Genome-Wide Association Study for Milk Protein Content in Romanian Dual-Purpose Cattle
Abstract
1. Introduction
2. Materials and Methods
- SNP call rate exceeding 90% as recommended by McClure et al. (2018), resulting in the removal of 18,972 SNPs [27];
- Minor allele frequency above 0.05 following Teng et al. (2023), eliminating 18,986 SNPs [28];
- Hardy–Weinberg equilibrium p-value > 1 × 10−6 based on Zhang et al. (2016), excluding 1333 SNPs [29];
- Elimination of 1113 duplicate SNP positions;
- Restriction to autosomal chromosomes (BTA1-29).
3. Results
3.1. Phenotype Distribution and Population Characteristics
3.2. Population Structure Analysis
3.3. Genome-Wide Association Analysis
3.4. Chromosomal Distribution and Functional Annotation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHCYL1 | S-Adenosylhomocysteine Hydrolase-Like 1 |
| BTA | Bos Taurus Autosome |
| CNCS-UEFISCDI | National Council for Scientific Research-Executive Agency for Higher Education, Research, Development, and Innovation Funding |
| CSN | Casein (CSN1S1, CSN1S2, CSN2, CSN3-casein gene variants) |
| DGAT1 | Diacylglycerol O-Acyltransferase 1 |
| DIM | Days In Milk |
| DKK2 | Dickkopf-related protein 2 |
| DNA | Deoxyribonucleic Acid |
| EA | Effect Allele |
| EAAT5 | Excitatory Amino Acid Transporter 5 |
| EDTA | Ethylenediaminetetraacetic Acid |
| ERK1/2 | Extracellular signal-Regulated Kinase 1/2 |
| GWAS | Genome-Wide Association Studies |
| IP3 | Inositol 1,4,5-trisphosphate |
| IQR | Interquartile Range |
| KLF6 | Krüppel-Like Factor 6 |
| LD | Linkage Disequilibrium |
| LIC | Livestock Improvement Corporation |
| MAF | Minor Allele Frequency |
| MAPK | Mitogen-Activated Protein Kinase |
| NZD | New Zealand Dollar |
| OA | Other Allele |
| PAEP | Progestagen-Associated Endometrial Protein (β-lactoglobulin) |
| PC | Principal Component |
| PCA | Principal Component Analysis |
| PNCDI | National Plan for Research, Development, and Innovation |
| PTHrP | Parathyroid Hormone-related Protein |
| Quantile-Quantile | |
| QTL | Quantitative Trait Loci |
| SE | Standard Error |
| SLC1A7 | Solute Carrier Family 1 Member 7 |
| SNP | Single Nucleotide Polymorphism |
| STAT5 | Signal Transducer and Activator of Transcription 5 |
| UMD | University of Maryland (bovine genome assembly) |
| ZMAT4 | Zinc finger Matrin-Type 4 |
References
- LIC. Economic Values in the New Zealand Dairy Industry. Livestock Improvement Corporation. 2025. Available online: https://www.lic.co.nz/about/supporting-our-industry/animal-evaluation/about-animal-evaluation/ (accessed on 22 September 2025).
- Huțu, I. Considerations on Lactation in Cattle under Romanian Formal Recording. Lucr. Științifice Med. Vet. Timișoara 2015, 48, 74–80. [Google Scholar]
- Lu, X.; Arbab, A.A.I.; Abdalla, I.M.; Liu, D.; Zhang, Z.; Xu, T.; Su, G.; Yang, Z. Genetic Parameter Estimation and Genome-Wide Association Study-Based Loci Identification of Milk-Related Traits in Chinese Holstein. Front. Genet. 2021, 12, 799664. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Poulsen, N.A.; Gebreyesus, G.; Larsen, L.B. Estimation of Genetic Parameters and Detection of Chromosomal Regions Affecting the Major Milk Proteins and Their Post-Translational Modifications in Danish Holstein and Danish Jersey Cattle. BMC Genet. 2016, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Schopen, G.C.B.; Heck, J.M.L.; Bovenhuis, H.; Visker, M.H.P.W.; van Valenberg, H.J.F.; van Arendonk, J.A.M. Genetic Parameters for Major Milk Proteins in Dutch Holstein-Friesians. J. Dairy Sci. 2009, 92, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, I.; Xiang, R.; Jenko, J.; Pausch, H.; Boussaha, M.; Schrooten, C.; Tribout, T.; Gjuvsland, A.B.; Boichard, D.; Nordbø, Ø.; et al. Meta-Analysis for Milk Fat and Protein Percentage Using Imputed Sequence Variant Genotypes in 94,321 Cattle from Eight Cattle Breeds. Genet. Sel. Evol. 2020, 52, 37. [Google Scholar] [CrossRef]
- Pegolo, S.; Mach, N.; Ramayo-Caldas, Y.; Schiavon, S.; Bittante, G.; Cecchinato, A. Integration of GWAS, Pathway and Network Analyses Reveals Novel Mechanistic Insights into the Synthesis of Milk Proteins in Dairy Cows. Sci. Rep. 2018, 8, 566. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Ilie, D.E.; Mizeranschi, A.E.; Mihali, C.V.; Neamț, R.I.; Goilean, G.V.; Georgescu, O.I.; Zaharie, D.; Carabaș, M.; Huțu, I. Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes 2021, 12, 1495. [Google Scholar] [CrossRef]
- Ilie, D.E.; Cean, A.; Cziszter, L.T.; Gavojdian, D.; Ivan, A.; Kusza, S. Microsatellite and Mitochondrial DNA Study of Native Eastern European Cattle Populations: The Case of the Romanian Grey. PLoS ONE 2015, 10, e0138736. [Google Scholar] [CrossRef]
- Davidescu, M.A.; Simeanu, D.; Gorgan, D.L.; Ciorpac, M.; Creangă, S. Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture 2022, 12, 2059. [Google Scholar] [CrossRef]
- Cziszter, L.T.; Gavojdian, D.; Neamț, R.I.; Neciu, F.C.; Saplacan, S.I.; Ilie, D.E. Comparative Study on Production, Reproduction and Functional Traits Between Fleckvieh and Braunvieh Cattle. Asian-Australas. J. Anim. Sci. 2017, 30, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Grădinaru, A.C.; Petrescu-Mag, I.V.; Oroian, F.C.; Balint, C.; Oltean, I. Milk Protein Polymorphism Characterization—A Modern Tool for Sustainable Conservation of Endangered Romanian Cattle Breeds in the Context of Traditional Breeding. Sustainability 2018, 10, 534. [Google Scholar] [CrossRef]
- Neamț, R.I.; Saplacan, G.; Acatincai, S.; Cziszter, L.T.; Gavojdian, D.; Ilie, D.E. The Influence of CSN3 and LGB Polymorphisms on Milk Production and Chemical Composition in Romanian Simmental Cattle. Acta Biochim. Pol. 2017, 64, 493–497. [Google Scholar] [CrossRef]
- Ilie, D.E.; Mizeranschi, A.E.; Mihali, C.V.; Neamț, R.I.; Cziszter, L.T.; Carabaș, M.; Grădinaru, A.C. Polymorphism of the Prolactin (PRL) Gene and Its Effect on Milk Production Traits in Romanian Cattle Breeds. Vet. Sci. 2023, 10, 275. [Google Scholar] [CrossRef] [PubMed]
- Panagiotou, O.A.; Evangelou, E.; Ioannidis, J.P. Genome-wide Significant Associations for Variants with Minor Allele Frequency of 5% or Less—An Overview: A HuGE Review. Annu. Rev. Genom. Hum. Genet. 2013, 14, 441–465. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 15 September 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation, R Package Version 1.1.4. 2023. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 1 October 2025).
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Hocking, T.; Schwendinger, B. data.table: Extension of ‘data.frame’, R Package Version 1.14.8. 2023. Available online: https://CRAN.R-project.org/package=data.table (accessed on 16 September 2025).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Grolemund, G.; Wickham, H. Dates and Times Made Easy with lubridate. J. Stat. Softw. 2011, 40, 1–25. [Google Scholar] [CrossRef]
- Zi-min, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. A Whole-Genome Assembly of the Domestic Cow, Bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef]
- Wickham, H.; Bryan, J. readxl: Read Excel Files, R Package Version 1.4.3. 2025. Available online: https://CRAN.R-project.org/package=readxl (accessed on 1 October 2025).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Lander, E.S.; Kruglyak, L. Genetic Dissection of Complex Traits: Guidelines for Interpreting and Reporting Linkage Results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef]
- McClure, M.C.; McCarthy, J.; Flynn, P.; McClure, J.C.; Dair, E.; O’Connell, D.K.; Kearney, J.F. SNP Data Quality Control in a National Beef and Dairy Cattle System and Highly Accurate SNP-Based Parentage Verification and Identification. Front. Genet. 2018, 9, 84. [Google Scholar] [CrossRef]
- Teng, J.; Wang, D.; Zhao, C.; Zhang, X.; Chen, Z.; Liu, J.; Sun, D.; Tang, H.; Wang, W.; Li, J.; et al. Longitudinal Genome-Wide Association Studies of Milk Production Traits in Holstein Cattle Using Whole-Genome Sequence Data Imputed from Medium-Density Chip Data. J. Dairy Sci. 2023, 106, 2535–2550. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Y.; Mukiibi, R.; Chen, L.; Vinsky, M.; Plastow, G.; Basarab, J.; Stothard, P.; Li, C. Pathway-Based Genome-Wide Association Studies for Two Meat Production Traits in Simmental Cattle. Sci. Rep. 2016, 6, 18389. [Google Scholar] [CrossRef]
- Coppa, L.; Khanal, P.; Pant, S.; Nakano, A.H.; Austin, K.J.; Cammack, K.M.; Lee, J.; Murdoch, B.M. Genome-Wide Association Study for Carcass Weight in Pasture-Finished Beef Cattle in Hawai’i. Front. Genet. 2023, 14, 1168150. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Ko, S.-R.; Le, V.T.; Jee, M.-G.; Jung, Y.J.; Kang, K.-K.; Cho, Y.-G. Development of SNP Markers from GWAS for Selecting Seed Coat and Aleurone Layers in Brown Rice (Oryza sativa L.). Genes 2022, 13, 1805. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. httr: Tools for Working with URLs and HTTP. R Package Version 1.4.7. 2023. Available online: https://CRAN.R-project.org/package=httr (accessed on 16 September 2025).
- Ooms, J. The jsonlite Package: A Practical and Consistent Mapping Between JSON Data and R Objects. arXiv 2014, arXiv:1403.2805. Available online: https://arxiv.org/abs/1403.2805 (accessed on 17 September 2025). [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar] [CrossRef]
- Wilke, C.O. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’, R Package Version 1.1.3. 2024. Available online: https://CRAN.R-project.org/package=cowplot (accessed on 17 September 2025).
- Auguie, B.; Antonov, A. gridExtra: Miscellaneous Functions for “Grid” Graphics, R Package Version 2.3. 2017. Available online: https://CRAN.R-project.org/package=gridExtra (accessed on 18 September 2025).
- Ooms, J. writexl: Export Data Frames to Excel ‘xlsx’ Format, R Package Version 1.5.0. 2025. Available online: https://CRAN.R-project.org/package=writexl (accessed on 18 September 2025).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Doherty, A.; Smith-Byrne, K.; Ferreira, T.; Holmes, M.V.; Holmes, C.; Pulit, S.L.; Lindgren, C.M. GWAS Identifies 14 Loci for Device-Measured Physical Activity and Sleep Duration. Nat. Commun. 2018, 9, 5257. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Y.; Zhou, Q.; Wang, R.; Xia, B.; Ma, D.; Luo, K.; Liu, Q. Testis-Specific Gene C7orf61 Is Involved in Mouse Sperm-Egg Fusion. Urol. J. 2024, 21, 348–355. [Google Scholar] [CrossRef]
- Liu, Y.; Han, B.; Zheng, W.; Peng, F.; Lin, Y.; Ren, J.; Cao, L.; Shen, Y.; Zhao, C.; Li, J. Identification of Genetic Associations and Functional SNPs of Bovine KLF6 Gene on Milk Production Traits in Chinese Holstein. BMC Genom. Data 2023, 24, 72. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Kim, J.; Ahn, S.E.; Lee, S.I.; Bazer, F.W.; Han, J.Y.; Song, G. AHCYL1 Is Mediated by Estrogen-Induced ERK1/2 MAPK Cell Signaling and MicroRNA Regulation to Affect Functional Aspects of the Avian Oviduct. PLoS ONE 2012, 7, e49204. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, J.N.; Wysolmerski, J.J. The Calcium-Sensing Receptor Regulates Mammary Gland Parathyroid Hormone-Related Protein Production and Calcium Transport. J. Clin. Investig. 2004, 113, 598–608. [Google Scholar] [CrossRef]
- Arriza, J.L.; Eliasof, S.; Kavanaugh, M.P.; Amara, S.G. Excitatory Amino Acid Transporter 5, a Retinal Glutamate Transporter Coupled to a Chloride Conductance. Proc. Natl. Acad. Sci. USA 1997, 94, 4155–4160. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, S.; Wang, M.; Zhai, W.; Xu, Z.; Wang, C.; Li, X.; Liu, J.; Li, F. Zinc Finger Homeodomain Factor Zfhx3 Is Essential for Mammary Lactogenic Differentiation by Maintaining Prolactin Signaling Activity. J. Biol. Chem. 2016, 291, 12809–12820. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Hui, T.; Shao, B.; Li, L.; Du, Z.; Lu, L.; Ye, L.; Li, S.; Li, Q.; Xiao, Q.; et al. Dickkopf-Related Protein 2 Induces G0/G1 Arrest and Apoptosis Through Suppressing Wnt/β-Catenin Signaling and Is Frequently Methylated in Breast Cancer. Oncotarget 2017, 8, 39443–39459. [Google Scholar] [CrossRef]
- Lopdell, T.J. Using QTL to Identify Genes and Pathways Underlying the Regulation and Production of Milk Components in Cattle. Animals 2023, 13, 911. [Google Scholar] [CrossRef]

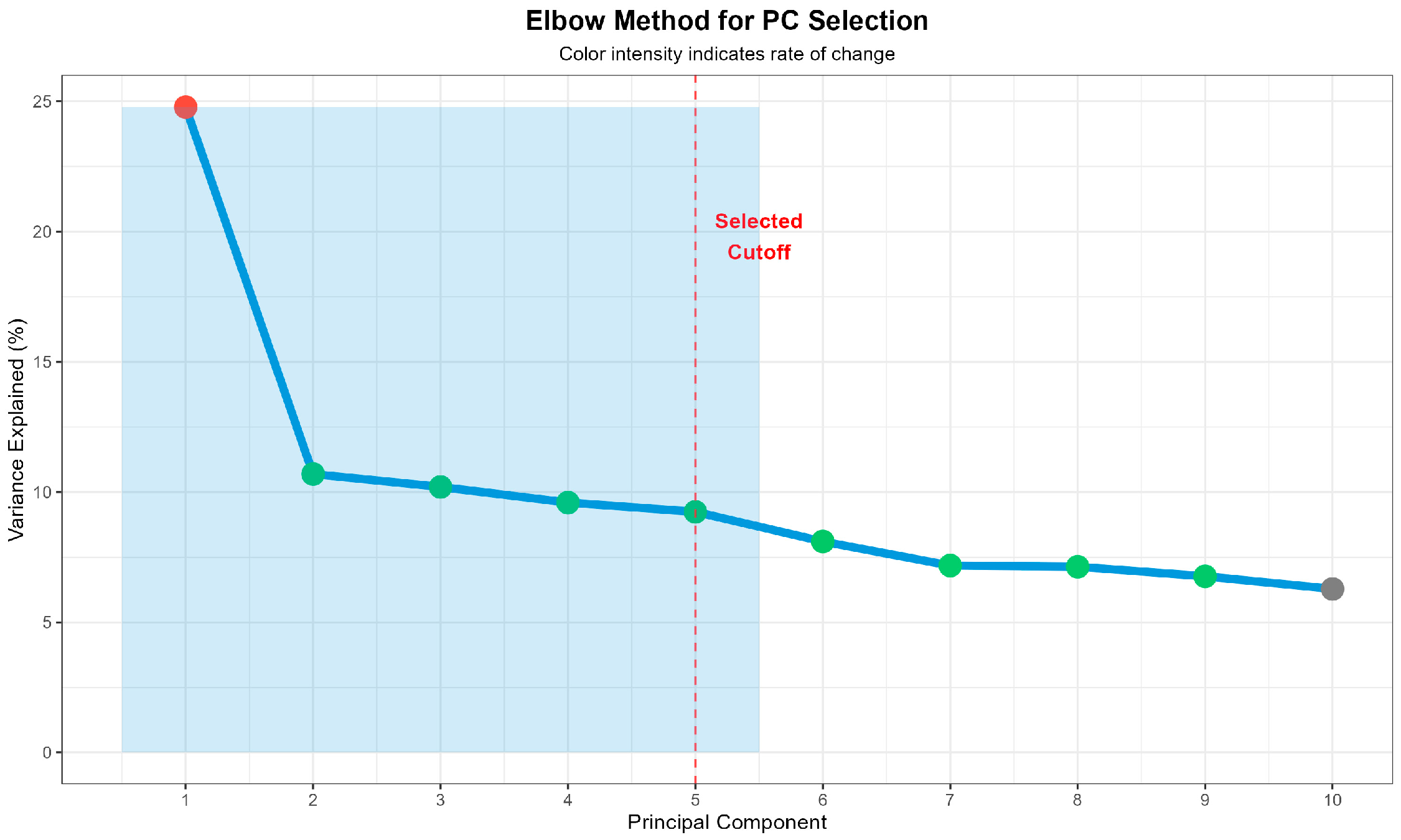

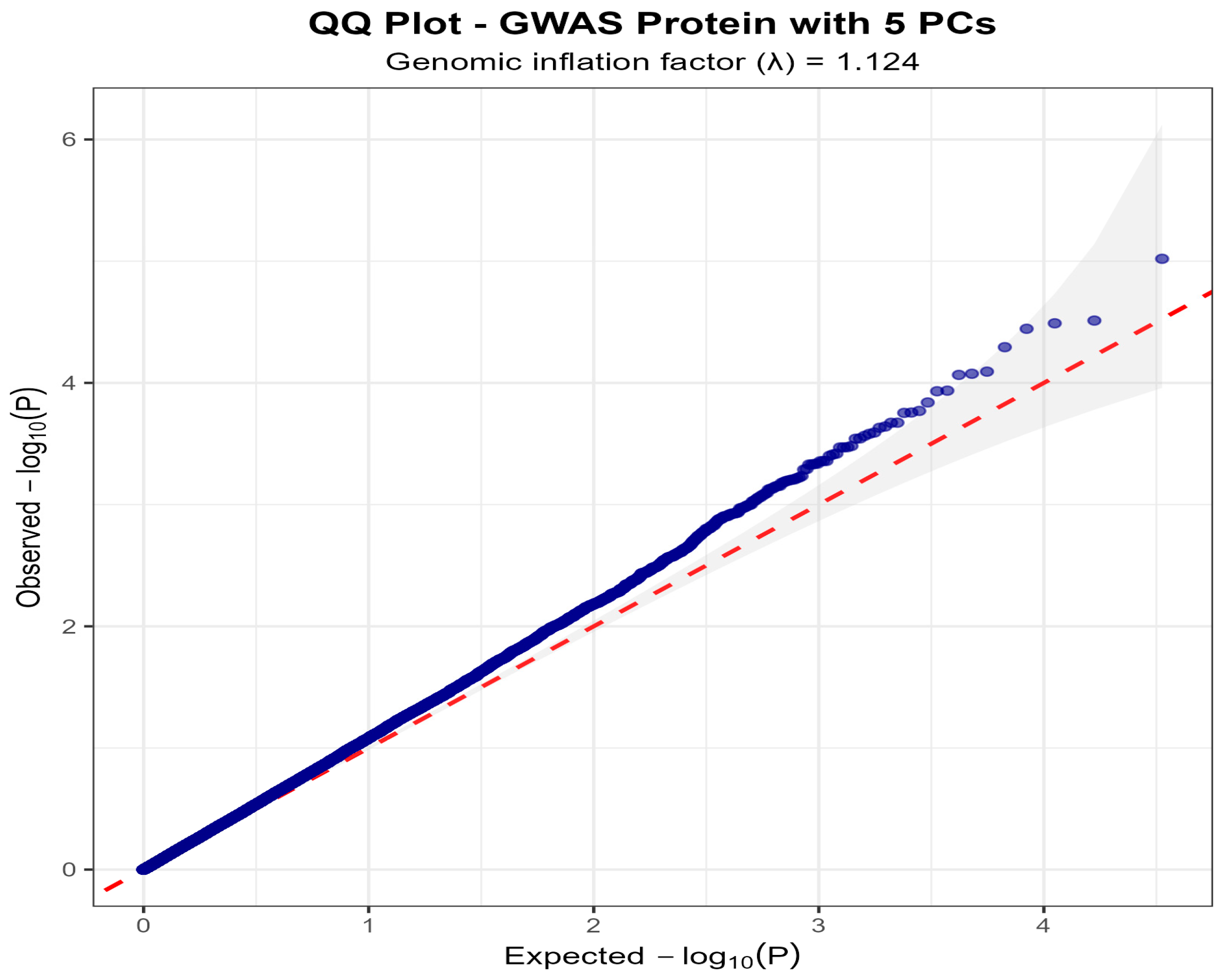
| Rank | SNP ID | Chr | Position (bp) | EA/OA | MAF | β (SE) | p-Value | Gene | Location |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AX-115120431 | 25 | 36,625,127 | A/C | 0.299 | −0.0301 (0.0067) | 9.56 × 10−6 | C7orf61 | Within gene |
| 2 | AX-106740068 | 27 | 35,144,687 | G/A | 0.264 | 0.0199 (0.0047) | 3.08 × 10−5 | ZMAT4 | 96.6 kb |
| 3 | AX-106725208 | 3 | 33,559,007 | G/A | 0.296 | −0.0190 (0.0045) | 3.24 × 10−5 | AHCYL1 | 2.7 kb |
| 4 | AX-106756277 | 13 | 44,876,436 | A/G | 0.178 | 0.0257 (0.0061) | 3.59 × 10−5 | KLF6 | 68.6 kb |
| 5 | AX-185117724 | 27 | 35,382,031 | C/T | 0.085 | 0.0292 (0.0071) | 5.09 × 10−5 | ZMAT4 | Within gene |
| 6 | AX-117081548 | 3 | 93,682,810 | G/T | 0.315 | −0.0193 (0.0048) | 8.08 × 10−5 | SLC1A7 | Within gene |
| 7 | AX-185118865 | 2 | 55,947,021 | C/C | 0.498 | 0.0173 (0.0043) | 8.41 × 10−5 | Intergenic | - |
| 8 | AX-106749405 | 6 | 19,518,197 | C/T | 0.411 | −0.0166 (0.0042) | 8.60 × 10−5 | DKK2 | 7.0 kb |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bratu, D.G.; Blaga, Ș.; Zanfira, B.C.; Mircu, C.; Spătaru, I.I.; Torda, I.; Mizeranschi, A.E.; Ilie, D.E.; Cziszter, L.T.; Vizitiu, D.A.; et al. Genome-Wide Association Study for Milk Protein Content in Romanian Dual-Purpose Cattle. Life 2025, 15, 1668. https://doi.org/10.3390/life15111668
Bratu DG, Blaga Ș, Zanfira BC, Mircu C, Spătaru II, Torda I, Mizeranschi AE, Ilie DE, Cziszter LT, Vizitiu DA, et al. Genome-Wide Association Study for Milk Protein Content in Romanian Dual-Purpose Cattle. Life. 2025; 15(11):1668. https://doi.org/10.3390/life15111668
Chicago/Turabian StyleBratu, Daniel George, Șerban Blaga, Bianca Cornelia Zanfira, Călin Mircu, Ioana Irina Spătaru, Iuliu Torda, Alexandru Eugeniu Mizeranschi, Daniela Elena Ilie, Ludovic Toma Cziszter, Dorin Alexandru Vizitiu, and et al. 2025. "Genome-Wide Association Study for Milk Protein Content in Romanian Dual-Purpose Cattle" Life 15, no. 11: 1668. https://doi.org/10.3390/life15111668
APA StyleBratu, D. G., Blaga, Ș., Zanfira, B. C., Mircu, C., Spătaru, I. I., Torda, I., Mizeranschi, A. E., Ilie, D. E., Cziszter, L. T., Vizitiu, D. A., Boldura, O. M., & Huțu, I. (2025). Genome-Wide Association Study for Milk Protein Content in Romanian Dual-Purpose Cattle. Life, 15(11), 1668. https://doi.org/10.3390/life15111668







