequences. Life, 15(6), 929. https://doi.org/10.3390/life15060929






