Abstract
The development of therapies that target cancer stem cells (CSCs) and bulk tumors is both crucial and urgent. Several signaling pathways, like Notch and Hedgehog (Hh), have been strongly associated with CSC stemness maintenance and metastasis. However, the extensive crosstalk present between these two signaling networks complicates the development of long-term therapies that also minimize adverse effects on healthy tissues and are not overcome by therapy resistance from CSCs. The present work aims to overview the roles of Notch and Hh in cancer outburst and the intersection of the two pathways with one another, as well as with other networks, such as Wnt/β-catenin, TGF, and JAK/STAT3, and to explore the shaping of the tumor microenvironment (TME) with specific influence on CSC development and maintenance.
1. Introduction
The Notch and Hedgehog (Hh) pathways are two of the most relevant regulators of cell differentiation, tissue patterning, and maintenance [1,2]. Highly conserved, their structural roles have long been established as controlling the body architecture during embryogenesis and tissue homeostasis throughout adulthood. In healthy conditions, Notch and Hh affect a diversity of cellular processes, such as proliferation, differentiation, and apoptosis, and are often controlling each other [3]. Notch signaling is crucial in the maturation of several tissues, like bone, muscle, heart, vasculature, liver, stomach, and the hematopoietic system. Specifically, Notch is crucial in mesenchymal progenitor cell (MPC) growth and formation by regulating the production of bone cells through specific receptors and ligands (NOTCH1, NOTCH2, Dll1, and JAG1) [4]. Moreover, it also has a spatial and temporary input into bone fraction healing, with MPC depletion and fracture nonunion in the absence of Notch signaling, as observed in mouse models [5]. During the cardiomyogenesis process, Notch signaling has an impactful contribution in promoting the maturation of the atrioventricular architecture with the formation of the heart valve and the development of the epithelial–mesenchymal transition (EMT) by downregulating VEGFR2, the main negative regulator of EMT in the atrioventricular chambers [6,7]. In addition, it was noticed that embryos lacking Notch1, Rbpj, Hey1/Heyl, or Hey2 proteins could not develop EMT, which leads to an invasion of endocardium cells in the cardiac jelly [8]. Subsequently, Notch signaling is also pivotal in vascular endothelial cell development. Several studies have shown that deficiencies in Notch-specific molecules, like Delta-like 4 (DLL4), NOTCH2, NOTCH3, and NOTCH4, impact normal development severely, with vascular dysplasia, aortic defects, and abnormal development of multiple organs [9,10]. Notch signaling was also observed to promote biliary duct cell differentiation, as well as the specific timing of the process, with Sox9 as a key downstream target [11,12]. NOTCH2 is also indispensable in liver regeneration processes, like differentiation of the progenitor cells of the liver tissue [13,14]. In the gastrointestinal tract, expression of Hes1 through Notch control guides the intestinal stem cells into epithelial cell differentiation [15]. Additionally, NOTCH receptors are involved in maintaining the antral cell stem population of the stomach, while the DLL 1 and 4 and JAG1 ligands regulate the differentiation of pancreatic progenitors of the α cells [16]. Lastly, the development of lymphocytes, macrophages, granulocytes, and dendritic cells and the maintenance of the stemness of hematopoietic stem cells are all Notch-dependent processes [17,18,19,20,21].
Hh signaling, on the other hand, drives pivotal downstream regulations of transcription factors that control the morphogenesis and spatial patterning of the organism, like the differentiation and mineralization of bone cells [22], activation of myosatocytes through Myf5 and MyoD expression for skeletal muscle repair [23], control of hair follicle stem cells and epidermal homeostasis [24], intestinal stem cell differentiation and proliferation [25], and liver regeneration, via EMT interaction, mediated by Hh [26].
Although distinct in their roles, the two pathways usually exhibit a bidirectional regulation, with frequent overlapping and crosstalk. For example, in the developmental stages of the spinal cord, the expression of the homeodomain expression factor (HD) and proneural basic helix-loop-helix proteins (bHLH) is critically controlled by the duration and level of the Hh signaling pathway, with indirect downstream effects on the Notch control and Notch-associated jagged gene expression and Fringe proteins [27,28]. Hh can also directly determine neuronal differentiation by activating the promoters of Hes1, a classic target gene of Notch, via Gli protein control [29]. On the other hand, Notch signaling helps maintain Hh responsiveness by maintaining the stability of essential proteins, like Gli transcription factors, and by keeping the progenitor cells receptive to Hh signaling [30,31], thus guiding the cell fate to specification in the central nervous system development.
Both pathways are closely regulated for the correct development of the embryo, and their improper regulation during this stage may lead to a predisposition for cancer onset by disrupting homeostasis and stem cell control [32]. Recent studies have shifted the focus from their roles in development to their contributions to cancer biology. Dysregulation of Notch and Hh signaling has been implicated in tumorigenesis, where abnormal activation or inhibition drives the progression of several tumors [3]. First, the defective signaling of Notch and Hh during embryogenesis can lead to an unbalanced population of stem cells and differentiated cells in several tissues, with later consequences into CSC development and maintenance of population through self-renewal, which then contributes to processes like neoplasia, metastasis, drug resistance, and high recurrence rate [3]. The survival of Notch–Hh-positive CSCs and their ability to evade the immune system has also been associated with the hypoxic tumor microenvironment (TME), which was proved to maintain the population of undifferentiated stem cells and also to reprogram the non-stem cancer cells into CSCs, with activating Notch-target genes [33]. Second, Notch and Hh defects can lead to tissue-specific cancer. For instance, mutations of Ptch and Smo, Hh components, can lead to the Gorlin syndrome (GS), which manifests as basal cell carcinoma [34]. Additionally, Notch-associated CSCs have been identified in the early stages of a large number of tumors, like breast [35], colorectal [36], pancreatic [37], brain [38], lung [39], and skin cancers [40], as well as hematological malignancies [41], exercising roles like tumor initiation, EMT, progression, and hypoxia maintenance. Lastly, Notch-Hh associated developmental defects can have a long-term impact on the microenvironment, with morphological and physiological shaping that favors chronic inflammation, CSCs maintenance, and tissue remodeling, setting the stage for prospective cancer development [42,43].
Notch and Hh signaling do not operate in isolation but have complex interplays with other signaling pathways, which exhibit significant roles in CSCs and cancer progression. Most particularly, the crosstalk between Notch and Wnt/β-catenin has an important influence in determining the balance between the stem cells and the specialized ones. The activation of Notch further activates Wnt to promote the proliferation of CSCs [44]. Additionally, Hh interacts with the PI3K/Akt pathway in pathological conditions. The defective activation of Hh enhances the Akt signaling, thus facilitating the survival and growth of CSCs [45]. Furthermore, both Notch and Hh exhibit significant crosstalk with the transforming growth factor-beta (TGF-β) in promoting EMT, together with Wnt/β-catenin, which is a crucial process involved in cancer metastasis and CSC maintenance [46].
The present article aims to explore the roles of Notch and Hh signaling in normal tissue development, highlight the aberrations encountered in the pathways that contribute to tumorigenesis, and focus on cancer stem cell (CSC) origin and maintenance within the TME. Additionally, it provides current insights into targeted cancer therapies and attempts to overcome treatment resistance.
2. Structure and Function of Notch Signaling
The activation of Notch signaling is a pathway that drives carcinogenesis and is also highly conserved, with crucial roles in determining cell fates, proliferation, differentiation, and apoptosis [47].
A particular feature of the Notch signaling cascade is the requirement for direct contact between adjacent cells, as both the ligands and receptors are found on the plasma membranes of the interacting cells (Figure 1). The receiving cells usually experience changes in cell fate due to the Notch signals. Such interactions are vital to processes such as hematopoiesis, immune regulation, neural stem cell survival, maturation of the colorectal epithelial, and breast development [48,49,50].
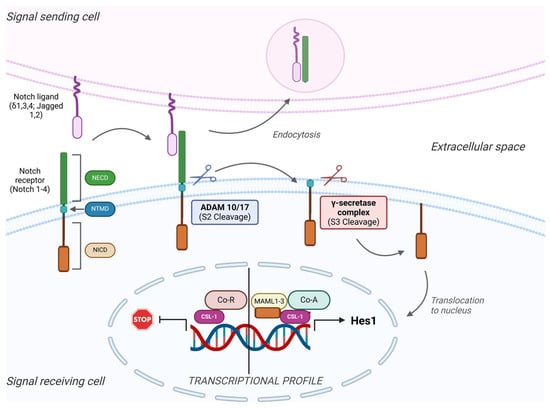
Figure 1.
Overview of the general mechanisms involved in Notch signaling. The Notch signaling pathway involves adjacent cells, and their communication involves four receptors (Notch 1–4) and five types of ligands (Delta 1, 3, and 4 and Jagged 1 and 2). When the ligand binds to the extracellular domain of the receptor (NECD), two subsequent cleavages occur. First, the metalloprotease complex ADAM 10/17 cuts the NECD from the rest of the receptor (S2 cleavage), followed by the S3 cleavage mediated by the γ-secretase complex. This proteolysis leads to the detachment of the intracellular domain (NICD) from the transmembrane domain (TMD) and its translocation into the nucleus. Once inside the nucleus (right), it attaches to the CSL, a DNA binding protein, together with other co-activator (Co-A) factors like Mastermind-like proteins (MAML 1–3), and activates the transcription and translation of target genes. On the other hand, when the NICD is not released from the membrane, co-repressors (Co-R) attach to the CSL protein and inhibit the activation of the target genes.
Structurally, four Notch receptors have been identified, each an extracellular heterodimer and a transmembrane domain held together by hydrogen bonds [51]. The extracellular segment contains a sequence of multiple epidermal growth factor-like domains, which mediate the interactions with Notch receptor–ligands [52]. The intracellular fragment of the receptor serves as a transcription factor. Notch receptors have five known ligands: Jagged-, Jagged-2, and Delta-like ligands (DLL1, DLL3, and DLL4), localized on the receptor-expressing neighboring cells [53].
Upon receptor–ligand binding, these ligands trigger an initial cleavage event at a particular site on the Notch receptor. This site is subsequently cleaved by the ADAM MMPs [54], releasing the ligand-bound Notch fragment into the extracellular space, where it undergoes endocytosis and degradation by the adjacent ligand-expressing cell. Another cleavage by the γ-secretase complex releases the Notch intracellular domain (NICD) [8], which then undergoes nuclear translocation [55,56]. In the absence of NICD, the CBF-1-Suppressor of Hairless/Lag1 (CSL-1) interacts with co-repressors, which block their transcriptional activity [57,58,59]. NICD binds to CSL-1 as a transcriptional co-activator, displacing co-repressors and enabling the binding of Mastermind-like proteins (MAML) and others, forming the Notch transcription complex. This complex activates the transcription of effector proteins, like the Hairy/Enhancer of Split (HES) family, the Hey family, and factors like c-Myc and cyclin D1 [59,60], which promote cell proliferation and other cell-specific functions.
Since developmental pathways such as Notch are critical for normal stem cell maintenance, they often dysregulate in cancer. Altered Notch signaling can hinder the normal differentiation of stem cells, contributing to the development of malignancies like T-cell acute lymphoblastic leukemia (T-ALL), glioblastoma multiforme, and cancers of the colon, pancreas, breast, ovary, and prostate [61,62,63,64,65,66,67,68]. Genetic mutations also disrupt normal Notch signaling. Activating mutations in Notch receptors are found in about 60% of T-ALLs, where the receptor becomes active independent of ligand binding or its degradation is delayed due to mutations in the C-terminal PEST domain [69], which usually marks NICD for degradation by the proteasome. In addition, around 10% of chronic lymphocytic and myeloid leukemias, as well as some breast adenocarcinomas, exhibit Notch mutations, which are associated with poorer survival rates [70]. In contrast, loss-of-function mutations in Notch occur in cancers like skin cancer, medullary thyroid cancer, cervical cancer, small cell lung cancer, and hepatocellular cancer (HCC). Notch may act as a tumor suppressor in these cases [71,72].
Other mechanisms that disrupt Notch signaling can contribute to cancer development. For example, Notch hyperactivity may arise from mutations in Numb, a Notch repressor, which is lost in about half the cases of breast cancer [73]. Additionally, the RNA-binding protein Musashi-1, which plays a role in translational inhibition, can cause Numb inactivation and Notch-1 overexpression. Reducing Musashi-1 levels decreases Notch-1 signaling, leading to increased apoptosis and suppressed tumor growth, possibly due to the role of Musashi-1 in maintaining stem-like properties in cancer [74,75,76].
3. CSC and Notch Inhibition
The significance of Notch signaling extends beyond its developmental role in normal stem cells to include its function in CSCs [77]. Elevated Notch activity and increased expression of Notch family genes in cancer stem suggest that Notch may drive rapid tumor growth and aggressive behavior [78]. This has led to the hypothesis that inhibiting the Notch pathway could be a promising therapeutic strategy in cancer treatment. Supporting this idea, Fan et al. demonstrated that blocking Notch signaling with γ-secretase inhibitors reduced the development of glioblastoma neurospheres in in vitro studies, decreased the expression of CSCs markers (CD133, NESTIN, BMI1, and OLIG2), and prevented tumor formation in mice injected with glioblastoma cells treated with γ-secretase inhibitors. This treatment also significantly prolonged the survival of mice with intracranial xenografts [79]. Another study using murine orthotopic xenografts derived from temozolomide-sensitive and resistant glioblastoma neurosphere lines found similar results: Notch inhibition led to tumor growth suppression, decreased stem cell marker expression, and induced glial differentiation. These effects persisted even after therapy concluded [80], highlighting Notch’s potential role in maintaining the CSC population.
The involvement of Notch in the EMT process, which is crucial for both embryonic development and cancer progression, may partially explain its role in stem cell renewal and maintenance [77]. EMT is characterized by a visible change in cell morphology from an epithelial to a fibroblast-like appearance, with associated loss of cell adhesion, cell polarity, and increased migration and invasiveness [81]. Significant CSC-like subpopulations are also present in the matrix of such modified cells [82,83,84,85]. Notch signaling promotes EMT by downregulating E-cadherin expression through the activation of Slug, thus initiating the β-catenin pathway, as observed in breast cancer cells [86]. Furthermore, TGF-β-induced EMT can be stopped by inhibiting Notch-1 signaling [87].
γ-secretase inhibitors act effectively against cells with high Notch activity, such as CSCs, but combining them with chemotherapy in order to target bulk tumor mass, which consists of non-stem CSCs, would increase the effectiveness of the anti-cancer strategy. This approach was tested in pancreatic cancer xenografts using a selective γ-secretase inhibitor, combined with gemcitabine, a first-line treatment for pancreatic ductal adenocarcinoma. While gemcitabine alone did not induce tumor regression or deplete pancreatic CSCs, the combination therapy led to significant tumor regression in three out of four xenografts, reduced CSCs, and had a more sustained effect without further treatment compared to gemcitabine alone. This mechanism promoted apoptosis, reduced cell proliferation, inhibited angiogenesis, and controlled metastatic advancement [88]. These findings are particularly relevant, as gemcitabine-resistant pancreatic cancer cells often exhibit increased Notch activity, which promotes EMT, stem cell marker expression, and chemotherapy resistance [89,90].
Targeting both the stem cell population and the bulk tumor cells is critical in advanced stages of cancer. Initial debulking with conventional chemotherapy maintains organ function, while targeting CSCs with selective inhibitors can help prevent relapse [91]. In breast cancer, a combination of the γ-secretase inhibitor (GSI) MK-0752 with docetaxel (a platinum-based chemotherapy) reduced tumor size more effectively than either agent alone and delayed tumor evolution. Notch inhibition by MK-0752 reduced the breast CSC population and enhanced the efficacy of docetaxel. A phase I clinical trial of this combination in 30 patients with advanced breast cancer showed a reduction in CD44(+)/CD24(-) and ALDH(+) stem cells and their mammosphere-forming capacity [92].
However, the potential adverse effects of GSIs should be considered for long-term use. Notch signaling regulates the balance of intestinal cell types, and GSI-mediated inhibition can cause severe goblet cell metaplasia, leading to diarrhea [93]. Additionally, the γ-secretase complex is involved in amyloid protein production in Alzheimer’s disease, as well as an increased incidence of skin cancers, possibly due to Notch’s tumor-suppressing role in epithelial tissues [69].
Beyond GSIs, other classes of Notch inhibitors are being developed, including monoclonal antibodies targeting Notch ligands and receptors, receptor-like decoys that bind and inactivate ligands, targeted peptides, natural compounds, and agents targeting proteins downstream of the Notch receptor [77]. Some phytochemicals, such as curcumin, sulforaphane, resveratrol, genistein, epigallocatechin gallate, and psoralidin, have been found to inhibit the NICD, preventing its nuclear translocation. These compounds can inhibit cancer growth, induce apoptosis, and reduce migration and invasion, affecting both cancer cells and CSCs [94,95].
As Notch-targeted therapies continue to diversify, advances in understanding the Notch signaling pathway may allow for more personalized cancer treatments. A genomic analysis of the Notch pathway in 16 glioma tumor-initiating cell populations identified genes such as NOTCH1, NOTCH3, MAML1, JAG2, HES1, and DLL-3, which were associated with active Notch signaling and enriched in tumors responsive to treatment. This suggests that GSIs may be particularly effective in glioblastomas with high Notch pathway activation [96]. Rizzo et al. have proposed that for personalized treatment, it is essential to determine not only the expression of specific Notch components and ligands but also whether targeting the entire Notch system or specific components is more effective for each tumor type. The study also emphasizes the need to assess the interaction between Notch and other signaling pathways to design rational combination therapies [59].
4. Notch Signaling and the TME
Although details about the various microenvironments supporting cancer stem-like cells in different cancer types are still limited, the occurrence and spread of cancer are heavily connected to the TME. Tumor-associated fibroblasts, endothelial cells, adipocytes, and certain immune cells are key components of this background, playing a significant role in regulating both normal and malignant stem cells through paracrine, juxtacrine, or other short-range signaling methods [97].
4.1. Endothelial Cells
Endothelial cells are crucial elements of the TME, especially around CSCs, creating what is known as the “vascular niche”. Studies have shown that glioblastoma stem cells are protected by this environment, which hosts endothelial cells [98]. Notch expression strongly influences both glioblastoma stem cells and tumor angiogenesis. In one study, Hovinga et al. proposed that Notch inhibition could significantly affect the behavior of endothelial cells within the tumoral niche, as well as the glioblastoma cells. On a three-dimensional glioblastoma explant model that mimics the tumor’s cytoarchitecture and stroma, Notch inhibition through GSIs led to a loss in the number of endothelial cells and also to the downregulation of Notch target genes (HES1 and HES5) and also eliminated the expression of the endothelial marker CD105. The important implication observed on tumor angiogenesis and the tumor niche, which consequently led to decreased neurosphere formation and a lower number of CD133(+) glioblastoma cells, highlighted the major function of endothelial cells in Notch signaling within glioblastoma. Additionally, by treating the tumors with a Notch inhibitor for five days prior to irradiation, the proliferation and self-renewal capacity of the tumor explants showed a significant reduction, which proves that this strategy works more efficiently than irradiation alone [99].
Lu et al. also managed to bring additional support for the vital role of the TME by studying its behavior in colorectal cancer. They cultured colorectal CSCs in a conditioned medium derived from freshly isolated endothelial cells and observed that the latter cells managed to enhance the stem-like phenotype of the cancer cells in a significant manner: Aldefluor-positive colorectal cancer cell population by 16-fold, the CD133 positive cells by 7-fold, and the sphere-forming capacity by over 6-fold. This mechanism was due to the paracrine capacity of endothelial cells to induce dedifferentiation on non-stem colorectal cancer cells by secreting Jagged-1. On top of that, the soluble Jagged-1 was depleted in the endothelial cell-conditioned medium by a type of antibody that recognized the N-terminus of Jagged-1, which reduced colorectal cancer cells’ sphere-forming ability. These findings bring additional support to the hypothesis that endothelial cells in the tumor niche promote the CSC phenotype by the enhancement of Notch signaling, independent of cell-to-cell contact [100].
4.2. Angiogenesis
Notch signaling is also involved in regulating cancer angiogenesis, with DDL4 as a key factor in suppressing tumor angiogenesis and its known high expression in tumor blood vessels. Angiogenesis is induced by two types of endothelial cells: first, the “Tip” VEGF-stimulated endothelial cells, which lead the migration during angiogenesis when an angiogenic stimulus occurs. VEGF is also involved in DLL-4 expression in Tip cells, which generates the activation of Notch signaling in the neighboring “Stalk” cells, preventing additional Tip cells from forming and ensuring a proper vascular structure. Inhibition of DLL-4 signaling induces uncontrollable vessel sprouting and creates a dense but nonfunctional angiogenic network, which reduces tumor growth and increases tumor hypoxia [101].
Endothelial cells may also originate from CSCs through trans-differentiation or vascular mimicry. First observed in melanoma cases, this phenomenon has been described in other types of cancer [102,103]. For example, glioma CSCs can undergo vascular mimicry under hypoxic conditions [104], and it was also observed that the vast majority of tumor endothelial cells can originate from these CSCs. Although studies proposed that Notch may influence the differentiation process, much still remains unknown [102].
4.3. Adipose-Derived Stem Cells (ADSCs)
Similarly to endothelial cells from the TME, ADSCs also have a key role in facilitating tumor growth and proliferation. In one study, it was observed that by co-culturing ADSCs with H358 lung cancer cells, EMT-like alterations occur in the latter cells. Notably, these EMT-like chances were blocked by GSIs, suggesting a link to Notch signaling [105].
4.4. Hypoxia
The TME is often hypoxic, especially in the tumor central areas that are affected by poor vascularization and necrosis and also in the invasive front, due to rapid cancerous cells [106]. Hypoxia has been associated with more aggressive tumors and increased levels of invasiveness and metastasis, likely because it develops CSCs [107] and diminishes their differentiation [108]. Hypoxia-inducible factor (HIF) expression is upregulated by hypoxia and also mediates many effects of low oxygen levels, like promoting angiogenesis, tumor growth and spreading, and EMT [109,110]. HIF1α is upregulated by Notch signaling [111], and the molecular HIF1α and Notch1 increase the stability and activity of Notch proteins [112].
In hypoxic conditions, Notch promotes the CSC phenotype and blocks their differentiation, which creates a more invasive and pro-survival characteristic [113]. However, these effects may, in turn, be blocked by Notch inhibitors [108,114] and may also promote E-cadherin expression and AKT phosphorylation inhibition [114]. At the hypoxic invasive front, Jagged-2 and Notch signaling activation are both elevated. Jagged-2 activation under hypoxia conditions induces EMT and maintains cell survival. In the bone marrow stroma, the activation of hypoxia-induced Jagged2 activation enhances CSC growth, which strengthens the importance of the Jagged-2 ligand in the progression of the tumor and metastasis. This is particularly explanatory to why bone tissue is such a frequent metastasis site, since it is known to be hypoxic [77].
5. Crosstalk with Other Signaling Pathways
The Notch pathway interacts with several other cancer-related signaling pathways, which is crucial for developing personalized cancer treatments. One such pathway is the PI3K/AKT/mTOR axis, known for regulating cell growth, proliferation, and survival. In many cancers, this pathway is hyperactive. Notch signaling enhances mTOR pathway activity by inducing the transcription of HES-1, a repressor of PTEN, a tumor suppressor that normally inhibits the PI3K/AKT/mTOR pathway. Since PTEN loss is common in cancer, Notch activation can further increase mTOR pathway activity. Blocking Notch signaling with GSIs can therefore reduce activity in both the Notch and mTOR pathways. However, as tumors progress, some cells may become resistant to GSIs due to mutational loss of PTEN, which shifts their “oncogene addiction” to the mTOR pathway, making them sensitive to AKT inhibitors instead [115]. Notch signaling can also upregulate the Insulin growth factor-1 (IGF-1) receptor [116], enhancing mTOR signaling independently of PTEN status. Also, inhibiting mTOR signaling may activate Notch signaling [117], though other studies have shown that AKT inhibition can suppress Notch activity [118]. This complex relationship provides a justification for using combinations of inhibitors targeting both Notch and mTOR pathways [115]. They share downstream effectors like Myc, which Notch promotes and which, in turn, activates the mTOR pathway [119], leading to increased survival of cancer cells by restricting p53-mediated apoptosis. Inhibiting Notch signaling can reduce cancer cell aggressiveness by affecting other pathways, such as mTOR and NF-κB.
The NF-κB pathway is another key regulator of cell survival, differentiation, and proliferation, and it controls the transcription of genes involved in immunity, inflammation, and cancer [120]. Notch and NF-κB signaling can interact at several points in the cascade. First, there is reciprocal transcriptional regulation: Notch signaling can promote the transcription of components of the NF-κB complex [121] and upregulate IκBα, an inhibitor of the NF-κB pathway, at the same time [122]. NF-κB, in turn, can promote the expression of Notch target genes, including members of the HES family [123], and activate Notch in a paracrine manner [124] by increasing the expression of Notch ligands like Jagged1 [125]. Additionally, NICD, the active form of Notch1, can bind directly to the NF-κB complex. Initially thought to inhibit NF-κB activity [126], recent studies suggest that NICD binding may actually prevent NF-κB from being exported out of the nucleus, thereby enhancing NF-κB-mediated gene transcription [127].
The RAS/RAF/MAPK signaling system transmits signals from tyrosine-kinase receptors like EGFR to regulate cell cycle progression, survival, and apoptosis. Notch and RAS networks often crosstalk in cancers such as pancreatic, colorectal, breast, and gliomas [128,129,130,131]. RAS signaling enhances the Notch pathway by upregulating the levels of the Notch ligand DLL-1 and elevating the expression and activity of NICD. Ras also increases presenilin-1, a component of the γ-secretase complex [132]. This crosstalk is fundamental in CSC behavior. In glioblastoma, cooperation between the Notch and RAS pathways leads to the development of CSCs-like populations that express higher levels of nestin, a glioblastoma stem cell marker [133].
Other signaling routes related to cancer may also interact with Notch. In colorectal cancer, Wnt-mediated β-catenin signaling upregulates the Notch ligand Jagged-1 [134] and increases Notch-2 expression [135]. However, in glioblastoma stem-like CD133(+) cells, the combination of Wnt activation and hypoxia-induced HIF-α activity induces neuronal differentiation and suppresses Notch signaling [136], reducing the stem-like population. This suppression may be linked to Wnt-mediated induction of Numb, which inhibits NICD activity [137]. Crosstalk also occurs between Notch and the Hh signaling network, which regulates embryonic development, stem cell maintenance, and cancer progression. The Hh pathway can induce Notch activity by upregulating the Jagged-2 ligand in cancer [138]. In breast CSCs, Notch, Hh, and Wnt together regulate stem cell proliferation and differentiation [139]. There are also direct interactions between Notch and the JAK/STAT signaling system and an antagonistic relationship between Notch and Her2/Neu expression and estrogen receptor (ER) status, which may be clinically significant for therapy [79,140].
Given these interactions between Notch and other cancer-related pathways, it is of great importance to study the combination therapies targeting Notch alongside PI3K/AKT/mTOR inhibitors, NF-κB deterrence, Her2/Neu targets, platinum-based chemotherapies, EGFR inhibitors, and Hh pathway suppressors [50].
6. Role of Hedgehog Signaling Pathway in CSCs
The Hh pathway, like the Notch and Wnt signaling pathways, is a highly conserved system that regulates cell proliferation and differentiation, especially during embryonic development. Initially identified in Drosophila, homologous genes have since been found in the mammalian genome, where they have crucial roles in several developmental processes, including cell survival and stem cell maintenance [141], but also in cancer onset.
The Hh pathway is named after its three ligands: Sonic, Indian, and Desert, which induce gene transcription in targeted cells. The signal-receiving cells express Patched receptors (PTCH1 and PTCH2 in vertebrates), which are 12-transmembrane proteins that usually suppress Smoothened (Smo), a receptor resembling G-protein-coupled receptors. Normally, PTCH receptors inhibit Smo activity, reducing the conversion of GLi transcription factors to their active form, leaving them to repress Hh pathway-regulated genes. When Hh ligands bind to PTCH receptors, they relive this suppression, activating Smo and increasing levels of active Gli transcription factors, which then promote the expression of target genes (Figure 2). These genes encode proteins lice c-Myc, cyclin D1, and Patched receptors, some of which participate in a negative feedback loop with Hh signaling [142].
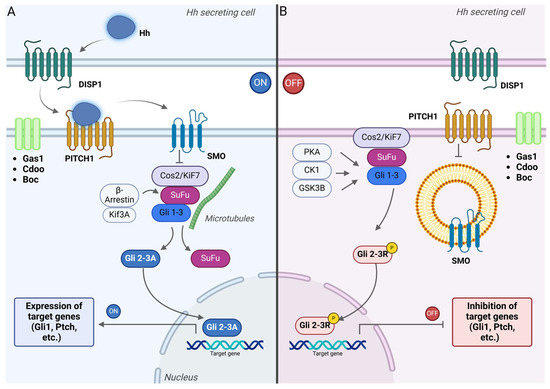
Figure 2.
Signal transduction in the Hedgehog (Hh) signaling pathway. (A) Hh ligand secretion depends on the transmembrane transporter Dispatched. When Hh ligands bind to PITCH receptors, they inhibit PITCH activity, which allows Smo proteins, which are similar to G-protein-coupled-receptors, to be phosphorylated and transported to the membrane of primary cilia in vertebrates. This prevents the phosphorylation of Gli proteins, promoting their conversion into Gli 2-3A activator form, and moves into the nucleus to activate the expression of Hh-related genes [142]. (B) When no Hh signal is secreted, PITCH inhibits the activation of Smo. The Gli 2-3 complex is retained in the cytosol by SuFu and undergoes partial phosphorylation under the influence of a protein kinase complex (PKA, CK1, and GSK3β). The Gli 2-3 repressor form is synthesized (Gli 2-3R) and translocated into the nucleus, where it binds to the DNA and inhibits the expression of the target genes.
The understanding of the molecular mechanism of Hh signaling led to the formulation of a few hypotheses. For instance, it was suggested that PTCH receptors may transport molecules into Hh-sensitive cells that can further regulate Smo activity [143]. But the precise localization of Hh proteins in specific cellular structures is critical for signaling. In vertebrates, the primary cilium is considered to be the exclusive cell site where Hh signal transduction takes place.
There are still unclear aspects about the molecular mechanisms involved in the regulation of the Hh pathway. After translation, Hh proteins undergo autocleavage and are modified with palmitoyl and cholesteryl groups at their N- and C-terminals [144]. The secretion of processed Hh proteins is regulated by various proteins, including the transmembrane transporter Dispatched, which moves Hh ligands across the secreting cell’s plasma membrane (Figure 1). Glypicans, MMPs, and enzymes involved in heparan sulfate biosynthesis [145] also influence the tissue distribution of Hh ligands [146]. The cholesterol moiety helps determine the form of Hh ligand release, which can vary between monomeric, multimeric, and encapsulated forms depending on auxiliary proteins [147]. Additionally, recent studies suggest a possible involvement of filopodia in Hh transport, though this remains unproven in vertebrates [148].
Several Hh-binding proteins and proteoglycans, like HIP, CDO, BOC, GAS1, and Glypican-3, are directly involved in the regulation of the Hh pathway at the PTCH level [149]. When the Hh ligands are absent, PTCH receptors are located in the primary cilia of vertebrate cells, while Smo proteins are displaced in the membrane of intracellular vesicles [150]. When Hh ligands bind to PTCH, the receptor moves from the cilia and is sequestered in vesicles for degradation, enabling Smo proteins to move to the primary cilium and interact with the Gli-processing system. However, Smo requires additional kinase-mediated phosphorylation to activate Hh signaling [151].
The GPCR-like nature of Smo proteins might also influence pathways such as Gαi, but the exact role in Hh signaling is still unclear. Activated Smo prevents the conversion of Gli proteins into repressors by kinases such as PKA, GSK3β, and CKIα, allowing them to be processed into transcriptional activators through post-translational modifications.
Gli-2 and Gli-3, which are zinc-finger transcription factors, undergo various modifications depending on the Hh signal presence, balancing the production of their activator and repressor forms [142]. In the absence of Hh ligands, CKIα, GSK3β, and PKA kinases phosphorylate Gli proteins. Consequently, βTrCP, an E3-ubiquitin ligase-recruiter, is attached [152], which further triggers the ubiquitination of Gli proteins and their partial degradation by the proteasome complex and the final GliR transcriptional repressors production. SuFu protein also binds to unprocessed Gli, preventing its movement into the nucleus and blocking gene transcription.
When Hh ligands bind to PTCH, Smo is phosphorylated by CKI and GPRK2 [153] and moves to the primary cilium membrane by lateral transport inside the cell membrane [154] or by the fusion between Smo-loaded vesicles with the cell membrane [155]. Smo’s accumulation releases Gli from SuFu [155], prevents Gli phosphorylation by kinases, and omits the formation of GliR, leading to the nuclear translocation of Gli proteins and initiation of target gene transcription.
Furthermore, non-canonical Hh signaling has been described in cancer, involving growth factors like EGF, TGFβ, or PDGFα, which can activate GLi-mediated transcription without Smo [156]. Additionally, a non-GLi-dependent mediated pathway was also associated with cancer development, but its specific involvement is yet to be understood [157].
7. Hedgehog Signaling Pathway and Its Role in Cancer Onset
The first evidence of Hh signaling involvement in cancer came from one study showing that the loss of PTCH receptor function leads to nevoid basal cell carcinoma syndrome (Gorlin syndrome) [34]. This autosomal dominant disorder is frequently associated with the outbreak of other neoplasms, like embryonal rhabdomyosarcoma, meningiomas, and odontogenic tumors. PTCH mutations are found in over 90% of sporadic basal cell carcinomas and around 20% of medulloblastomas. Additionally, a ligand-independent alternative of the canonical Hh pathway is formed as a result of loss-of-function mutations in SuFu, activating mutations in Smo, and increased GLi expression. In the ligand-dependent form, Hh ligands originate from tumor cells themselves, creating an autocrine signaling loop seen in certain gastrointestinal, pancreatic, lung, and breast cancers [158]. More commonly, Hh signaling occurs through paracrine interactions between tumor cells and the surrounding environment, activating stromal cells to release growth factors that stimulate mitogenic pathways in nearby cancer cells [159]. The most common examples where paracrine signaling is observed are pancreatic, colon, breast, ovarian, and prostate cancers [158,160]; chronic lymphocytic leukemia [161]; and multiple myeloma [162]. It was noticed that these tumor cells display no cilia protrusions, which might be the starting point of cancer development, since Hh signaling becomes impossible [163]. A reverse paracrine mechanism has also been identified in hematologic malignancies, where stromal cells secrete Hh ligands that enhance tumor cell survival by upregulating Bcl-2, inhibiting apoptosis [161].
Despite these findings, the role of ligand-dependent Hh signaling in cancer remains unclear due to the complex interplay of factors like tumor type, stroma, and crosstalk with other pathways. Hh signaling may serve different roles across cancers: as a tumor driver in basal cell carcinoma, a promoter in small cell lung cancer [164], and a modulator of tumor growth that influences CSCs and residual tumor populations post-treatment.
The Hh pathway also contributes to EMT, a key process in cancer metastasis, by promoting the transcription of proteins like SNAI1, which suppress E-cadherin and enhance cell migration [165]. Disrupting Hh signaling has been shown to inhibit EMT in pancreatic cancer cell lines treated with cyclopamine [166]. Hh signaling can also suppress Wnt signaling, indirectly facilitating EMT by preventing tumor cells from retaining their epithelial state through Wnt’s juxtacrine signals in the TME [167,168].
Apoptosis regulation is another aspect influenced by Hh signaling. Knockdown of Gli-3 via siRNA enhanced the effects of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) by upregulating the TRAIL receptor and death receptor 4 (DR4) and increasing sensitivity to TRAIL binding [169]. Additionally, cyclopamine-induced inhibition of Smo increased Fas receptor expression and blocked UVB-induced basal cell carcinoma development [170].
Cyclopamine is a terpene alkaloid extracted from wild corn ily (Veratrum californicum), with non-toxic effects on non-cancerous cells, as observed in vitro on nh-skp-FB0012 foreskin fibroblasts cells [171], or on human astrocytes [172]. Glioblastoma is another type of cancer with strong resistance to chemotherapy, like common temozolomide and radiotherapy, due to CSCs. Monotherapies and combined therapies of Cyclopamine and/or Temozolomide were administered in vitro on GBM95, GBM02, and GBM03 human glioblastoma cell lines. Alone, it managed to reduce the viability of the cancerous cells, and, combined with the chemotherapeutic, it helped in sensitizing the cells to the medication and inducing apoptosis [171]. Cyclopamine also induced the downregulation of Gli-1 expression in a dose-dependent manner in the EC9706 human esophagus cancer cell line, thus suppressing the Hh signaling pathway [173].
The Gli-1 pathway was also observed to be downregulated as an effect of another natural inhibitor, genistein, of MCF-7 breast cancer cells and also in xenografted naked mice with MCf-7 cells. The level of protein expression of both Gli-1 and Smo was significantly reduced by genistein treatment, as well as the number of breast CSCs [174]. Genistein (4′,5,7-trihydroxyisoflavone) is an isoflavone phytoestrogen present in several soybean foods [175], with known antitumor effects. The exploitation of natural sources that target the Hh signaling pathway has become a hot topic in finding treatments for several types of cancer, as monotherapies and also combined with standard medications, and have shown significant action in inhibiting the stemness of the tumors while having no toxic effect on healthy cells and tissues [176,177]. Despite the promising results reported so far, more studies are required on natural inhibitors, especially on understanding more clearly the mechanism of action in different types of cancer, as well as possible interactions with common therapeuticals, due to their known high pleiotropy and low bioavailability [176].
8. Hedgehog Signaling (Hh) and CSCs
Hh signaling has been found to significantly influence the CSC population in various types of cancer, including pancreatic [178], breast [179,180], gliomas [181], gastric [182], and colon cancer [183,184]. In several of these, Hh signaling contributes to aggressive tumor phenotypes that are highly resistant to treatment. For example, in gastric cancer, Song et al. identified a subset of cells expressing CD44, CD24, and CD133, which exhibited higher proliferation rates and resistance to chemotherapy. These cells showed elevated Hh signaling activity [182].
In HCC, Hh signaling has been linked to chemoresistance and invasiveness, largely due to its role in promoting EMT. Treatment with cyclopamine, an Hh pathway inhibitor, reduced the population of CD133+/EpCAM+ HCC CSCs [185]. In pancreatic cancer, combining cyclopamine with gemcitabine resulted in decreased CSCs markers and was associated with tumor regression, highlighting potential therapeutic combinations between Hh inhibitors and standard cancer treatments [186]. Beyond gastrointestinal cancers, similar effects of Hh signaling on CSCs have been observed. In glioma cells, Hh activity induced the expression of genes such as Nanog, which regulate CSC properties [187]. Nanog signaling also appeared to enhance Hh signaling, potentially creating a feedback loop that promotes stemness in glioma CSCs.
In prostate and glioblastoma CSCs, Hh signaling inhibition with erismodegib was shown to prevent EMT, with some of these effects mediated by changes in microRNAs and proteins [188,189]. There is evidence that Hh signaling specifically drives the proliferation of CSCs while having little effect on non-stem cancer cells [190,191].
Hh signaling also contributes to radio- and chemoresistance, primarily driven by CSCs [192,193].
In breast cancer, chemoresistance was found to be associated with P-glycoprotein activity, which correlates with Hh signaling in breast CSCs. Finally, simultaneous inhibition of Notch and Hh signaling sensitized prostate cancer tumor-initiating cells to docetaxel, a chemotherapy drug, by inhibiting AKT and downregulating the anti-apoptotic protein Bcl-2, highlighting the potential of targeting multiple pathways in CSC-driven cancers [194].
9. Hh and Crosstalk with Other Signaling Pathways
The Hh signaling pathway interacts with multiple other signaling networks, although the exact mechanism of these interactions remains poorly understood. Some pathways, such as RAS/RAF/MEK/ERK, PI3K/AKT/mTOR, EGFR, and Notch, which are all involved in regulating cell proliferation, growth, and survival, have better-characterized relationships with the Hh axis [195]. In various cancers, such as colon, pancreatic, and lung cancer, K-RAS has been shown to work in conjugation with Hh signaling [196]. In particular, for constitutive K-RAS activation, a hallmark of certain aggressive cancers like pancreatic cancer models, the absence of Smo proteins did not appear to affect Gli-dependent gene expression [197,198]. In melanomas, inhibition of RAS signaling caused Gli proteins to accumulate in the tumor cell cytoplasm, suggesting that K-RAS may play a role in the nuclear translocation of Gli, a process that is not yet fully understood [199].
Crosstalk with the mTOR pathway also occurs at various points and may contribute to cancer development in specific contexts.
The redundancy and interaction between Hh signaling and other pathways have potential therapeutic implications. In glioblastoma, for example, the simultaneous inhibition of both the Notch and Hh pathways significantly increased the toxicity of the chemotherapy drug temozolomide in the CSC population compared to inhibiting either pathway alone. This highlights the importance of targeting multiple pathways to improve treatment efficacy in certain cancers [200].
10. Conclusions
Recent advances in our understanding of Hh signaling, both in normal physiological processes and in cancer, have significantly influenced the development of new treatment strategies for various cancer types.
The Notch signaling pathway is a highly conserved signaling pathway with essential roles in regulating cellular processes like cell fate, proliferation, differentiation, and apoptosis. Since it has such a large implication in these functions, any dysregulation of the pathway can lead to human cancer and also affect cell populations, like the CSCs. As it has already been proven that CSCs dictate the level of aggressiveness of the tumoral cells and their resistance to therapies, targeting the Notch signaling system with molecular strategies could be a very efficient problem-solving strategy in the control and elimination of the CSCs.
Recent advances in our understanding of molecular signaling pathways, both in normal physiological processes and cancer, have significantly influenced the development of new treatment strategies for various cancer types. While there are still unanswered questions regarding the precise molecular mechanisms of Hh signaling and its deregulation in cancer, it is clear that Hh signaling plays a crucial role in oncogenesis. Targeting this pathway holds great promise for enhancing efforts to combat the key characteristics of cancer, including uncontrolled cell proliferation, tumor aggressiveness, and metastasis, all of which are hallmarks of CSCs.
Author Contributions
S.I. wrote the manuscript with M.N., S.B. and D.D. supervised the work and corrected the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
S.I. is funded by an internal grant of the Iuliu Hatieganu University—School of Doctoral Studies. S.I. and S.B. are also supported by a grant awarded by the Romanian National Ministry of Research, Innovation and Digitalization, a bilateral collaboration grant between Romania and Moldova (PN-IV-P8-8.3-ROMD-2023-0036). He is funded by a national grant of the Romanian Research Ministry—PNRR 2024-2026 (PNRR/2022/C9/MCID/18, Contract No. 760278/26.03.2024).
Data Availability
Any inquiries regarding supporting data availability of this study should be directed to the corresponding author.
Acknowledgments
All authors gratefully acknowledge the support of the Iuliu Hatieganu University in Cluj Napoca.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Penton, A.L.; Leonard, L.D.; Spinner, N.B. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 2012, 23, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.T.; Huang, P. Complex crosstalk of Notch and Hedgehog signalling during the development of the central nervous system. Cell. Mol. Life Sci. 2021, 78, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Xu, M.; Yang, J.; Ma, X. The role of Hedgehog and Notch signaling pathway in cancer. Mol. Biomed. 2022, 3, 44. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Jesse, A.M.; Kohn, A.; Gunnell, L.M.; Honjo, T.; Zuscik, M.J.; O’Keefe, R.J.; Hilton, M.J. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 2010, 137, 1461–1471. [Google Scholar] [CrossRef]
- Hankenson, K.D.; Gagne, K.; Shaughnessy, M. Extracellular signaling molecules to promote fracture healing and bone regeneration. Adv. Drug Deliv. Rev. 2015, 94, 3–12. [Google Scholar] [CrossRef]
- MacGrogan, D.; Nus, M.; de la Pompa, J.L. Notch signaling in cardiac development and disease. Curr. Top. Dev. Biol. 2010, 92, 333–365. [Google Scholar] [CrossRef]
- Luxán, G.; D’Amato, G.; MacGrogan, D.; de la Pompa, J.L. Endocardial Notch Signaling in Cardiac Development and Disease. Circ. Res. 2016, 118, e1–e18. [Google Scholar] [CrossRef]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertrán, E.; Pérez-Pomares, J.M.; Díez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisúa-Belmonte, J.C.; et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes. Dev. 2004, 18, 99–115. [Google Scholar] [CrossRef]
- Lobov, I.B.; Renard, R.A.; Papadopoulos, N.; Gale, N.W.; Thurston, G.; Yancopoulos, G.D.; Wiegand, S.J. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc. Natl. Acad. Sci. USA 2007, 104, 3219–3224. [Google Scholar] [CrossRef]
- Baeten, J.T.; Lilly, B. Differential Regulation of NOTCH2 and NOTCH3 Contribute to Their Unique Functions in Vascular Smooth Muscle Cells. J. Biol. Chem. 2015, 290, 16226–16237. [Google Scholar] [CrossRef]
- Sparks, E.E.; Huppert, K.A.; Brown, M.A.; Washington, M.K.; Huppert, S.S. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology 2010, 51, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Panikkar, A.; Xu, J.; Antoniou, A.; Raynaud, P.; Lemaigre, F.; Stanger, B.Z. Notch signaling controls liver development by regulating biliary differentiation. Development 2009, 136, 1727–1739. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.; Raynaud, P.; Cordi, S.; Zong, Y.; Tronche, F.; Stanger, B.Z.; Jacquemin, P.; Pierreux, C.E.; Clotman, F.; Lemaigre, F.P. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology 2009, 136, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Espinoza, L.; Dowbaj, A.M.; Kohler, T.N.; Strauss, B.; Sarlidou, O.; Belenguer, G.; Pacini, C.; Martins, N.P.; Dobie, R.; Wilson-Kanamori, J.R.; et al. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell 2021, 28, 1907–1921.e8. [Google Scholar] [CrossRef]
- Jensen, J.; Pedersen, E.E.; Galante, P.; Hald, J.; Heller, R.S.; Ishibashi, M.; Kageyama, R.; Guillemot, F.; Serup, P.; Madsen, O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000, 24, 36–44. [Google Scholar] [CrossRef]
- Seymour, P.A.; Collin, C.A.; Egeskov-Madsen, A.R.; Jørgensen, M.C.; Shimojo, H.; Imayoshi, I.; de Lichtenberg, K.H.; Kopan, R.; Kageyama, R.; Serup, P. Jag1 Modulates an Oscillatory Dll1-Notch-Hes1 Signaling Module to Coordinate Growth and Fate of Pancreatic Progenitors. Dev. Cell 2020, 52, 731–747.e8. [Google Scholar] [CrossRef]
- Wendorff, A.A.; Koch, U.; Wunderlich, F.T.; Wirth, S.; Dubey, C.; Brüning, J.C.; MacDonald, H.R.; Radtke, F. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 2010, 33, 671–684. [Google Scholar] [CrossRef]
- López-López, S.; Romero de Ávila, M.J.; Hernández de León, N.C.; Ruiz-Marcos, F.; Baladrón, V.; Nueda, M.L.; Laborda, J.; García-Ramírez, J.J.; Monsalve, E.M.; Díaz-Guerra, M.J.M. NOTCH4 Exhibits Anti-Inflammatory Activity in Activated Macrophages by Interfering With Interferon-γ and TLR4 Signaling. Front. Immunol. 2021, 12, 734966. [Google Scholar] [CrossRef]
- Radke, A.L.; Reynolds, L.E.; Melo, R.C.; Dvorak, A.M.; Weller, P.F.; Spencer, L.A. Mature human eosinophils express functional Notch ligands mediating eosinophil autocrine regulation. Blood 2009, 113, 3092–3101. [Google Scholar] [CrossRef]
- Lewis, K.L.; Caton, M.L.; Bogunovic, M.; Greter, M.; Grajkowska, L.T.; Ng, D.; Klinakis, A.; Charo, I.F.; Jung, S.; Gommerman, J.L.; et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 2011, 35, 780–791. [Google Scholar] [CrossRef]
- Butler, J.M.; Nolan, D.J.; Vertes, E.L.; Varnum-Finney, B.; Kobayashi, H.; Hooper, A.T.; Seandel, M.; Shido, K.; White, I.A.; Kobayashi, M.; et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 2010, 6, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Chen, T.; Wang, Y.; Tian, R.; Zhang, L.; Song, P.; Yang, S.; Zhu, Y.; Guo, X.; Huang, Y.; et al. Mesenchymal stem cells overexpressing Ihh promote bone repair. J. Orthop. Surg. Res. 2014, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Koleva, M.; Kappler, R.; Vogler, M.; Herwig, A.; Fulda, S.; Hahn, H. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol. Life Sci. 2005, 62, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Tanaka, N. Roles of the Hedgehog Signaling Pathway in Epidermal and Hair Follicle Development, Homeostasis, and Cancer. J. Dev. Biol. 2017, 5, 12. [Google Scholar] [CrossRef]
- Frey, M.R. Sonic Hedgehog: Powering up Intestinal Regeneration? Cell. Mol. Gastroenterol. Hepatol. 2023, 16, 650–651. [Google Scholar] [CrossRef]
- Peng, T.; Frank, D.B.; Kadzik, R.S.; Morley, M.P.; Rathi, K.S.; Wang, T.; Zhou, S.; Cheng, L.; Lu, M.M.; Morrisey, E.E. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 2015, 526, 578–582. [Google Scholar] [CrossRef]
- Dessaud, E.; Ribes, V.; Balaskas, N.; Yang, L.L.; Pierani, A.; Kicheva, A.; Novitch, B.G.; Briscoe, J.; Sasai, N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010, 8, e1000382. [Google Scholar] [CrossRef]
- Yeo, S.Y.; Chitnis, A.B. Jagged-mediated Notch signaling maintains proliferating neural progenitors and regulates cell diversity in the ventral spinal cord. Proc. Natl. Acad. Sci. USA 2007, 104, 5913–5918. [Google Scholar] [CrossRef]
- Wall, D.S.; Mears, A.J.; McNeill, B.; Mazerolle, C.; Thurig, S.; Wang, Y.; Kageyama, R.; Wallace, V.A. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 2009, 184, 101–112. [Google Scholar] [CrossRef]
- Jacobs, C.T.; Huang, P. Notch signalling maintains Hedgehog responsiveness via a Gli-dependent mechanism during spinal cord patterning in zebrafish. Elife 2019, 8, e49252. [Google Scholar] [CrossRef]
- Ringuette, R.; Atkins, M.; Lagali, P.S.; Bassett, E.A.; Campbell, C.; Mazerolle, C.; Mears, A.J.; Picketts, D.J.; Wallace, V.A. A Notch-Gli2 axis sustains Hedgehog responsiveness of neural progenitors and Müller glia. Dev. Biol. 2016, 411, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Karamboulas, C.; Ailles, L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Lai, A.G. Aberrations in Notch-Hedgehog signalling reveal cancer stem cells harbouring conserved oncogenic properties associated with hypoxia and immunoevasion. Br. J. Cancer 2019, 121, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Gorlin, R.J. Nevoid basal-cell carcinoma syndrome. Medicine 1987, 66, 98–113. [Google Scholar] [CrossRef]
- Lombardo, Y.; Faronato, M.; Filipovic, A.; Vircillo, V.; Magnani, L.; Coombes, R.C. Nicastrin and Notch4 drive endocrine therapy resistance and epithelial to mesenchymal transition in MCF7 breast cancer cells. Breast Cancer Res. 2014, 16, R62. [Google Scholar] [CrossRef]
- Arcaroli, J.J.; Tai, W.M.; McWilliams, R.; Bagby, S.; Blatchford, P.J.; Varella-Garcia, M.; Purkey, A.; Quackenbush, K.S.; Song, E.-K.; Pitts, T.M.; et al. A NOTCH1 gene copy number gain is a prognostic indicator of worse survival and a predictive biomarker to a Notch1 targeting antibody in colorectal cancer. Int. J. Cancer 2016, 138, 195–205. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Abou-Antoun, T.J.; Hale, J.S.; Lathia, J.D.; Dombrowski, S.M. Brain Cancer Stem Cells in Adults and Children: Cell Biology and Therapeutic Implications. Neurotherapeutics 2017, 14, 372–384. [Google Scholar] [CrossRef]
- Donnem, T.; Andersen, S.; Al-Shibli, K.; Al-Saad, S.; Busund, L.T.; Bremnes, R.M. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: Coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer 2010, 116, 5676–5685. [Google Scholar] [CrossRef]
- Asnaghi, L.; Ebrahimi, K.B.; Schreck, K.C.; Bar, E.E.; Coonfield, M.L.; Bell, W.R.; Handa, J.; Merbs, S.L.; Harbour, J.W.; Eberhart, C.G. Notch signaling promotes growth and invasion in uveal melanoma. Clin. Cancer Res. 2012, 18, 654–665. [Google Scholar] [CrossRef]
- García-Peydró, M.; Fuentes, P.; Mosquera, M.; García-León, M.J.; Alcain, J.; Rodríguez, A.; García de Miguel, P.; Menéndez, P.; Weijer, K.; Spits, H.; et al. The NOTCH1/CD44 axis drives pathogenesis in a T cell acute lymphoblastic leukemia model. J. Clin. Investig. 2018, 128, 2802–2818. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.J.; Brandt, W.D.; Heth, J.A.; Muraszko, K.M.; Fan, X.; Bar, E.E.; Eberhart, C.G. Lateral inhibition of Notch signaling in neoplastic cells. Oncotarget 2015, 6, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Meurette, O.; Mehlen, P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018, 34, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef]
- Liu, R.; Yu, Y.; Wang, Q.; Zhao, Q.; Yao, Y.; Sun, M.; Zhuang, J.; Sun, C.; Qi, Y. Interactions between hedgehog signaling pathway and the complex tumor microenvironment in breast cancer: Current knowledge and therapeutic promises. Cell Commun. Signal. 2024, 22, 432. [Google Scholar] [CrossRef]
- Aval, S.F.; Lotfi, H.; Sheervalilou, R.; Zarghami, N. Tuning of major signaling networks (TGF-β, Wnt, Notch and Hedgehog) by miRNAs in human stem cells commitment to different lineages: Possible clinical application. Biomed. Pharmacother. 2017, 91, 849–860. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Dontu, G.; Jackson, K.W.; McNicholas, E.; Kawamura, M.J.; Abdallah, W.M.; Wicha, M.S. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004, 6, R605–R615. [Google Scholar] [CrossRef]
- Androutsellis-Theotokis, A.; Leker, R.R.; Soldner, F.; Hoeppner, D.J.; Ravin, R.; Poser, S.W.; Rueger, M.A.; Bae, S.-K.; Kittappa, R.; McKay, R.D.G. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 2006, 442, 823–826. [Google Scholar] [CrossRef]
- Pannuti, A.; Foreman, K.; Rizzo, P.; Osipo, C.; Golde, T.; Osborne, B.; Miele, L. Targeting Notch to target cancer stem cells. Clin. Cancer Res. 2010, 16, 3141–3152. [Google Scholar] [CrossRef]
- Fleming, R.J. Structural conservation of Notch receptors and ligands. Semin. Cell Dev. Biol. 1998, 9, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Rebay, I.; Fleming, R.J.; Fehon, R.G.; Cherbas, L.; Cherbas, P.; Artavanis-Tsakonas, S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell 1991, 67, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Mumm, J.S.; Schroeter, E.H.; Saxena, M.T.; Griesemer, A.; Tian, X.; Pan, D.J.; Ray, W.J.; Kopan, R. A Ligand-Induced Extracellular Cleavage Regulates γ-Secretase-like Proteolytic Activation of Notch1. Mol. Cell 2000, 5, 197–206. [Google Scholar] [CrossRef]
- Struhl, G.; Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 1999, 398, 522–525. [Google Scholar] [CrossRef]
- Fortini, M.E.; Artavanis-Tsakonas, S. The suppressor of hairless protein participates in notch receptor signaling. Cell 1994, 79, 273–282. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Zhou, S.; Chen, L.; Young, D.B.; Hayward, S.D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 23–28. [Google Scholar] [CrossRef]
- Nagel, A.C.; Krejci, A.; Tenin, G.; Bravo-Patiño, A.; Bray, S.; Maier, D.; Preiss, A. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell Biol. 2005, 25, 10433–10441. [Google Scholar] [CrossRef]
- Rizzo, P.; Osipo, C.; Foreman, K.; Golde, T.; Osborne, B.; Miele, L. Rational targeting of Notch signaling in cancer. Oncogene 2008, 27, 5124–5131. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Banerjee, S.; Sarkar, F.H. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer. Res. 2008, 28, 3621–3630. [Google Scholar]
- Stylianou, S.; Clarke, R.B.; Brennan, K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006, 66, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, M.; Kawaguchi, T.; Nigro, J.M.; Feuerstein, B.G.; Berger, M.S.; Miele, L.; Pieper, R.O. Contribution of Notch signaling activation to human glioblastoma multiforme. J. Neurosurg. 2007, 106, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Purow, B.W.; Haque, R.M.; Noel, M.W.; Su, Q.; Burdick, M.J.; Lee, J.; Sundaresan, T.; Pastorino, S.; Park, J.K.; Mikolaenko, I.; et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005, 65, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Santagata, S.; Demichelis, F.; Riva, A.; Varambally, S.; Hofer, M.D.; Kutok, J.L.; Kim, R.; Tang, J.; Montie, J.E.; Chinnaiyan, A.M.; et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004, 64, 6854–6857. [Google Scholar] [CrossRef] [PubMed]
- Real, P.J.; Ferrando, A.A. NOTCH inhibition and glucocorticoid therapy in T-cell acute lymphoblastic leukemia. Leukemia 2009, 23, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Maitra, A.; Ghosh, B.; Zechner, U.; Argani, P.; Iacobuzio-Donahue, C.A.; Sriuranpong, V.; Iso, T.; Meszoely, I.M.; Wolfe, M.S.; et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 2003, 3, 565–576. [Google Scholar] [CrossRef]
- Hopfer, O.; Zwahlen, D.; Fey, M.F.; Aebi, S. The Notch pathway in ovarian carcinomas and adenomas. Br. J. Cancer 2005, 93, 709–718. [Google Scholar] [CrossRef]
- Qiao, L.; Wong, B.C. Role of Notch signaling in colorectal cancer. Carcinogenesis 2009, 30, 1979–1986. [Google Scholar] [CrossRef]
- South, A.P.; Cho, R.J.; Aster, J.C. The double-edged sword of Notch signaling in cancer. Semin. Cell Dev. Biol. 2012, 23, 458–464. [Google Scholar] [CrossRef]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Dotto, G.P. Notch tumor suppressor function. Oncogene 2008, 27, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, Y.; Wu, K.C.; Liu, J.; Zhao, Y.Q.; Pan, Y.L.; Du, R.; Zheng, G.R.; Xiong, Y.M.; Xu, H.L.; et al. RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Exp. Cell Res. 2010, 316, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Serresi, M.; Santolini, E.; Capra, M.; Hulleman, E.; Galimberti, V.; Zurrida, S.; Maisonneuve, P.; Viale, G.; Di Fiore, P.P. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004, 167, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Götte, M.; Greve, B.; Kelsch, R.; Müller-Uthoff, H.; Weiss, K.; Kharabi Masouleh, B.; Sibrowski, W.; Kiesel, L.; Buchweitz, O. The adult stem cell marker Musashi-1 modulates endometrial carcinoma cell cycle progression and apoptosis via Notch-1 and p21WAF1/CIP1. Int. J. Cancer 2011, 129, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yu, H.; Linnoila, R.I.; Li, L.; Li, D.; Mo, B.; Okano, H.; Penalva, L.O.; Glazer, R.I. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget 2013, 4, 739–750. [Google Scholar] [CrossRef]
- Wang, X.Y.; Penalva, L.O.; Yuan, H.; Linnoila, R.I.; Lu, J.; Okano, H.; Glazer, R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 2010, 9, 221. [Google Scholar] [CrossRef]
- Espinoza, I.; Pochampally, R.; Xing, F.; Watabe, K.; Miele, L. Notch signaling: Targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013, 6, 1249–1259. [Google Scholar] [CrossRef]
- Mimeault, M.; Batra, S.K. Altered gene products involved in the malignant reprogramming of cancer stem/progenitor cells and multitargeted therapies. Mol. Asp. Med. 2014, 39, 3–32. [Google Scholar] [CrossRef]
- Fan, X.; Khaki, L.; Zhu, T.S.; Soules, M.E.; Talsma, C.E.; Gul, N.; Koh, C.; Zhang, J.; Li, Y.M.; Maciaczyk, J.; et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells 2010, 28, 5–16. [Google Scholar] [CrossRef]
- Chu, Q.; Orr, B.A.; Semenkow, S.; Bar, E.E.; Eberhart, C.G. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clin. Cancer Res. 2013, 19, 3224–3233. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial-mesenchymal transitions in cancer onset and progression. Bull. Acad. Natl. Med. 2009, 193, 1969–1978; discussion 1978–1969. [Google Scholar] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Hollier, B.G.; Evans, K.; Mani, S.A. The epithelial-to-mesenchymal transition and cancer stem cells: A coalition against cancer therapies. J. Mammary Gland. Biol. Neoplasia 2009, 14, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Santisteban, M.; Reiman, J.M.; Asiedu, M.K.; Behrens, M.D.; Nassar, A.; Kalli, K.R.; Haluska, P.; Ingle, J.N.; Hartmann, L.C.; Manjili, M.H.; et al. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res. 2009, 69, 2887–2895. [Google Scholar] [CrossRef]
- Kong, D.; Banerjee, S.; Ahmad, A.; Li, Y.; Wang, Z.; Sethi, S.; Sarkar, F.H. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS ONE 2010, 5, e12445. [Google Scholar] [CrossRef]
- Leong, K.G.; Niessen, K.; Kulic, I.; Raouf, A.; Eaves, C.; Pollet, I.; Karsan, A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 2007, 204, 2935–2948. [Google Scholar] [CrossRef]
- Zavadil, J.; Cermak, L.; Soto-Nieves, N.; Böttinger, E.P. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004, 23, 1155–1165. [Google Scholar] [CrossRef]
- Yabuuchi, S.; Pai, S.G.; Campbell, N.R.; de Wilde, R.F.; De Oliveira, E.; Korangath, P.; Streppel, M.M.; Rasheed, Z.A.; Hidalgo, M.; Maitra, A.; et al. Notch signaling pathway targeted therapy suppresses tumor progression and metastatic spread in pancreatic cancer. Cancer Lett. 2013, 335, 41–51. [Google Scholar] [CrossRef]
- Shah, A.N.; Summy, J.M.; Zhang, J.; Park, S.I.; Parikh, N.U.; Gallick, G.E. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 2007, 14, 3629–3637. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Kong, D.; Banerjee, S.; Ahmad, A.; Azmi, A.S.; Ali, S.; Abbruzzese, J.L.; Gallick, G.E.; Sarkar, F.H. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009, 69, 2400–2407. [Google Scholar] [CrossRef]
- Schott, A.F.; Landis, M.D.; Dontu, G.; Griffith, K.A.; Layman, R.M.; Krop, I.; Paskett, L.A.; Wong, H.; Dobrolecki, L.E.; Lewis, M.T.; et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013, 19, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.T.; Manfra, D.; Poulet, F.M.; Zhang, Q.; Josien, H.; Bara, T.; Engstrom, L.; Pinzon-Ortiz, M.; Fine, J.S.; Lee, H.-J.J.; et al. Chronic Treatment with the γ-Secretase Inhibitor LY-411,575 Inhibits β-Amyloid Peptide Production and Alters Lymphopoiesis and Intestinal Cell Differentiation. J. Biol. Chem. 2004, 279, 12876–12882. [Google Scholar] [CrossRef] [PubMed]
- Dandawate, P.; Padhye, S.; Ahmad, A.; Sarkar, F.H. Novel strategies targeting cancer stem cells through phytochemicals and their analogs. Drug Deliv. Transl. Res. 2013, 3, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Das, T.P.; Damodaran, C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br. J. Cancer 2013, 109, 2587–2596. [Google Scholar] [CrossRef]
- Saito, N.; Fu, J.; Zheng, S.; Yao, J.; Wang, S.; Liu, D.D.; Yuan, Y.; Sulman, E.P.; Lang, F.F.; Colman, H.; et al. A high Notch pathway activation predicts response to γ secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells 2014, 32, 301–312. [Google Scholar] [CrossRef]
- Palomero, T.; Sulis, M.L.; Cortina, M.; Real, P.J.; Barnes, K.; Ciofani, M.; Caparros, E.; Buteau, J.; Brown, K.; Perkins, S.L.; et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007, 13, 1203–1210. [Google Scholar] [CrossRef]
- Albini, A.; Sporn, M.B. The tumour microenvironment as a target for chemoprevention. Nat. Rev. Cancer 2007, 7, 139–147. [Google Scholar] [CrossRef]
- Gilbertson, R.J.; Rich, J.N. Making a tumour’s bed: Glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer 2007, 7, 733–736. [Google Scholar] [CrossRef]
- Hovinga, K.E.; Shimizu, F.; Wang, R.; Panagiotakos, G.; Van Der Heijden, M.; Moayedpardazi, H.; Correia, A.S.; Soulet, D.; Major, T.; Menon, J.; et al. Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells 2010, 28, 1019–1029. [Google Scholar] [CrossRef]
- Lu, J.; Ye, X.; Fan, F.; Xia, L.; Bhattacharya, R.; Bellister, S.; Tozzi, F.; Sceusi, E.; Zhou, Y.; Tachibana, I.; et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 2013, 23, 171–185. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, F.; Wang, A.; Zheng, S.; Lu, Y. Dll4-Notch signaling in regulation of tumor angiogenesis. J. Cancer Res. Clin. Oncol. 2014, 140, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, M.J.; Seftor, R.E.; Seftor, E.A.; Gruman, L.M.; Lee, L.M.; Nickoloff, B.J.; Miele, L.; Sheriff, D.D.; Schatteman, G.C. Transendothelial function of human metastatic melanoma cells: Role of the microenvironment in cell-fate determination. Cancer Res. 2002, 62, 665–668. [Google Scholar] [PubMed]
- Hendrix, M.J.; Seftor, E.A.; Hess, A.R.; Seftor, R.E. Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat. Rev. Cancer 2003, 3, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Pallini, R.; Biffoni, M.; Todaro, M.; Invernici, G.; Cenci, T.; Maira, G.; Parati, E.A.; Stassi, G.; Larocca, L.M.; et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 2010, 468, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Yoo, S.H.; Kim, S.H. Adipose-derived stem cells induced EMT-like changes in H358 lung cancer cells. Anticancer. Res. 2013, 33, 4421–4430. [Google Scholar]
- Horrée, N.; van Diest, P.J.; Sie-Go, D.M.; Heintz, A.P. The invasive front in endometrial carcinoma: Higher proliferation and associated derailment of cell cycle regulators. Hum. Pathol. 2007, 38, 1232–1238. [Google Scholar] [CrossRef]
- Marie-Egyptienne, D.T.; Lohse, I.; Hill, R.P. Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: Potential role of hypoxia. Cancer Lett. 2013, 341, 63–72. [Google Scholar] [CrossRef]
- Yeung, T.M.; Gandhi, S.C.; Bodmer, W.F. Hypoxia and lineage specification of cell line-derived colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4382–4387. [Google Scholar] [CrossRef]
- Haase, V.H. Oxygen regulates epithelial-to-mesenchymal transition: Insights into molecular mechanisms and relevance to disease. Kidney Int. 2009, 76, 492–499. [Google Scholar] [CrossRef]
- Poon, E.; Harris, A.L.; Ashcroft, M. Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert. Rev. Mol. Med. 2009, 11, e26. [Google Scholar] [CrossRef]
- Soares, R.; Balogh, G.; Guo, S.; Gärtner, F.; Russo, J.; Schmitt, F. Evidence for the notch signaling pathway on the role of estrogen in angiogenesis. Mol. Endocrinol. 2004, 18, 2333–2343. [Google Scholar] [CrossRef] [PubMed]
- Sahlgren, C.; Gustafsson, M.V.; Jin, S.; Poellinger, L.; Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 6392–6397. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.V.; Zheng, X.; Pereira, T.; Gradin, K.; Jin, S.; Lundkvist, J.; Ruas, J.L.; Poellinger, L.; Lendahl, U.; Bondesson, M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 2005, 9, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Okuda, H.; Watabe, M.; Kobayashi, A.; Pai, S.K.; Liu, W.; Pandey, P.R.; Fukuda, K.; Hirota, S.; Sugai, T.; et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30, 4075–4086. [Google Scholar] [CrossRef]
- Medyouf, H.; Gusscott, S.; Wang, H.; Tseng, J.C.; Wai, C.; Nemirovsky, O.; Trumpp, A.; Pflumio, F.; Carboni, J.; Gottardis, M.; et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J. Exp. Med. 2011, 208, 1809–1822. [Google Scholar] [CrossRef]
- Shepherd, C.; Banerjee, L.; Cheung, C.W.; Mansour, M.R.; Jenkinson, S.; Gale, R.E.; Khwaja, A. PI3K/mTOR inhibition upregulates NOTCH-MYC signalling leading to an impaired cytotoxic response. Leukemia 2013, 27, 650–660. [Google Scholar] [CrossRef]
- Calzavara, E.; Chiaramonte, R.; Cesana, D.; Basile, A.; Sherbet, G.V.; Comi, P. Reciprocal regulation of Notch and PI3K/Akt signalling in T-ALL cells in vitro. J. Cell. Biochem. 2008, 103, 1405–1412. [Google Scholar] [CrossRef]
- Chan, S.M.; Weng, A.P.; Tibshirani, R.; Aster, J.C.; Utz, P.J. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood 2007, 110, 278–286. [Google Scholar] [CrossRef]
- Meurette, O.; Stylianou, S.; Rock, R.; Collu, G.M.; Gilmore, A.P.; Brennan, K. Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells. Cancer Res. 2009, 69, 5015–5022. [Google Scholar] [CrossRef]
- Campbell, K.J.; Perkins, N.D. Regulation of NF-kappaB function. Biochem. Soc. Symp. 2006, 165–180. [Google Scholar] [CrossRef]
- Cheng, P.; Zlobin, A.; Volgina, V.; Gottipati, S.; Osborne, B.; Simel, E.J.; Miele, L.; Gabrilovich, D.I. Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells. J. Immunol. 2001, 167, 4458–4467. [Google Scholar] [CrossRef] [PubMed]
- Oakley, F.; Mann, J.; Ruddell, R.G.; Pickford, J.; Weinmaster, G.; Mann, D.A. Basal expression of IkappaBalpha is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J. Biol. Chem. 2003, 278, 24359–24370. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.T.; Cariappa, A.; Liu, H.; Muir, B.; Sgroi, D.; Boboila, C.; Pillai, S. Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J. Immunol. 2007, 179, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.H.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef]
- Bash, J.; Zong, W.X.; Banga, S.; Rivera, A.; Ballard, D.W.; Ron, Y.; Gélinas, C. Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 1999, 18, 2803–2811. [Google Scholar] [CrossRef]
- Guan, E.; Wang, J.; Laborda, J.; Norcross, M.; Baeuerle, P.A.; Hoffman, T. T cell leukemia-associated human Notch/translocation-associated Notch homologue has I kappa B-like activity and physically interacts with nuclear factor-kappa B proteins in T cells. J. Exp. Med. 1996, 183, 2025–2032. [Google Scholar] [CrossRef]
- Shin, H.M.; Minter, L.M.; Cho, O.H.; Gottipati, S.; Fauq, A.H.; Golde, T.E.; Sonenshein, G.E.; Osborne, B.A. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006, 25, 129–138. [Google Scholar] [CrossRef]
- Hanlon, L.; Avila, J.L.; Demarest, R.M.; Troutman, S.; Allen, M.; Ratti, F.; Rustgi, A.K.; Stanger, B.Z.; Radtke, F.; Adsay, V.; et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010, 70, 4280–4286. [Google Scholar] [CrossRef]
- Veenendaal, L.M.; Kranenburg, O.; Smakman, N.; Klomp, A.; Borel Rinkes, I.H.; van Diest, P.J. Differential Notch and TGFβ signaling in primary colorectal tumors and their corresponding metastases. Cell. Oncol. 2008, 30, 1–11. [Google Scholar] [CrossRef]
- Xu, P.; Qiu, M.; Zhang, Z.; Kang, C.; Jiang, R.; Jia, Z.; Wang, G.; Jiang, H.; Pu, P. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J. Neurooncol. 2010, 97, 41–51. [Google Scholar] [CrossRef]
- Mittal, S.; Subramanyam, D.; Dey, D.; Kumar, R.V.; Rangarajan, A. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol. Cancer 2009, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Weijzen, S.; Rizzo, P.; Braid, M.; Vaishnav, R.; Jonkheer, S.M.; Zlobin, A.; Osborne, B.A.; Gottipati, S.; Aster, J.C.; Hahn, W.C.; et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 2002, 8, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.H.; Holland, E.C. Notch signaling enhances nestin expression in gliomas. Neoplasia 2006, 8, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Rodilla, V.; Villanueva, A.; Obrador-Hevia, A.; Robert-Moreno, A.; Fernández-Majada, V.; Grilli, A.; López-Bigas, N.; Bellora, N.; Albà, M.M.; Torres, F.; et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Ungerbäck, J.; Elander, N.; Grünberg, J.; Sigvardsson, M.; Söderkvist, P. The Notch-2 gene is regulated by Wnt signaling in cultured colorectal cancer cells. PLoS ONE 2011, 6, e17957. [Google Scholar] [CrossRef] [PubMed]
- Rampazzo, E.; Persano, L.; Pistollato, F.; Moro, E.; Frasson, C.; Porazzi, P.; Della Puppa, A.; Bresolin, S.; Battilana, G.; Indraccolo, S.; et al. Wnt activation promotes neuronal differentiation of glioblastoma. Cell Death Dis. 2013, 4, e500. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. NUMB is a break of WNT-Notch signaling cycle. Int. J. Mol. Med. 2006, 18, 517–521. [Google Scholar] [CrossRef]
- Katoh, M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 2007, 3, 30–38. [Google Scholar] [CrossRef]
- Zhao, X.; Malhotra, G.K.; Lele, S.M.; Lele, M.S.; West, W.W.; Eudy, J.D.; Band, H.; Band, V. Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate. Proc. Natl. Acad. Sci. USA 2010, 107, 14146–14151. [Google Scholar] [CrossRef]
- Kamakura, S.; Oishi, K.; Yoshimatsu, T.; Nakafuku, M.; Masuyama, N.; Gotoh, Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 2004, 6, 547–554. [Google Scholar] [CrossRef]
- Ingham, P.W.; Placzek, M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat. Rev. Genet. 2006, 7, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature 2002, 418, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; Spek, C.A.; Peppelenbosch, M.P. Hedgehog: An unusual signal transducer. Bioessays 2004, 26, 387–394. [Google Scholar] [CrossRef]
- Gallet, A. Hedgehog morphogen: From secretion to reception. Trends Cell Biol. 2011, 21, 238–246. [Google Scholar] [CrossRef]
- Danesin, C.; Agius, E.; Escalas, N.; Ai, X.; Emerson, C.; Cochard, P.; Soula, C. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: Involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J. Neurosci. 2006, 26, 5037–5048. [Google Scholar] [CrossRef]
- Thérond, P.P. Release and transportation of Hedgehog molecules. Curr. Opin. Cell Biol. 2012, 24, 173–180. [Google Scholar] [CrossRef]
- Bischoff, M.; Gradilla, A.-C.; Seijo, I.; Andrés, G.; Rodríguez-Navas, C.; González-Méndez, L.; Guerrero, I. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat. Cell Biol. 2013, 15, 1269–1281. [Google Scholar] [CrossRef]
- Rubin, L.L.; de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006, 5, 1026–1033. [Google Scholar] [CrossRef]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef]
- Jia, J.; Tong, C.; Wang, B.; Luo, L.; Jiang, J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 2004, 432, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Struhl, G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 1998, 391, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sasai, N.; Ma, G.; Yue, T.; Jia, J.; Briscoe, J.; Jiang, J. Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011, 9, e1001083. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, L.; Scott, M.P.; Rohatgi, R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol. 2009, 187, 365–374. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Z.; Walsh, C.T.; McMahon, A.P. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl. Acad. Sci. USA 2009, 106, 2623–2628. [Google Scholar] [CrossRef]
- Shevde, L.A.; Samant, R.S. Nonclassical hedgehog-GLI signaling and its clinical implications. Int. J. Cancer 2014, 135, 1–6. [Google Scholar] [CrossRef]
- Mille, F.; Thibert, C.; Fombonne, J.; Rama, N.; Guix, C.; Hayashi, H.; Corset, V.; Reed, J.C.; Mehlen, P. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat. Cell Biol. 2009, 11, 739–746. [Google Scholar] [CrossRef]
- Scales, S.J.; de Sauvage, F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009, 30, 303–312. [Google Scholar] [CrossRef]
- Amakye, D.; Jagani, Z.; Dorsch, M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013, 19, 1410–1422. [Google Scholar] [CrossRef]
- Yauch, R.L.; Gould, S.E.; Scales, S.J.; Tang, T.; Tian, H.; Ahn, C.P.; Marshall, D.; Fu, L.; Januario, T.; Kallop, D.; et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008, 455, 406–410. [Google Scholar] [CrossRef]
- Hegde, G.V.; Peterson, K.J.; Emanuel, K.; Mittal, A.K.; Joshi, A.D.; Dickinson, J.D.; Kollessery, G.J.; Bociek, R.G.; Bierman, P.; Vose, J.M.; et al. Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: A potential new therapeutic target. Mol. Cancer Res. 2008, 6, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Dierks, C.; Grbic, J.; Zirlik, K.; Beigi, R.; Englund, N.P.; Guo, G.R.; Veelken, H.; Engelhardt, M.; Mertelsmann, R.; Kelleher, J.F.; et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nat. Med. 2007, 13, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Seeley, E.S.; Carrière, C.; Goetze, T.; Longnecker, D.S.; Korc, M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 2009, 69, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Martelotto, L.G.; Peifer, M.; Sos, M.L.; Karnezis, A.N.; Mahjoub, M.R.; Bernard, K.; Conklin, J.F.; Szczepny, A.; Yuan, J.; et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat. Med. 2011, 17, 1504–1508. [Google Scholar] [CrossRef]
- Li, X.; Deng, W.; Nail, C.D.; Bailey, S.K.; Kraus, M.H.; Ruppert, J.M.; Lobo-Ruppert, S.M. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene 2006, 25, 609–621. [Google Scholar] [CrossRef]
- Hay, E.D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995, 154, 8–20. [Google Scholar] [CrossRef]
- He, J.; Sheng, T.; Stelter, A.A.; Li, C.; Zhang, X.; Sinha, M.; Luxon, B.A.; Xie, J. Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J. Biol. Chem. 2006, 281, 35598–35602. [Google Scholar] [CrossRef]
- Gonzalez, D.M.; Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal 2014, 7, re8. [Google Scholar] [CrossRef]
- Kurita, S.; Mott, J.L.; Almada, L.L.; Bronk, S.F.; Werneburg, N.W.; Sun, S.Y.; Roberts, L.R.; Fernandez-Zapico, M.E.; Gores, G.J. GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis. Oncogene 2010, 29, 4848–4858. [Google Scholar] [CrossRef]
- Athar, M.; Li, C.; Tang, X.; Chi, S.; Zhang, X.; Kim, A.L.; Tyring, S.K.; Kopelovich, L.; Hebert, J.; Epstein, E.H., Jr.; et al. Inhibition of smoothened signaling prevents ultraviolet B-induced basal cell carcinomas through regulation of Fas expression and apoptosis. Cancer Res. 2004, 64, 7545–7552. [Google Scholar] [CrossRef]
- Carballo, G.B.; Matias, D.; Ribeiro, J.H.; Pessoa, L.S.; Arrais-Neto, A.M.; Spohr, T. Cyclopamine sensitizes glioblastoma cells to temozolomide treatment through Sonic hedgehog pathway. Life Sci. 2020, 257, 118027. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, S.A.; Grinenko, N.F.; Antonova, O.M.; Kurapov, P.B.; Shepeleva, I.I.; Chekhonin, V.P. Relationship between Hedgehog Signaling Pathway and Drug Resistance of Poorly Differentiated Gliomas. Bull. Exp. Biol. Med. 2018, 164, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, R.; Wang, Z.; Chen, S.; Chen, S.; Guo, G.; Liu, Z. Cyclopamine Suppresses Human Esophageal Carcinoma Cell Growth by Inhibiting Glioma-Associated Oncogene Protein-1, a Marker of Human Esophageal Carcinoma Progression. Med. Sci. Monit. 2019, 25, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Fan, S.; Wang, H.; Mao, J.; Shi, Y.; Ibrahim, M.M.; Ma, W.; Yu, X.; Hou, Z.; Wang, B.; et al. Genistein decreases the breast cancer stem-like cell population through Hedgehog pathway. Stem Cell. Res. Ther. 2013, 4, 146. [Google Scholar] [CrossRef]
- Coward, L.; Barnes, N.C.; Setchell, K.D.R.; Barnes, S. Genistein, daidzein, and their.beta.-glycoside conjugates: Antitumor isoflavones in soybean foods from American and Asian diets. J. Agric. Food Chem. 1993, 41, 1961–1967. [Google Scholar] [CrossRef]
- Spagnuolo, C.; Russo, G.L.; Orhan, I.E.; Habtemariam, S.; Daglia, M.; Sureda, A.; Nabavi, S.F.; Devi, K.P.; Loizzo, M.R.; Tundis, R.; et al. Genistein and cancer: Current status, challenges, and future directions. Adv. Nutr. 2015, 6, 408–419. [Google Scholar] [CrossRef]
- Taipale, J.; Chen, J.K.; Cooper, M.K.; Wang, B.; Mann, R.K.; Milenkovic, L.; Scott, M.P.; Beachy, P.A. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 2000, 406, 1005–1009. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66, 6063–6071. [Google Scholar] [CrossRef]
- Fiaschi, M.; Rozell, B.; Bergström, A.; Toftgård, R. Development of mammary tumors by conditional expression of GLI1. Cancer Res. 2009, 69, 4810–4817. [Google Scholar] [CrossRef]
- Clement, V.; Sanchez, P.; de Tribolet, N.; Radovanovic, I.; Ruiz i Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Yue, W.; Wei, B.; Wang, N.; Li, T.; Guan, L.; Shi, S.; Zeng, Q.; Pei, X.; Chen, L. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PLoS ONE 2011, 6, e17687. [Google Scholar] [CrossRef] [PubMed]
- Varnat, F.; Zacchetti, G.; Ruiz i Altaba, A. Hedgehog pathway activity is required for the lethality and intestinal phenotypes of mice with hyperactive Wnt signaling. Mech. Dev. 2010, 127, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Xu, X.F.; Xu, L.; Niu, P.Q.; Wang, F.; Hu, G.Y.; Wang, X.P.; Guo, C.Y. Cyclopamine blocked the growth of colorectal cancer SW116 cells by modulating some target genes of Gli1 in vitro. Hepatogastroenterology 2011, 58, 1511–1518. [Google Scholar] [CrossRef]
- Chen, X.; Lingala, S.; Khoobyari, S.; Nolta, J.; Zern, M.A.; Wu, J. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 2011, 55, 838–845. [Google Scholar] [CrossRef]
- Jimeno, A.; Feldmann, G.; Suárez-Gauthier, A.; Rasheed, Z.; Solomon, A.; Zou, G.M.; Rubio-Viqueira, B.; García-García, E.; López-Ríos, F.; Matsui, W.; et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol. Cancer Ther. 2009, 8, 310–314. [Google Scholar] [CrossRef]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.N.; Borges, I.; Ruiz i Altaba, A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef]
- Fu, J.; Rodova, M.; Nanta, R.; Meeker, D.; Van Veldhuizen, P.J.; Srivastava, R.K.; Shankar, S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013, 15, 691–706. [Google Scholar] [CrossRef]
- Nanta, R.; Kumar, D.; Meeker, D.; Rodova, M.; Van Veldhuizen, P.J.; Shankar, S.; Srivastava, R.K. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by regulating Bmi-1 and microRNA-128. Oncogenesis 2013, 2, e42. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, A.; Jamieson, C.H.; Fereshteh, M.; Abrahamsson, A.; Blum, J.; Kwon, H.Y.; Kim, J.; Chute, J.P.; Rizzieri, D.; et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature 2009, 458, 776–779. [Google Scholar] [CrossRef]
- Read, T.A.; Fogarty, M.P.; Markant, S.L.; McLendon, R.E.; Wei, Z.; Ellison, D.W.; Febbo, P.G.; Wechsler-Reya, R.J. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell 2009, 15, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Olive, K.P.; Jacobetz, M.A.; Davidson, C.J.; Gopinathan, A.; McIntyre, D.; Honess, D.; Madhu, B.; Goldgraben, M.A.; Caldwell, M.E.; Allard, D.; et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009, 324, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Sims-Mourtada, J.; Izzo, J.G.; Apisarnthanarax, S.; Wu, T.T.; Malhotra, U.; Luthra, R.; Liao, Z.; Komaki, R.; van der Kogel, A.; Ajani, J.; et al. Hedgehog: An attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin. Cancer Res. 2006, 12, 6565–6572. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Domenech, J.; Vidal, S.J.; Rodriguez-Bravo, V.; Castillo-Martin, M.; Quinn, S.A.; Rodriguez-Barrueco, R.; Bonal, D.M.; Charytonowicz, E.; Gladoun, N.; de la Iglesia-Vicente, J.; et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell 2012, 22, 373–388. [Google Scholar] [CrossRef]
- Brechbiel, J.; Miller-Moslin, K.; Adjei, A.A. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat. Rev. 2014, 40, 750–759. [Google Scholar] [CrossRef]
- Lauth, M. RAS and Hedgehog—Partners in crime. Front. Biosci. 2011, 16, 2259–2270. [Google Scholar] [CrossRef]
- Ji, Z.; Mei, F.C.; Xie, J.; Cheng, X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 2007, 282, 14048–14055. [Google Scholar] [CrossRef]
- Nolan-Stevaux, O.; Lau, J.; Truitt, M.L.; Chu, G.C.; Hebrok, M.; Fernández-Zapico, M.E.; Hanahan, D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes. Dev. 2009, 23, 24–36. [Google Scholar] [CrossRef]
- Stecca, B.; Mas, C.; Clement, V.; Zbinden, M.; Correa, R.; Piguet, V.; Beermann, F.; Ruiz i Altaba, A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. USA 2007, 104, 5895–5900. [Google Scholar] [CrossRef]
- Ulasov, I.V.; Nandi, S.; Dey, M.; Sonabend, A.M.; Lesniak, M.S. Inhibition of Sonic hedgehog and Notch pathways enhances sensitivity of CD133+ glioma stem cells to temozolomide therapy. Mol. Med. 2011, 17, 103–112. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).