One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023–July 2024)
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Sample Processing and RNA Extraction
2.3. Detection and Quantification of SARS-CoV-2
2.4. Sequencing of SARS-CoV-2 Variants in Wastewater Samples
2.5. Bioinformatics Analysis of SARS-CoV-2 Variants in Wastewater Samples
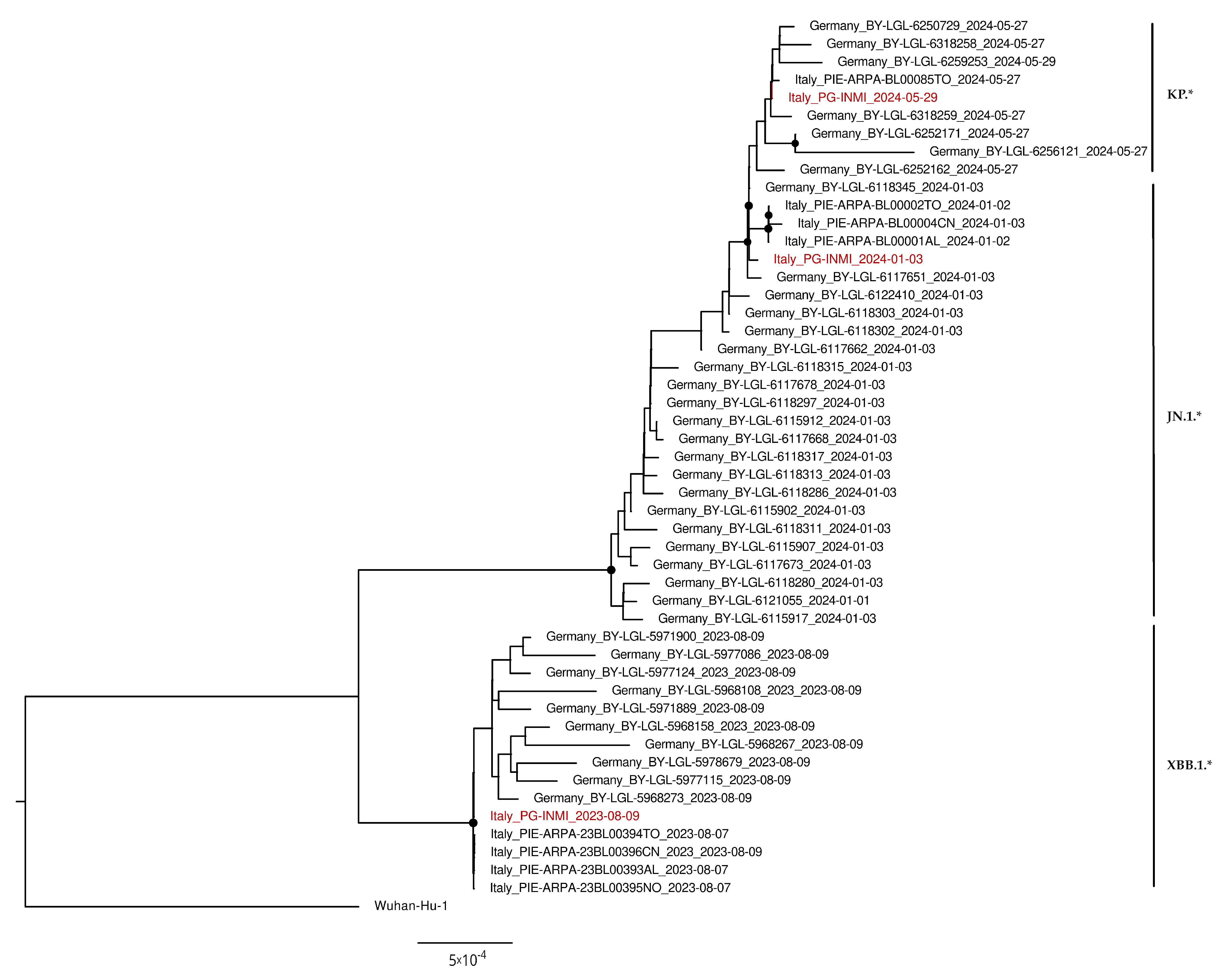
2.6. Phylogenetic Analysis
2.7. Assessment of SARS-CoV-2 Variants in Clinical Samples
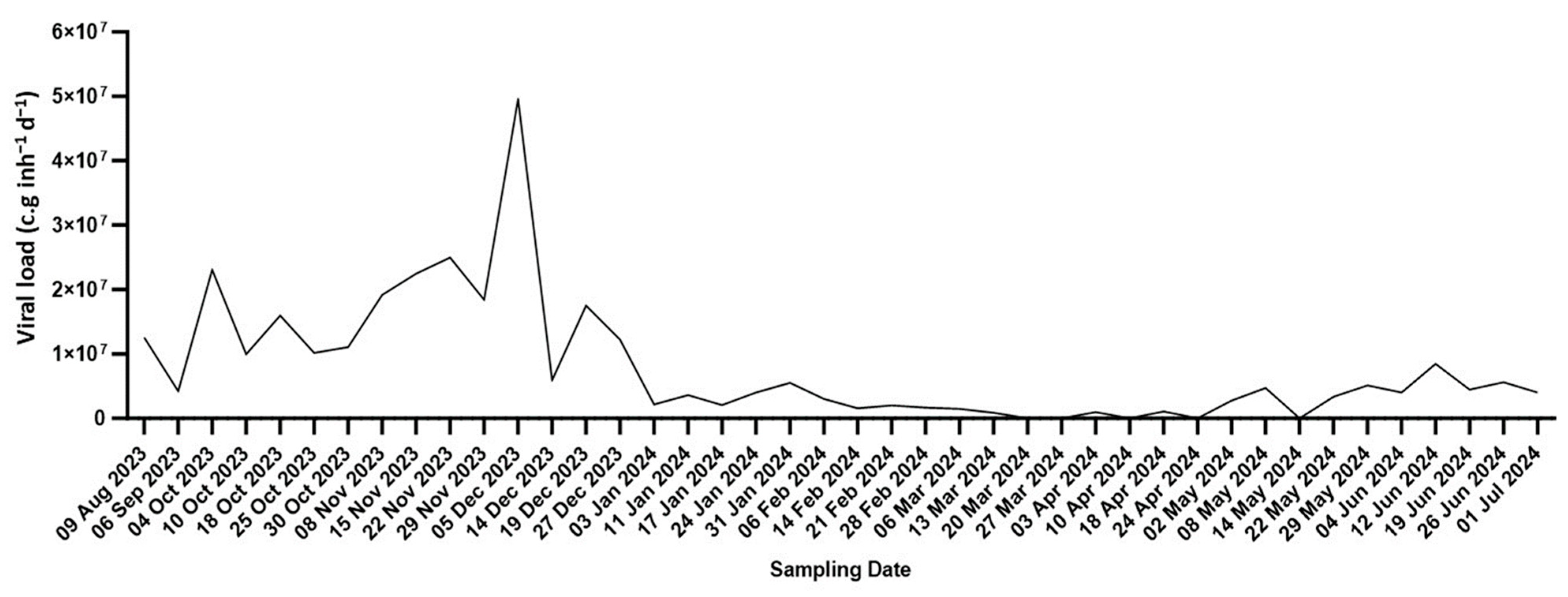
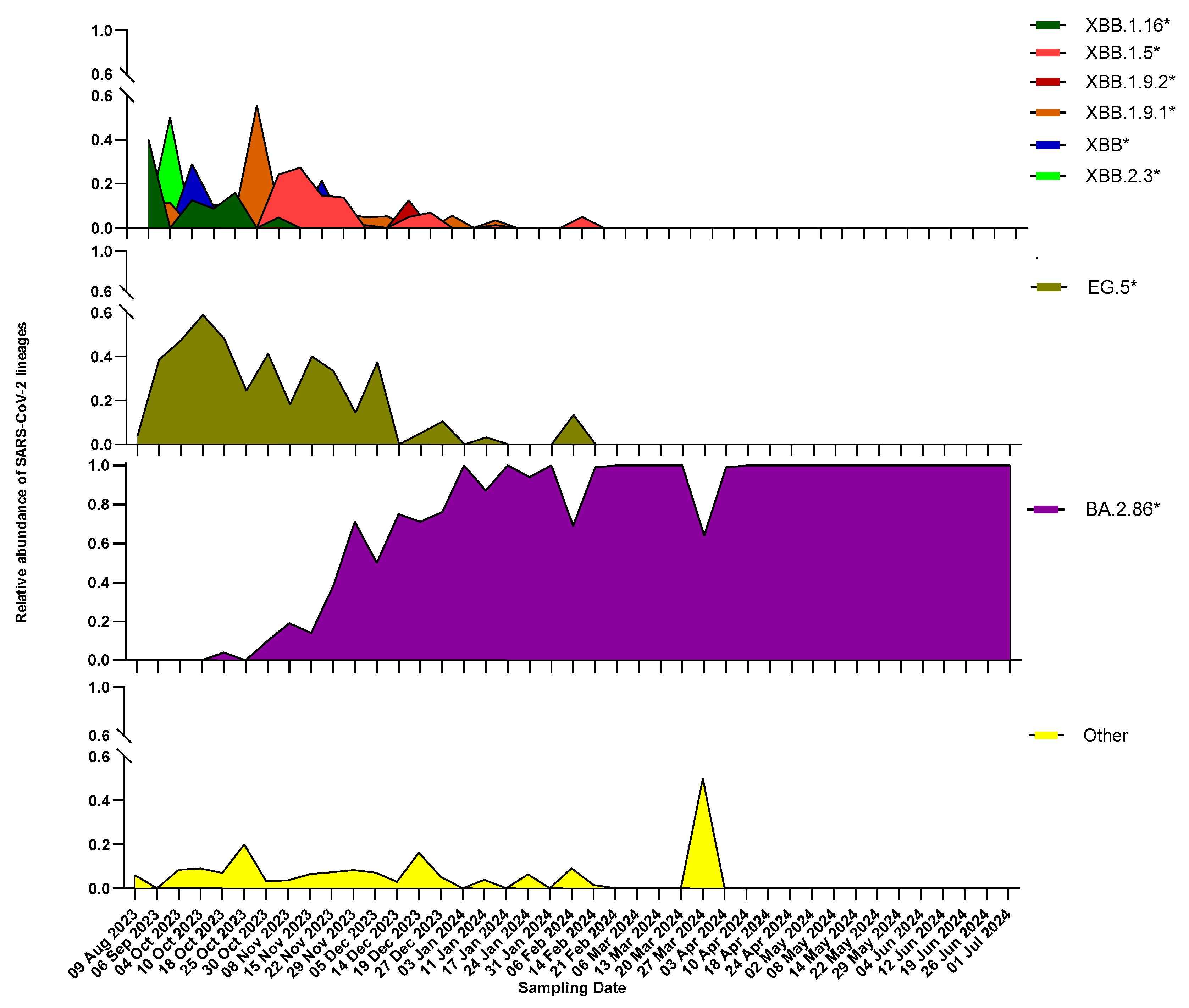
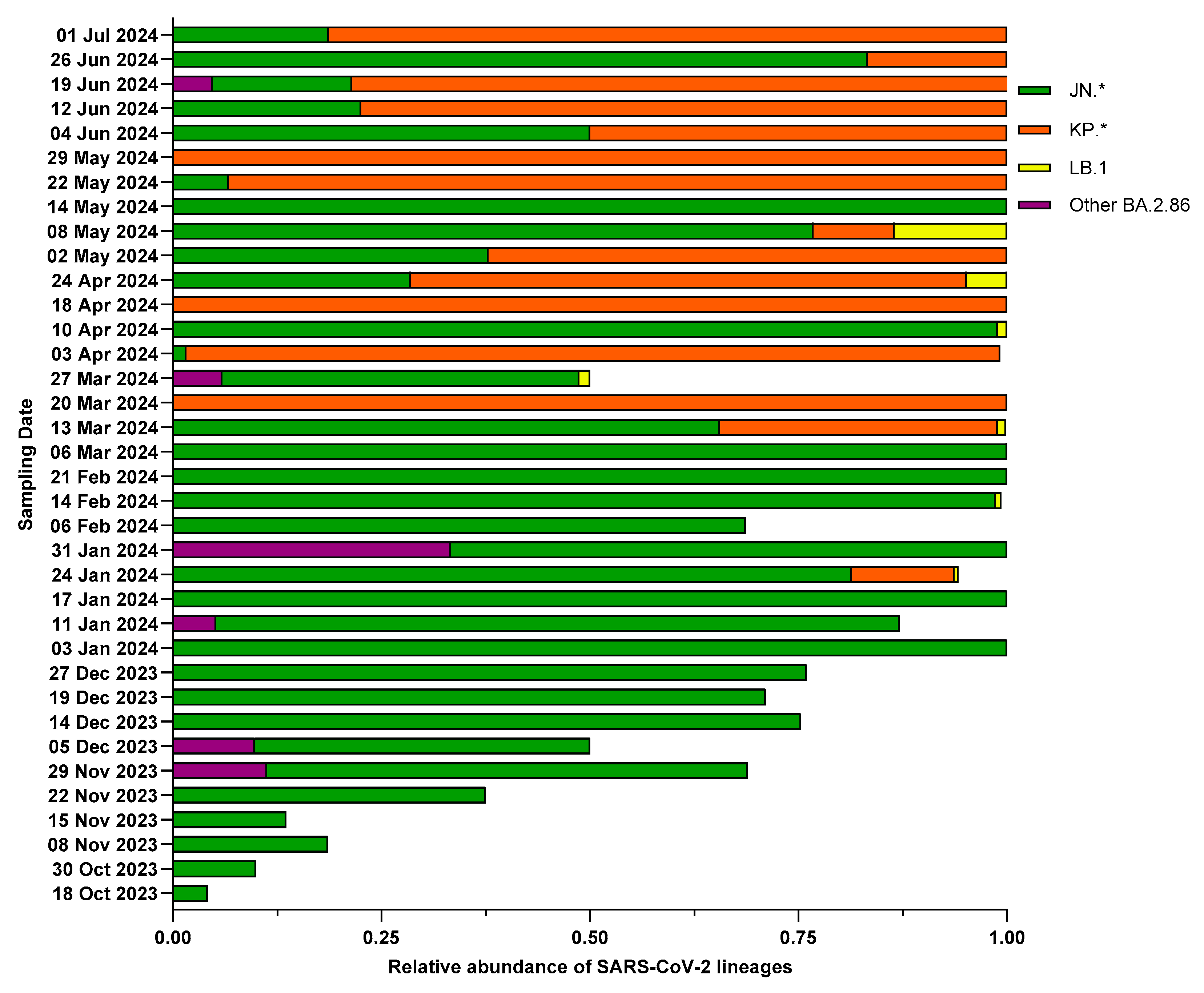
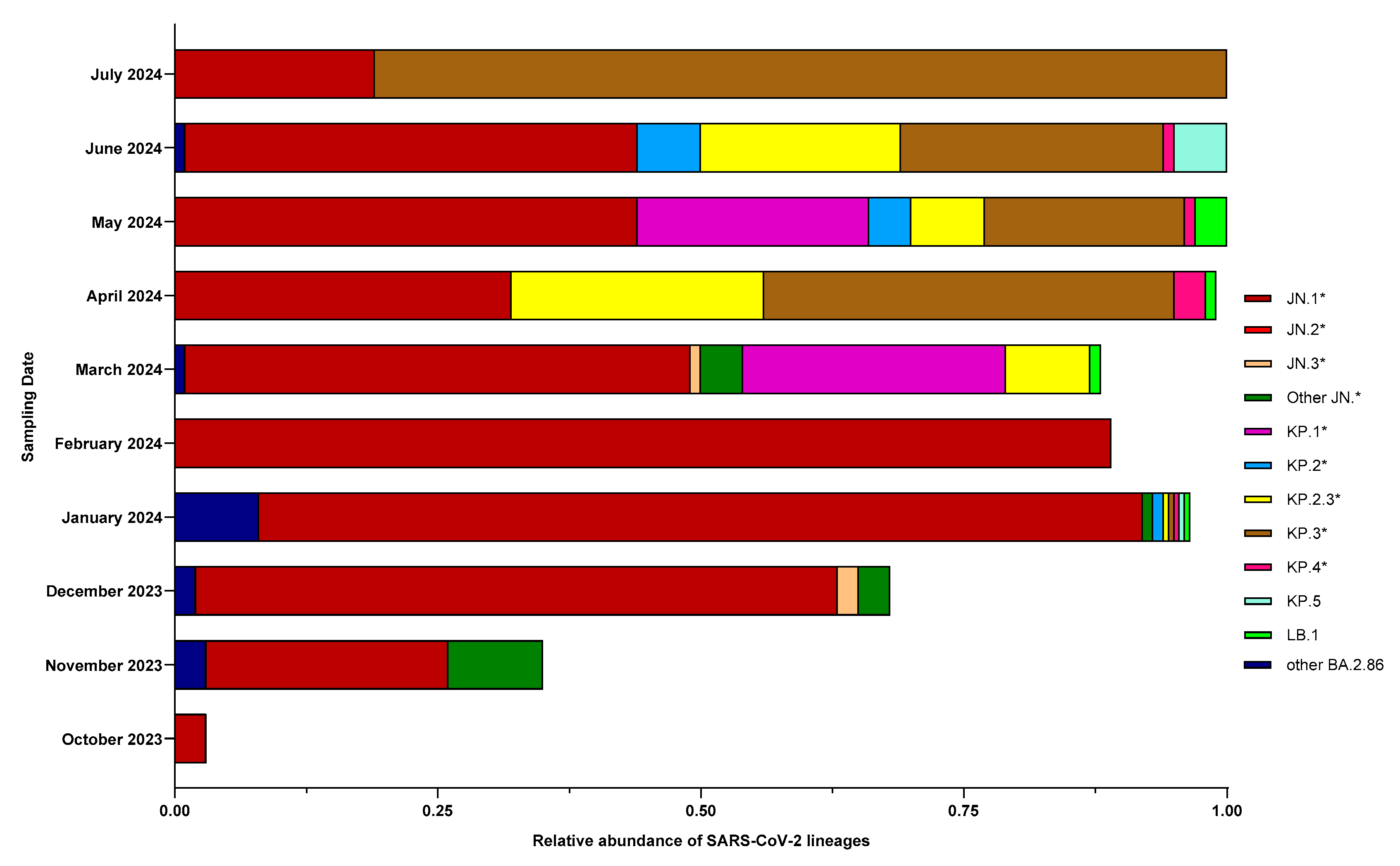
3. Results
3.1. Estimation of Circulating SARS-CoV-2 Quantities
3.2. Overview of SARS-CoV-2 Circulating Lineages
3.3. Characterization of BA.2.86 and Its Descendants
3.4. Phylogenetic Analysis
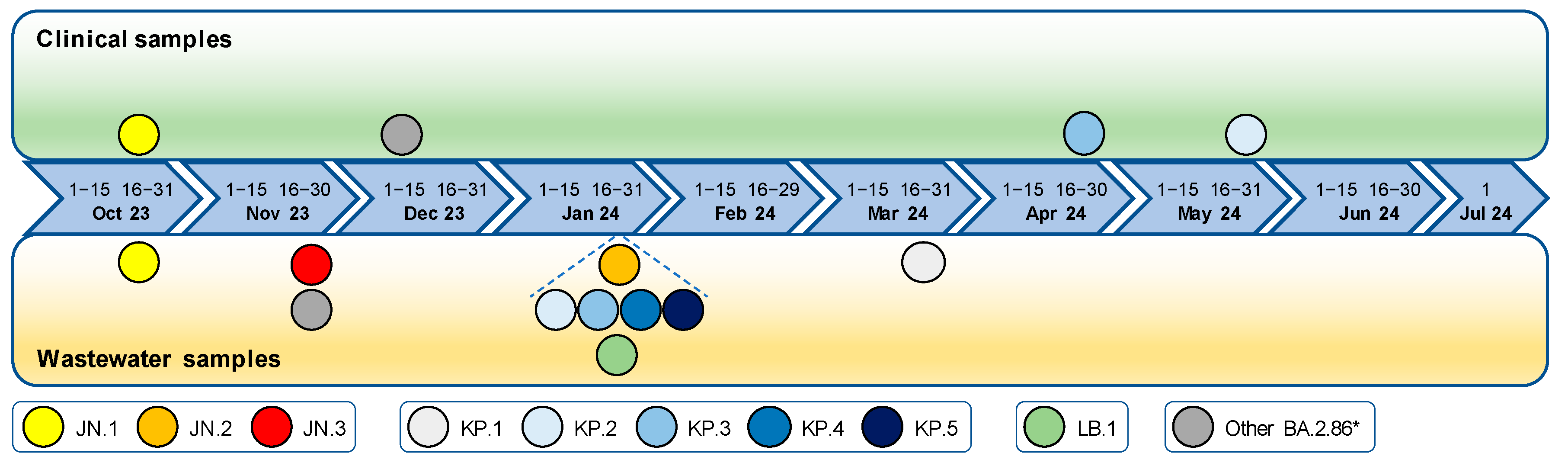
3.5. Detection Timeline of SARS-CoV-2 Lineages in Wastewater Compared to Clinical Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manirambona, E.; Okesanya, O.J.; Olaleke, N.O.; Oso, T.A.; Lucero-Prisno, D.E. Evolution and Implications of SARS-CoV-2 Variants in the Post-Pandemic Era. Discov. Public Health 2024, 21, 16. [Google Scholar] [CrossRef]
- Aggiornamento Dei Dati. Del Bollettino Della Sorveglianza Integrata COVID-19 in Italia. Available online: https://www.epicentro.iss.it/coronavirus/aggiornamenti (accessed on 24 April 2025).
- Shanmugam, B.K.; Alqaydi, M.; Abdisalam, D.; Shukla, M.; Santos, H.; Samour, R.; Petalidis, L.; Oliver, C.M.; Brudecki, G.; Bin Salem, S.; et al. A Narrative Review of High Throughput Wastewater Sample Processing for Infectious Disease Surveillance: Challenges, Progress, and Future Opportunities. Int. J. Env. Res. Public Health 2024, 21, 1432. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xu, J.; Guo, J.; Zhang, P.; Guo, X.; Zuo, Z.; Gao, L.; Jia, Z.; Xue, P.; Wang, J. An Epidemiologic Surveillance Study Based on Wastewater and Respiratory Specimens Reveals Influenza a Virus Prevalence and Mutations in Taiyuan, China during 2023–2024. BMC Infect. Dis. 2024, 24, 1286. [Google Scholar] [CrossRef]
- Annan, J.; Henderson, R.; Gray, M.; Clark, R.G.; Sarin, C.; Black, K. A Review of Wastewater-Based Epidemiology for the SARS-CoV-2 Virus in Rural, Remote, and Resource-Constrained Settings Internationally: Insights for Implementation, Research, and Policy for First Nations in Canada. Int. J. Env. Res. Public Health 2024, 21, 1429. [Google Scholar] [CrossRef]
- Farkas, K.; Williams, R.C.; Hillary, L.S.; Garcia-Delgado, A.; Jameson, E.; Kevill, J.L.; Wade, M.J.; Grimsley, J.M.S.; Jones, D.L. Harnessing the Power of Next-Generation Sequencing in Wastewater-Based Epidemiology and Global Disease Surveillance. Food Environ. Virol. 2025, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, E.E.; Okwuraiwe, A.; Kunle-Ope, C.N.; Igbasi, U.T.; Onyejepu, N.; Osuolale, K.; Shaibu, J.O.; Ojogbede, A.; Abuh, D.; Afocha, E.; et al. Surveillance of Public Health Pathogens in Lagos Wastewater Canals: A Cross-Sectional Study. BMC Public Health 2024, 24, 3590. [Google Scholar] [CrossRef]
- Nkambule, S.; Street, R.; Surujlal-Naicker, S.; Johnson, R.; Mathee, A. Wastewater Surveillance for SARS-CoV-2 during a Mass Sporting Event in the City of Cape Town, Western Cape. Front. Public Health 2024, 12, 1462629. [Google Scholar] [CrossRef]
- Jones, G.; Nelson, A.; Chadwick, D.R.; Cobley, S.; Jones, D.L.; Perrett, S.; Perry, W.B.; Weightman, A.J.; Williams, R.C.; Thomas, D.R. Evaluation of Wastewater Surveillance for SARS-CoV-2 in a Prison Population: A Mixed-Methods Approach. Front. Public Health 2024, 12, 1462186. [Google Scholar] [CrossRef]
- Haldar, T.; Katarmal, P.; Roy, B.; Koratkar, S. Dengue and Chikungunya Virus Dynamics, Identification, and Monitoring in Wastewater. Env. Monit. Assess. 2024, 196, 1166. [Google Scholar] [CrossRef]
- Girón-Guzmán, I.; Sánchez, G.; Pérez-Cataluña, A. Tracking Epidemic Viruses in Wastewaters. Microb. Biotechnol. 2024, 17, e70020. [Google Scholar] [CrossRef]
- Grassly, N.C.; Shaw, A.G.; Owusu, M. Global Wastewater Surveillance for Pathogens with Pandemic Potential: Opportunities and Challenges. Lancet Microbe 2024, 6, 100939. [Google Scholar] [CrossRef] [PubMed]
- Saravia, C.J.; Pütz, P.; Wurzbacher, C.; Uchaikina, A.; Drewes, J.E.; Braun, U.; Bannick, C.G.; Obermaier, N. Wastewater-Based Epidemiology: Deriving a SARS-CoV-2 Data Validation Method to Assess Data Quality and to Improve Trend Recognition. Front. Public Health 2024, 12, 1497100. [Google Scholar] [CrossRef] [PubMed]
- Masachessi, G.; Castro, G.M.; de los Angeles Marinzalda, M.; Cachi, A.M.; Sicilia, P.; Prez, V.E.; Martínez, L.C.; Giordano, M.O.; Pisano, M.B.; Ré, V.E.; et al. Unveiling the Silent Information of Wastewater-Based Epidemiology of SARS-CoV-2 at Community and Sanitary Zone Levels: Experience in Córdoba City, Argentina. J. Water Health 2024, 22, 2171–2183. [Google Scholar] [CrossRef]
- Schenk, H.; Rauch, W.; Zulli, A.; Boehm, A.B. SARS-CoV-2 Surveillance in US Wastewater: Leading Indicators and Data Variability Analysis in 2023–2024. PLoS ONE 2024, 19, e0313927. [Google Scholar] [CrossRef]
- Parkins, M.D.; Lee, B.E.; Acosta, N.; Bautista, M.; Hubert, C.R.J.; Hrudey, S.E.; Frankowski, K.; Pang, X.L. Wastewater-Based Surveillance as a Tool for Public Health Action: SARS-CoV-2 and Beyond. Clin. Microbiol. Rev. 2024, 37, e0010322. [Google Scholar] [CrossRef]
- Solis-Moreira, J. Study: New Wastewater Surveillance Method Detected SARS-CoV-2 Variants of Concern Up to 2 Weeks Before Clinical Tests. JAMA 2022, 328, 914–915. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus Network (CoViNet). Available online: https://www.who.int/groups/who-coronavirus-network (accessed on 24 April 2025).
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 Variant: A New Chapter in the COVID-19 Pandemic. Lancet 2021, 398, 2126–2128. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Akter, S.; Rashid, A.A.; Khair, S.; Alam, A.S.M.R.U. Unique Mutations in SARS-CoV-2 Omicron Subvariants’ Non-Spike Proteins: Potential Impacts on Viral Pathogenesis and Host Immune Evasion. Microb. Pathog. 2022, 170, 105699. [Google Scholar] [CrossRef]
- Lyngse, F.P.; Kirkeby, C.T.; Denwood, M.; Christiansen, L.E.; Mølbak, K.; Møller, C.H.; Skov, R.L.; Krause, T.G.; Rasmussen, M.; Sieber, R.N.; et al. Household Transmission of SARS-CoV-2 Omicron Variant of Concern Subvariants BA.1 and BA.2 in Denmark. Nat. Commun. 2022, 13, 5760. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron Lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Ntoumi, F.; Kremsner, P.G.; Lee, S.S.; Meyer, C.G. Emergence and Geographic Dominance of Omicron Subvariants XBB/XBB.1.5 and BF.7—the Public Health Challenges. Int. J. Infect. Dis. 2023, 128, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Mizuma, K.; Nasser, H.; Deguchi, S.; Padilla-Blanco, M.; Oda, Y.; Uriu, K.; Tolentino, J.E.M.; Tsujino, S.; Suzuki, R.; et al. Virological Characteristics of the SARS-CoV-2 BA.2.86 Variant. Cell Host Microbe 2024, 32, 170–180.e12. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.C.; Castro, J.; Lambrou, A.S.; Rose, E.B.; Cook, P.W.; Batra, D.; Cubenas, C.; Hughes, L.J.; MacCannell, D.R.; Mandal, P.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron XBB and JN.1 Lineages—United States, May 2023–September 2024. MMWR Morb. Mortal. Wkly. Rep. 2024, 73, 938–945. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater Sequencing Reveals Early Cryptic SARS-CoV-2 Variant Transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Bar-Or, I.; Weil, M.; Indenbaum, V.; Bucris, E.; Bar-Ilan, D.; Elul, M.; Levi, N.; Aguvaev, I.; Cohen, Z.; Shirazi, R.; et al. Detection of SARS-CoV-2 Variants by Genomic Analysis of Wastewater Samples in Israel. Sci. Total Environ. 2021, 789, 148002. [Google Scholar] [CrossRef]
- Yousif, M.; Rachida, S.; Taukobong, S.; Ndlovu, N.; Iwu-Jaja, C.; Howard, W.; Moonsamy, S.; Mhlambi, N.; Gwala, S.; Levy, J.I.; et al. SARS-CoV-2 Genomic Surveillance in Wastewater as a Model for Monitoring Evolution of Endemic Viruses. Nat. Commun. 2023, 14, 6325. [Google Scholar] [CrossRef]
- Gupta, P.; Liao, S.; Ezekiel, M.; Novak, N.; Rossi, A.; LaCross, N.; Oakeson, K.; Rohrwasser, A. Wastewater Genomic Surveillance Captures Early Detection of Omicron in Utah. Microbiol. Spectr. 2023, 11, e0039123. [Google Scholar] [CrossRef]
- Oloye, F.F.; Xie, Y.; Asadi, M.; Cantin, J.; Challis, J.K.; Brinkmann, M.; McPhedran, K.N.; Kristian, K.; Keller, M.; Sadowski, M.; et al. Rapid Transition between SARS-CoV-2 Variants of Concern Delta and Omicron Detected by Monitoring Municipal Wastewater from Three Canadian Cities. Sci. Total Environ. 2022, 841, 156741. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Veneri, C.; Mancini, P.; Bonanno Ferraro, G.; Brandtner, D.; Lucentini, L.; Bonadonna, L.; Rossi, M.; Grigioni, M.; et al. The Rapid Spread of SARS-CoV-2 Omicron Variant in Italy Reflected Early through Wastewater Surveillance. Sci. Total Environ. 2022, 837, 155767. [Google Scholar] [CrossRef]
- La Rosa, G.; Brandtner, D.; Bonanno Ferraro, G.; Veneri, C.; Mancini, P.; Iaconelli, M.; Lucentini, L.; Del Giudice, C.; Orlandi, L.; Suffredini, E.; et al. Wastewater Surveillance of SARS-CoV-2 Variants in October–November 2022 in Italy: Detection of XBB.1, BA.2.75 and Rapid Spread of the BQ.1 Lineage. Sci. Total Environ. 2023, 873, 162339. [Google Scholar] [CrossRef]
- Veneri, C.; Brandtner, D.; Mancini, P.; Bonanno Ferraro, G.; Iaconelli, M.; Suffredini, E.; Petrillo, M.; Leoni, G.; Paracchini, V.; Gawlik, B.M.; et al. Tracking the Spread of the BA.2.86 Lineage in Italy Through Wastewater Analysis. Food Environ. Virol. 2024, 16, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Guilbaud, R.; Levert, M.; Fagour, N.; Le Hingrat, Q.; Descamps, D.; Tarantola, A.; Grellet, S.; Londinsky, N.; Moskovoy, J.M.; et al. BA.2.86 Variant Emergence and Spread Dynamics through Wastewater Monitoring in Paris, France. Sci. Total Environ. 2024, 917, 170355. [Google Scholar] [CrossRef] [PubMed]
- Erster, O.; Bar-Or, I.; Azar, R.; Assraf, H.; Kabat, A.; Mannasse, B.; Moshayoff, V.; Fleishon, S.; Preis, S.A.; Yishai, R.; et al. Incursion of SARS-CoV-2 BA.2.86.1 Variant into Israel: National-Scale Wastewater Surveillance Using a Novel Quantitative Real-Time PCR Assay. Sci. Total Environ. 2024, 933, 173164. [Google Scholar] [CrossRef]
- Wannigama, D.L.; Amarasiri, M.; Phattharapornjaroen, P.; Hurst, C.; Modchang, C.; Chadsuthi, S.; Anupong, S.; Miyanaga, K.; Cui, L.; Werawatte, W.K.C.P.; et al. Wastewater-Based Epidemiological Surveillance of SARS-CoV-2 New Variants BA.2.86 and Offspring JN.1 in South and Southeast Asia. J. Travel Med. 2024, 31, taae040. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Gongora, C.; Berg, C.; Rehn, M.; Varg, J.E.; Dillner, L.; Latorre-Margalef, N.; Székely, A.J.; Andersson, E.; Movert, E. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage through Integrated Genomic Surveillance of Wastewater and COVID-19 Cases in Sweden, Weeks 31 to 38 2023. Eurosurveillance 2023, 28, 2300595. [Google Scholar] [CrossRef]
- Du, C.; Peng, Y.; Lyu, Z.; Yue, Z.; Fu, Y.; Yao, X.; Tang, J.; Luo, G.; Gao, C.; Fang, S.; et al. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage Through Wastewater Surveillance Using a Mediator Probe PCR Assay-Shenzhen City, Guangdong Province, China, 2023. China CDC Wkly. 2024, 6, 332–338. [Google Scholar] [CrossRef]
- SARS-CoV-2 Variants of Concern as of 25 April 2025. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 28 April 2025).
- La Rosa, G.; Bonadonna, L.; Suffredini, E. Protocollo Della Sorveglianza Di SARS-CoV-2 in Reflui Urbani (SARI)-Rev. 3. 2021. Available online: https://zenodo.org/record/5758725 (accessed on 2 January 2025).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An Amplicon-Based Sequencing Framework for Accurately Measuring Intrahost Virus Diversity Using PrimalSeq and IVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Freyja. UShER Global Phylogenetic Tree. Available online: https://github.com/andersen-lab/Freyja (accessed on 17 July 2024).
- Pango Lineages. Available online: https://cov-lineages.org (accessed on 22 July 2024).
- Rueca, M.; Giombini, E.; Messina, F.; Bartolini, B.; Di Caro, A.; Capobianchi, M.R.; Gruber, C.E.M. The Easy-to-Use SARS-CoV-2 Assembler for Genome Sequencing: Development Study. JMIR Bioinform. Biotechnol. 2022, 3, e31536. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.C.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.; Neher, R. Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Bicchieraro, G.; Ciurnelli, R.; Graziani, A.; Wong, A.Y.W.; Camilloni, B.; Mencacci, A.; Spaccapelo, R. SARS-CoV-2 Molecular Evolution: A Focus on Omicron Variants in Umbria, Italy. Microorganisms 2024, 12, 1330. [Google Scholar] [CrossRef]
- Hayman, D.T.S.; Adisasmito, W.B.; Almuhairi, S.; Behravesh, C.B.; Bilivogui, P.; Bukachi, S.A.; Casas, N.; Becerra, N.C.; Charron, D.F.; Chaudhary, A.; et al. Developing One Health Surveillance Systems. One Health 2023, 17, 100617. [Google Scholar] [CrossRef]
- Hill, R.; Stentiford, G.G.; Walker, D.I.; Baker-Austin, C.; Ward, G.; Maskrey, B.H.; van Aerle, R.; Verner-Jeffreys, D.; Peeler, E.; Bass, D. Realising a Global One Health Disease Surveillance Approach: Insights from Wastewater and Beyond. Nat. Commun. 2024, 15, 5324. [Google Scholar] [CrossRef]
- Tandukar, S.; Thakali, O.; Baral, R.; Tiwari, A.; Haramoto, E.; Tuladhar, R.; Joshi, D.R.; Sherchan, S.P. Application of Wastewater-Based Epidemiology for Monitoring COVID-19 in Hospital and Housing Wastewaters. Sci. Total Environ. 2024, 931, 171877. [Google Scholar] [CrossRef] [PubMed]
- Ofori, B.; Agoha, R.K.; Bokoe, E.K.; Armah, E.N.A.; Misita Morang’a, C.; Sarpong, K.A.N. Leveraging Wastewater-Based Epidemiology to Monitor the Spread of Neglected Tropical Diseases in African Communities. Infect. Dis. 2024, 56, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, K.A.; Wade, M.J.; Barnes, K.G.; Street, R.A.; Paterson, S. Wastewater-Based Epidemiology as a Public Health Resource in Low- and Middle-Income Settings. Environ. Pollut. 2024, 351, 124045. [Google Scholar] [CrossRef]
- Rasmussen, M.; Moller, F.T.; Gunalan, V.; Baig, S.; Bennedbak, M.; Christiansen, L.E.; Cohen, A.S.; Ellegaard, K.; Fomsgaard, A.; Franck, K.T.; et al. First Cases of SARS-CoV-2 BA.2.86 in Denmark, 2023. Eurosurveillance 2023, 28, 2300460. [Google Scholar] [CrossRef]
- Matra, S.; Ghode, H.; Rajput, V.; Pramanik, R.; Malik, V.; Rathore, D.; Kumar, S.; Kadam, P.; Tupekar, M.; Kamble, S.; et al. Wastewater Surveillance of Open Drains for Mapping the Trajectory and Succession of SARS-CoV-2 Lineages in 23 Cities of Maharashtra State (India) during June 2022 to May 2023. Heliyon 2025, 11, e42534. [Google Scholar] [CrossRef] [PubMed]
- Bartel, A.; Grau, J.H.; Bitzegeio, J.; Werber, D.; Linzner, N.; Schumacher, V.; Garske, S.; Liere, K.; Hackenbeck, T.; Rupp, S.I.; et al. Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses 2024, 16, 102. [Google Scholar] [CrossRef]
- Wannigama, D.L.; Amarasiri, M.; Phattharapornjaroen, P.; Hurst, C.; Modchang, C.; Chadsuthi, S.; Anupong, S.; Miyanaga, K.; Cui, L.; Fernandez, S.; et al. Tracing the New SARS-CoV-2 Variant BA.2.86 in the Community through Wastewater Surveillance in Bangkok, Thailand. Lancet Infect. Dis. 2023, 23, e464–e466. [Google Scholar] [CrossRef]
- Li, L.; Haak, L.; Carine, M.; Pagilla, K.R. Temporal Assessment of SARS-CoV-2 Detection in Wastewater and Its Epidemiological Implications in COVID-19 Case Dynamics. Heliyon 2024, 10, e29462. [Google Scholar] [CrossRef]
| Clinical Samples | Wastewater Samples | ||||||
|---|---|---|---|---|---|---|---|
| Lineage | Number (%) | First Detection | Last Detection | Number (%) | First Detection | Last Detection | |
| XBB* | 150 (63.6) | 02 August 23 | 11 January 24 | 17 (41.5) | 09 August 23 | 06 February 24 | |
| XBB.1.16 | 13 (5.5) | 02 August 23 | 15 December 23 | 5 (12.2) | 09 August 23 | 30 October 23 | |
| XBB.1.5 | 22 (9.3) | 18 August 23 | 17 December 23 | 10 (24.4) | 09 August 23 | 06 February 24 | |
| XBB.2.3 | 5 (2.1) | 23 August 23 | 07 October 23 | 6 (14.6) | 09 August 23 | 08 November 23 | |
| XBB.1.9.2 | 3 (1.3) | 11 September 23 | 16 November 23 | 5 (12.2) | 09 August 23 | 14 December 23 | |
| XBB.1.9.1 | 9 (3.8) | 15 September 23 | 04 December 23 | 14 (34.1) | 09 August 23 | 06 February 24 | |
| EG.5 | 97 (41.1) | 07 August 23 | 11 January 24 | 16 (39.0) | 09 August 23 | 06 February 24 | |
| Other XBB* | 1 (0.4) | 26 October 23 | 26 October 23 | 12 (29.3) | 09 August 23 | 11 January 24 | |
| BA.2.86* | 78 (33.1) | 16 October 23 | 27 June 24 | 36 (87.8) | 18 October 23 | 01 July 24 | |
| JN.1 | 71 (30.1) | 16 October 23 | 27 June 24 | 33 (80.5) | 18 October 23 | 01 July 24 | |
| JN.2 | 0 (0.0) | n.d. | n.d. | 1 (2.4) | 24 January 24 | 24 January 24 | |
| JN.3 | 0 (0.0) | n.d. | n.d. | 4 (9.8) | 29 November 23 | 27 March 24 | |
| KP.1 | 0 (0.0) | n.d. | n.d. | 3 (7.3) | 20 March 24 | 29 May 24 | |
| KP.2 | 1 (0.4) | 31 May 24 | 31 May 24 | 7 (17.1) | 24 January 24 | 19 June 24 | |
| KP.3 | 3 (1.3) | 27 April 24 | 14 May 24 | 10 (24.4) | 24 January 24 | 01 July 24 | |
| KP.4 | 0 (0.0) | n.d. | n.d. | 4 (9.8) | 24 January 24 | 12 June 24 | |
| KP.5 | 0 (0.0) | n.d. | n.d. | 2 (4.9) | 24 January 24 | 04 June 24 | |
| LB.1 | 0 (0.0) | n.d. | n.d. | 7 (17.1) | 24 January 24 | 08 May 24 | |
| Other BA.2.86* | 3 (1.3) | 09 December 23 | 17 January 24 | 6 (14.6) | 29 November 23 | 19 June 24 | |
| Other | 8 (3.4) | 10 September 23 | 21 May 24 | 21 (51.2) | 09 September 23 | 27 March 24 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nappo, A.; Petricciuolo, M.; Berno, G.; Carnevali, A.; Gruber, C.E.M.; Bicchieraro, G.; Spaccapelo, R.; Rueca, M.; Carletti, F.; Spezia, P.G.; et al. One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023–July 2024). Life 2025, 15, 850. https://doi.org/10.3390/life15060850
Nappo A, Petricciuolo M, Berno G, Carnevali A, Gruber CEM, Bicchieraro G, Spaccapelo R, Rueca M, Carletti F, Spezia PG, et al. One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023–July 2024). Life. 2025; 15(6):850. https://doi.org/10.3390/life15060850
Chicago/Turabian StyleNappo, Alessandra, Maya Petricciuolo, Giulia Berno, Agnese Carnevali, Cesare Ernesto Maria Gruber, Giulia Bicchieraro, Roberta Spaccapelo, Martina Rueca, Fabrizio Carletti, Pietro Giorgio Spezia, and et al. 2025. "One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023–July 2024)" Life 15, no. 6: 850. https://doi.org/10.3390/life15060850
APA StyleNappo, A., Petricciuolo, M., Berno, G., Carnevali, A., Gruber, C. E. M., Bicchieraro, G., Spaccapelo, R., Rueca, M., Carletti, F., Spezia, P. G., Veneri, C., La Rosa, G., Suffredini, E., Focosi, D., Chillemi, G., Federici, E., & Maggi, F. (2025). One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023–July 2024). Life, 15(6), 850. https://doi.org/10.3390/life15060850










