1. Introduction
The statistical laws of distribution, i.e., Maxwell–Boltzmann [
1,
2], Planck [
3], Bose–Einstein [
4], and Fermi–Dirac [
5,
6], played a significant role in the development of physics. It is timely to ask if the recent achievements of genomic research collected in big databases and statistically modeled could influence modern biology and science as well. A statistical model is associated with an attempt at a mathematical description of the components of the analyzed phenomenon and constitutes an important confirmation of its understanding. The aim of the work is to find the most general characteristics and trends describing comparative genomics.
Seminal studies from the late 20th century, such as the development of the BLAST algorithm [
7] and the foundational work by Henikoff and Henikoff (1992) [
8], provided essential groundwork for the development of comparative genomics, particularly within the field of bioinformatics. These studies were crucial for understanding the methods and tools that have shaped modern comparative analyses. They improved database searches and the accuracy of protein sequence alignments, which are core techniques in comparative genomics. Later papers in this area include the initial comparative analysis of the mouse [
9] and domestic dog genome [
10] or the human–chimpanzee genome comparison [
11], which are foundational for comparing large genomic datasets, with an understanding of evolutionary relationships and genetic function.
The aim of this study is to analyze the distribution of gene counts and genome sizes within a population of sequenced genomes from organisms representing the three domains of life—Bacteria, Archaea, and Eukaryota—using data collected in the National Center for Biotechnology Information (NCBI [
12]) genome database [
13].
Basic statistics in functional genomics, such as the distribution of gene counts across known genomes, reveal considerable variation influenced by factors including organismal domain, environmental conditions, genome architecture, and evolutionary history. The vast amount of empirical data available in genomic repositories, analyzed through comparative genomics, provides a detailed understanding of these distributions, highlighting both general trends and notable exceptions [
14,
15,
16].
Typical bacterial genomes contain between about 1000 and 6000 protein-coding genes, with a median of about 3500 [
17]. Archaea genomes contain between about 1000 and 5000 genes, with a median near 2500 [
18]. Eukaryote genomes are usually above 5000 genes centering around different values, e.g., 10,000 for single-cell organisms [
19], 20,000 for animals [
20], and 35,000 for plants [
19]. In prokaryotes, gene number distribution is unimodal, positively skewed, and centered near 3500 genes with a tail extending toward larger genomes. Eukaryotic gene counts exhibit broader and often multimodal distributions, primarily due to lineage-specific genome duplications and gene losses [
20].
In the presented paper, the analysis was based on the determination of a probability density function describing the likelihood of finding any genome failing within a specific range of values of the number of genes, normalized per unit length of this range. The advantage of a proper density function over a histogram is that its values are independent of the sample size.
It was checked that the simple model of genome origination and development as geometric progress is not enough for proper evaluation of the density function and leads to an “ultragenome catastrophe.” In subsequent analyses, accurate results were achieved by modeling the distribution as a linear combination of five Poisson functions, complemented by a single-step function with exponential decay and a constant-level component. This approach allowed us to interpret the observed genome frequency patterns as the outcome of a multi-path evolutionary process, with pathways varying in their rates of change, potential for differentiation, and, in some cases, culminating in gradual elimination. We also show that a simple transformation of gene number statistics can approximately describe genome log (size) statistics. Applied simple regression reveals the characteristic size of genes dominating in the smaller genomes of all domains of life. Extending the regression model to the nonlinear area of the gene number–genome size dependence, we proposed the size-driven mechanism of creation (origination and adding) of a new gene. As presented, the extension contains some speculative elements; it ought to be treated as a conceptual proposal waiting for additional biological validation. The model defines the two distinguished types of genes: extensive ones, which increase genome size when attached, and intensive ones, which do not change the genome size when they are emerging. This provided a compelling explanation for the observed shape of the relationships in the experimental dataset. The theoretical model with fitted parameters could also predict the total fractions of extensive and intensive genes. Finally, based on a successful comparison between estimated ratios and experimental data, we were able to associate extensive genes with protein-coding genes and pseudogenes, and intensive genes with non-coding genes. In general, statistics are often perceived as a tool for lumping all variables into a single analysis. In this study, we aimed to challenge that perception by applying a more nuanced approach.
4. Discussion
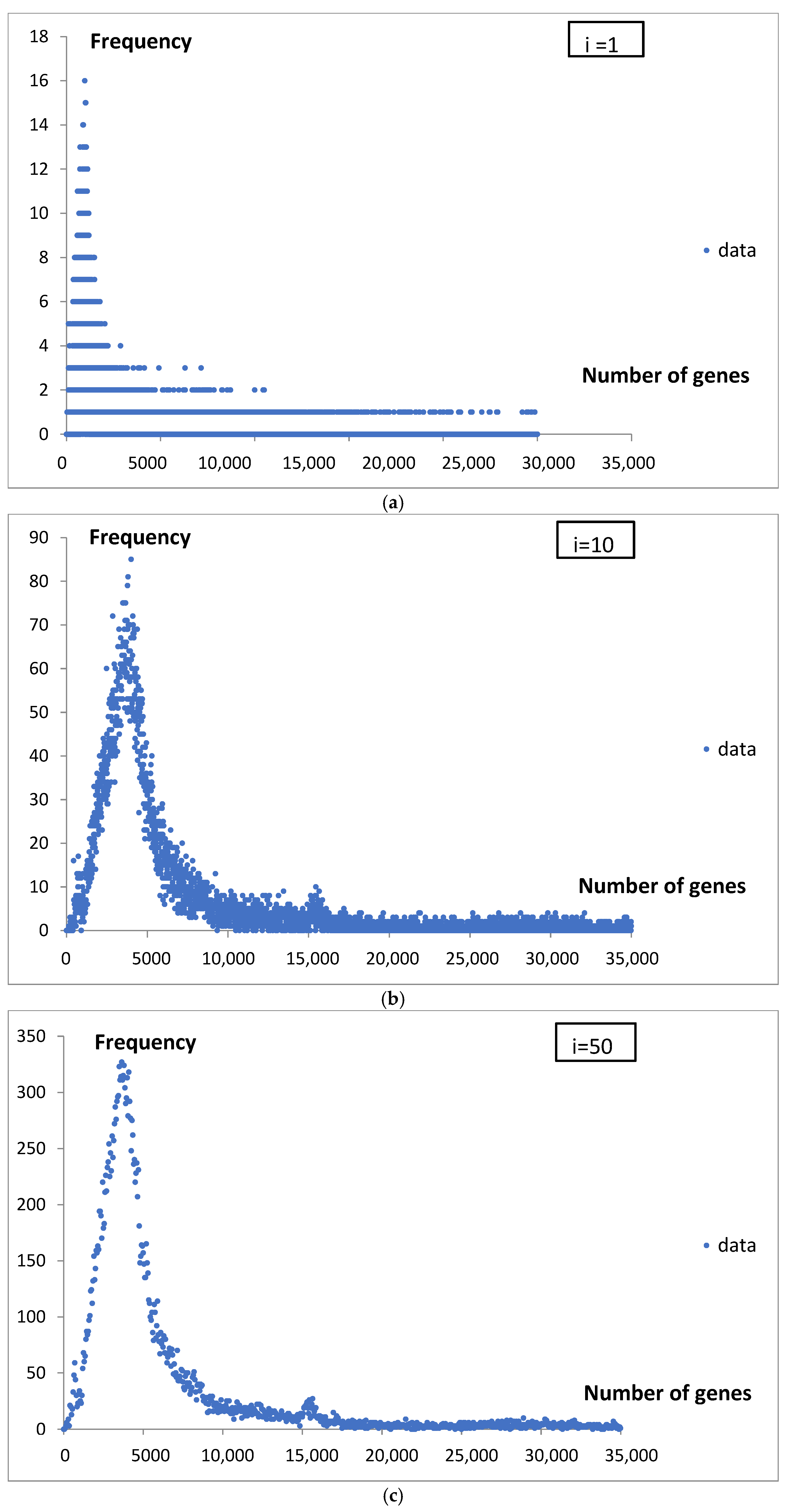
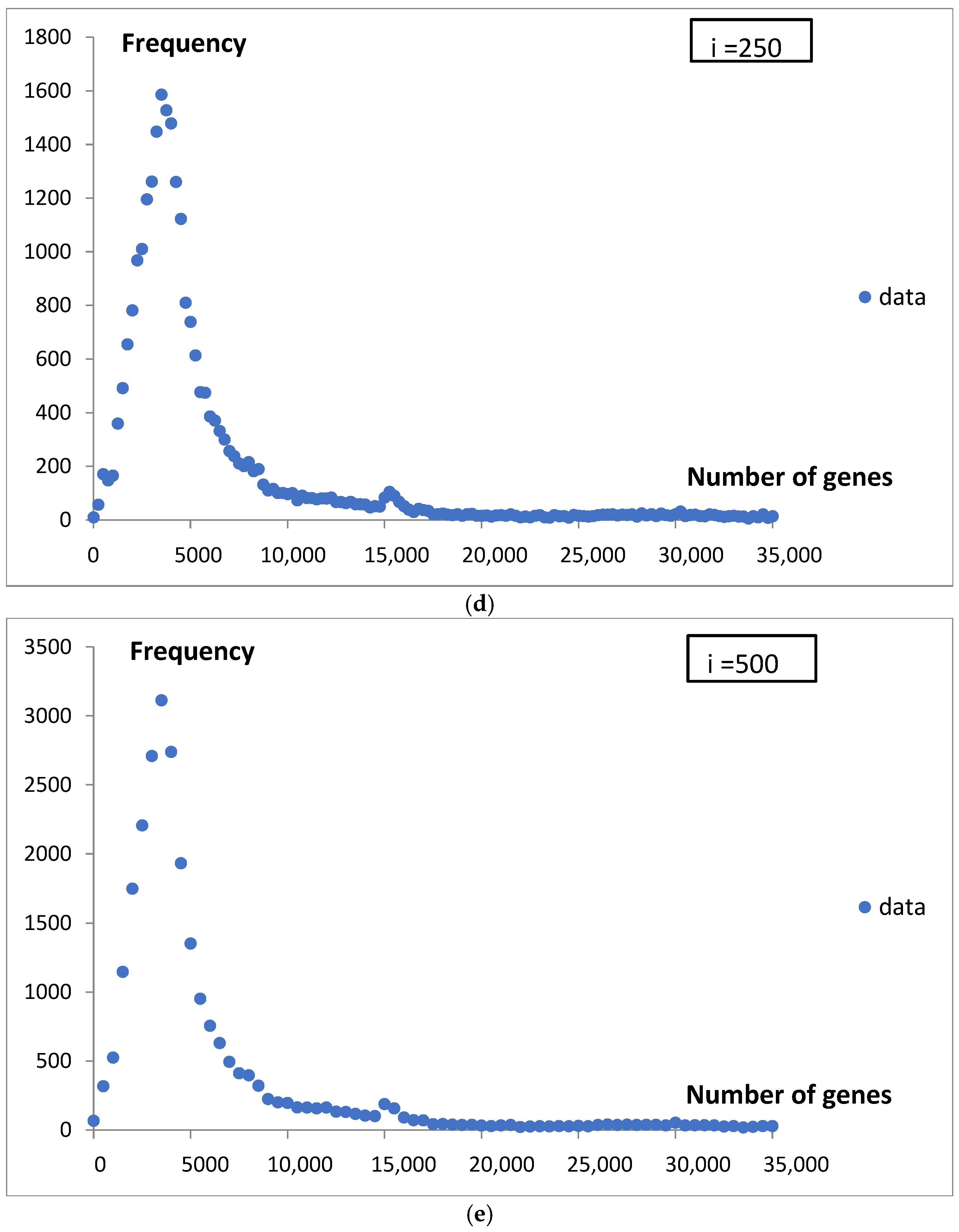
When we analyze genome statistics from large bioinformatics databases, it seems to us that, due to the significant scatter, different sizes of probes, and lack of a mathematical description of the shape, we cannot say much about individual cases, let alone the processes involved in them. Grouping data into classes and determining probability density introduces a standard that can facilitate their discussion and possible comparison with other studies. We illustrated this in
Figure 1a–e and
Figure 2.
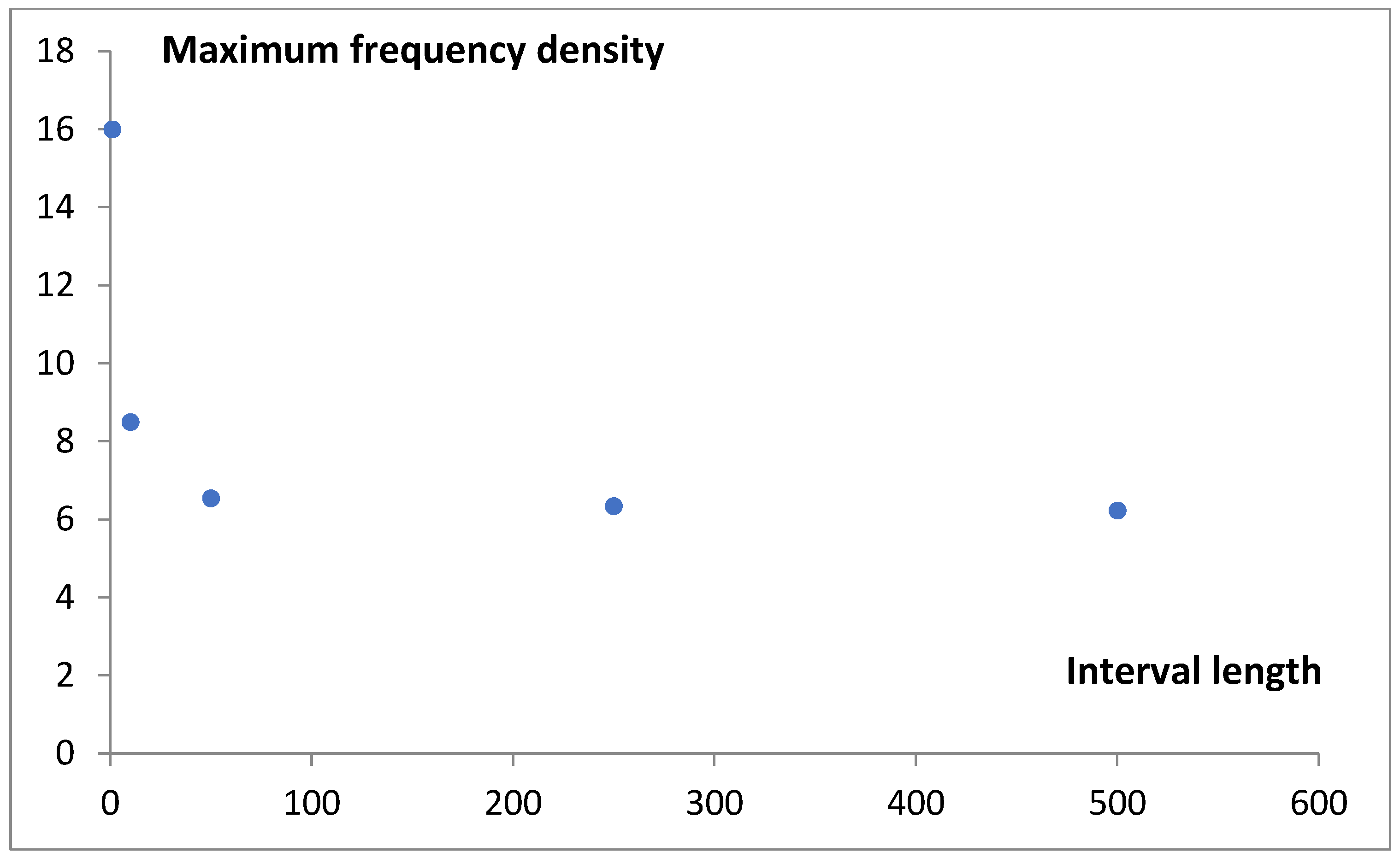
Although the values obtained using Rice’s rule (h
R = 59) and the Freedman–Diaconis rule (h
FD = 225) support the third and fourth data points in
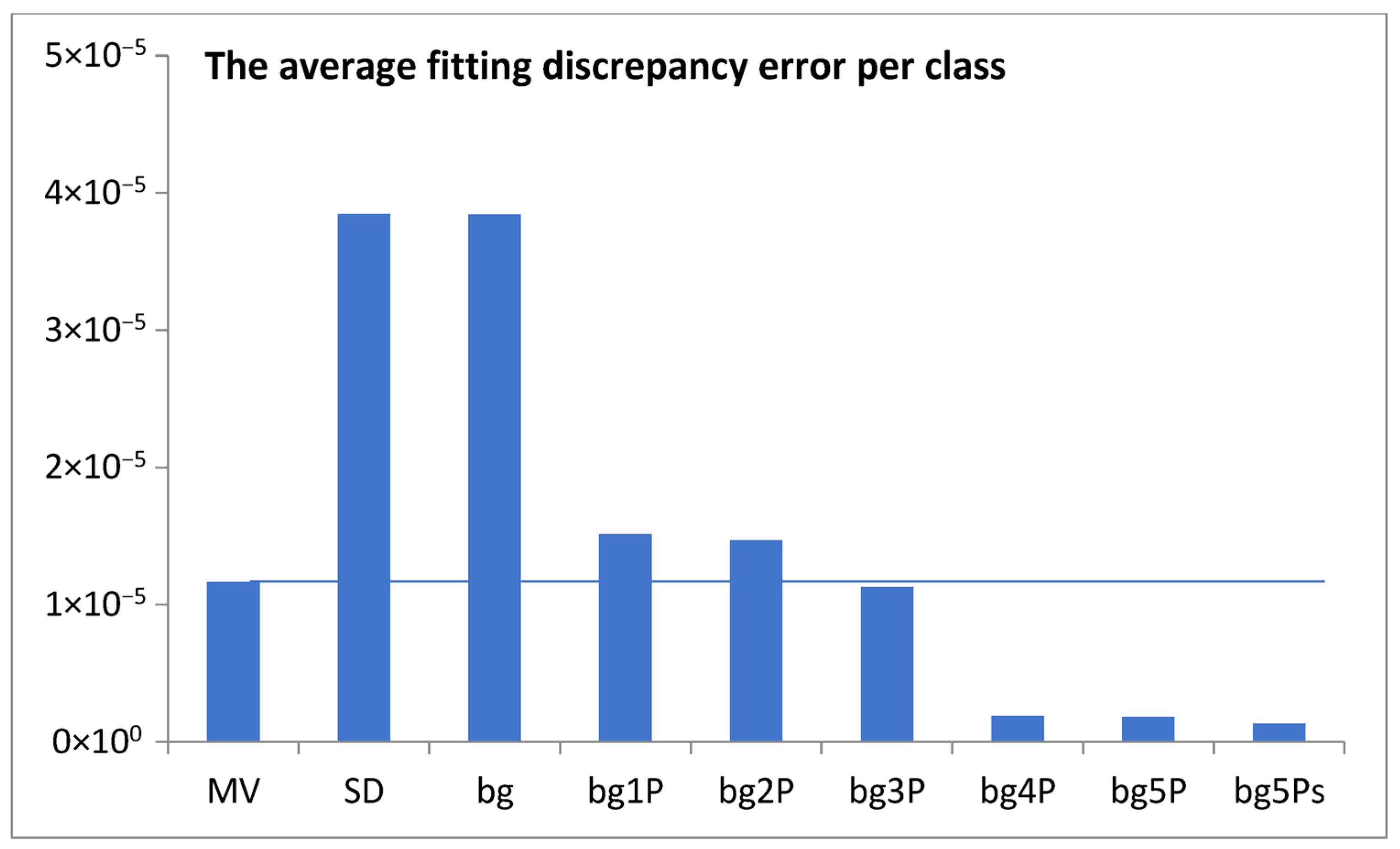
Figure 2, we choose to use a bin width of 500 genes in the histogram analysis. This choice slightly smoothed the data curves, allowing us to focus on the most significant features of the analyzed functions. The subsequent mathematical modeling aimed to deepen our understanding of the nature of the observed statistical distributions.
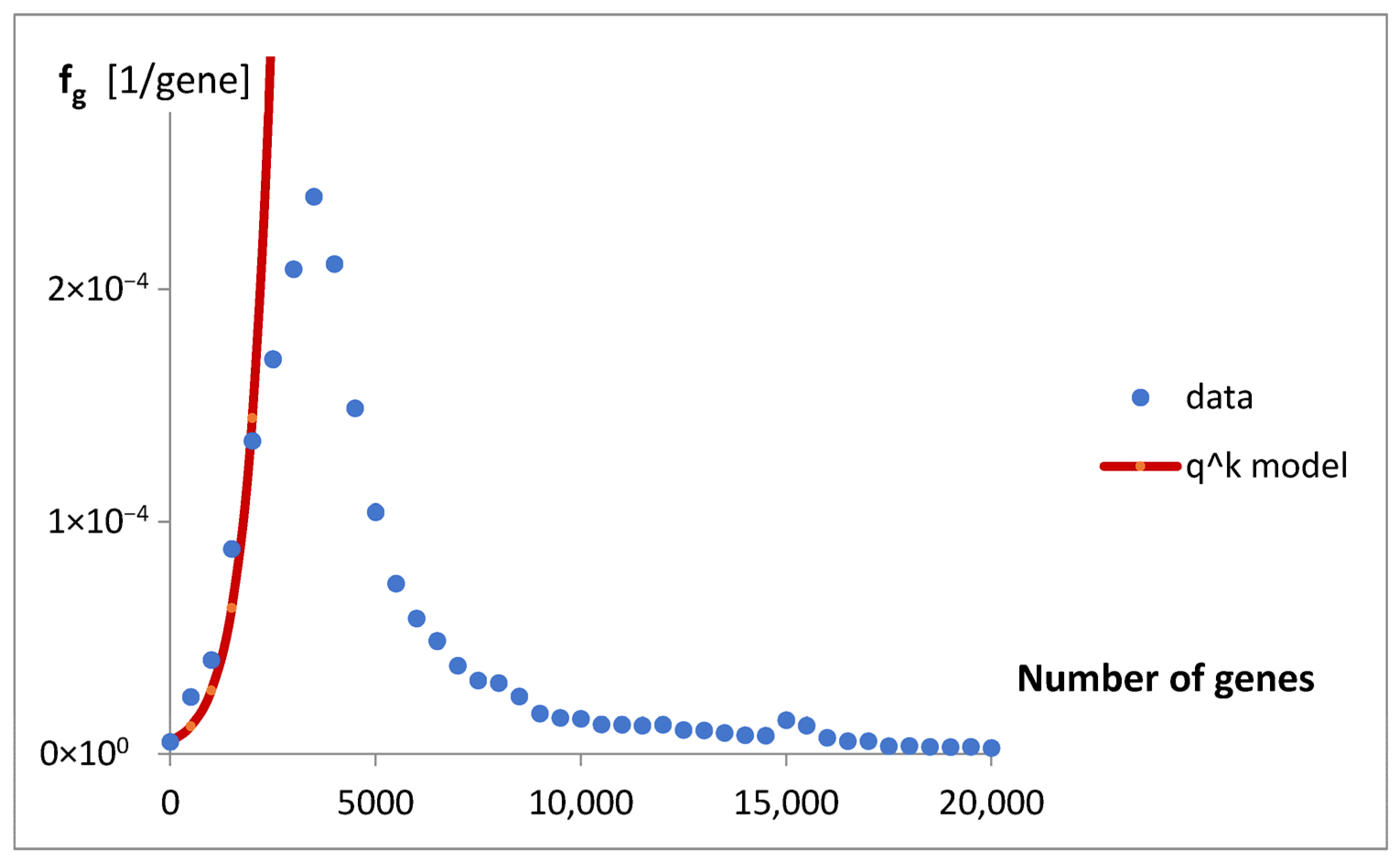
In the case of genomes analyzed in this paper, to free ourselves from the catastrophic predictions of the geometric model (
Figure 3), we sought statistics that would have an inherent ability to limit themselves.
The Poisson distribution models a series of discrete events occurring within a fixed time interval, where λ represents the average number of events (rate) in that interval. The events occur at random, with their exact timing being both independent and memoryless. It decays with the number of events. The exact value of the interval is not important. It may even be infinite.
According to our idea, events can represent genome transitions into subsequent classes containing genomes with an increased number of genes. Thus, the number of genes is treated as a kind of pseudo-time. In our model, on average, the 500 additional genes are significant when considering the latter state. Thus, a chosen group of N genomes, evolving at the rate λ, are currently observed according to a Poisson distribution formed throughout the entire evolutionary period. Moreover, we believe that all genomes initially evolved together, but over time, some groups of them broke off and moved on at different speeds.
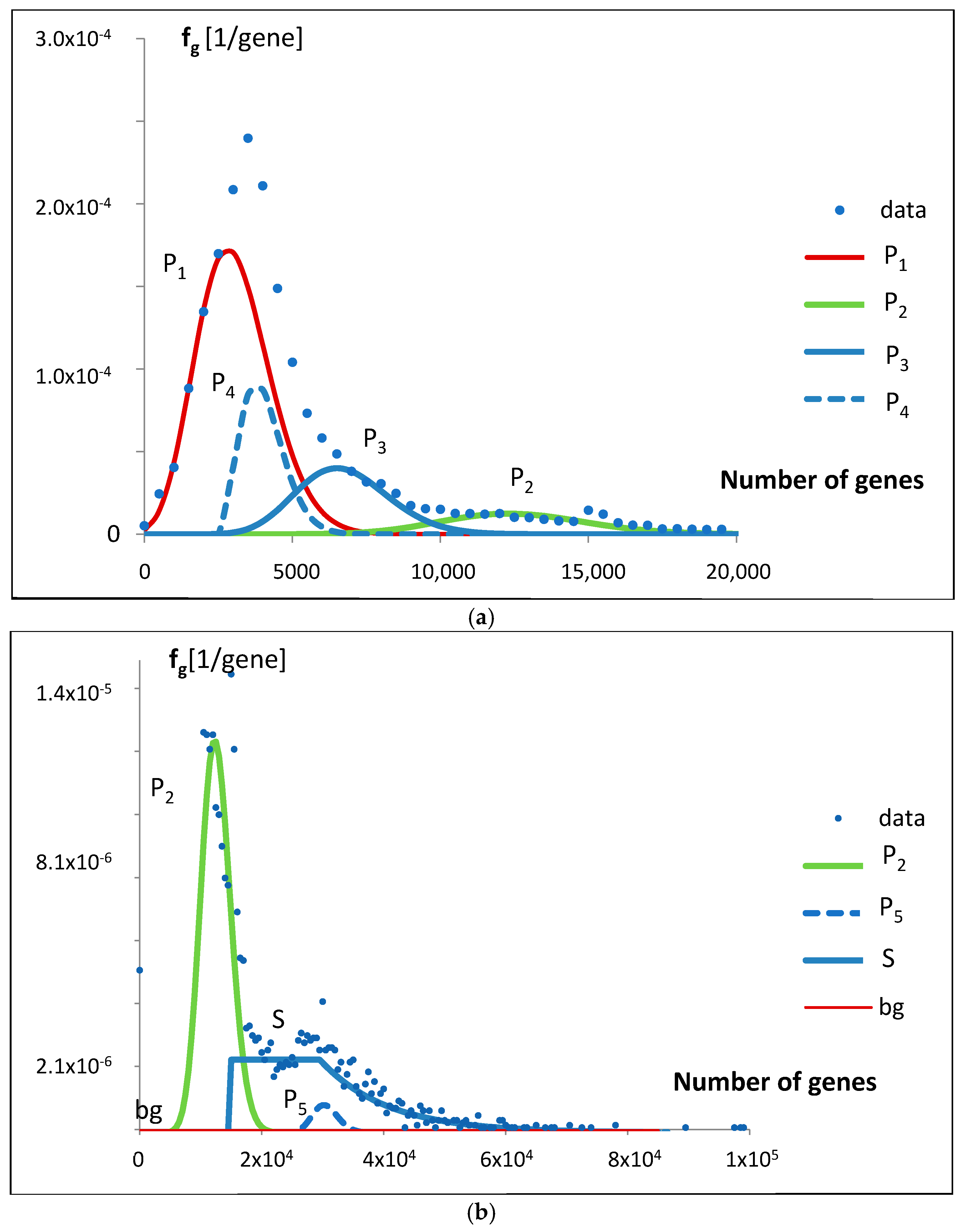
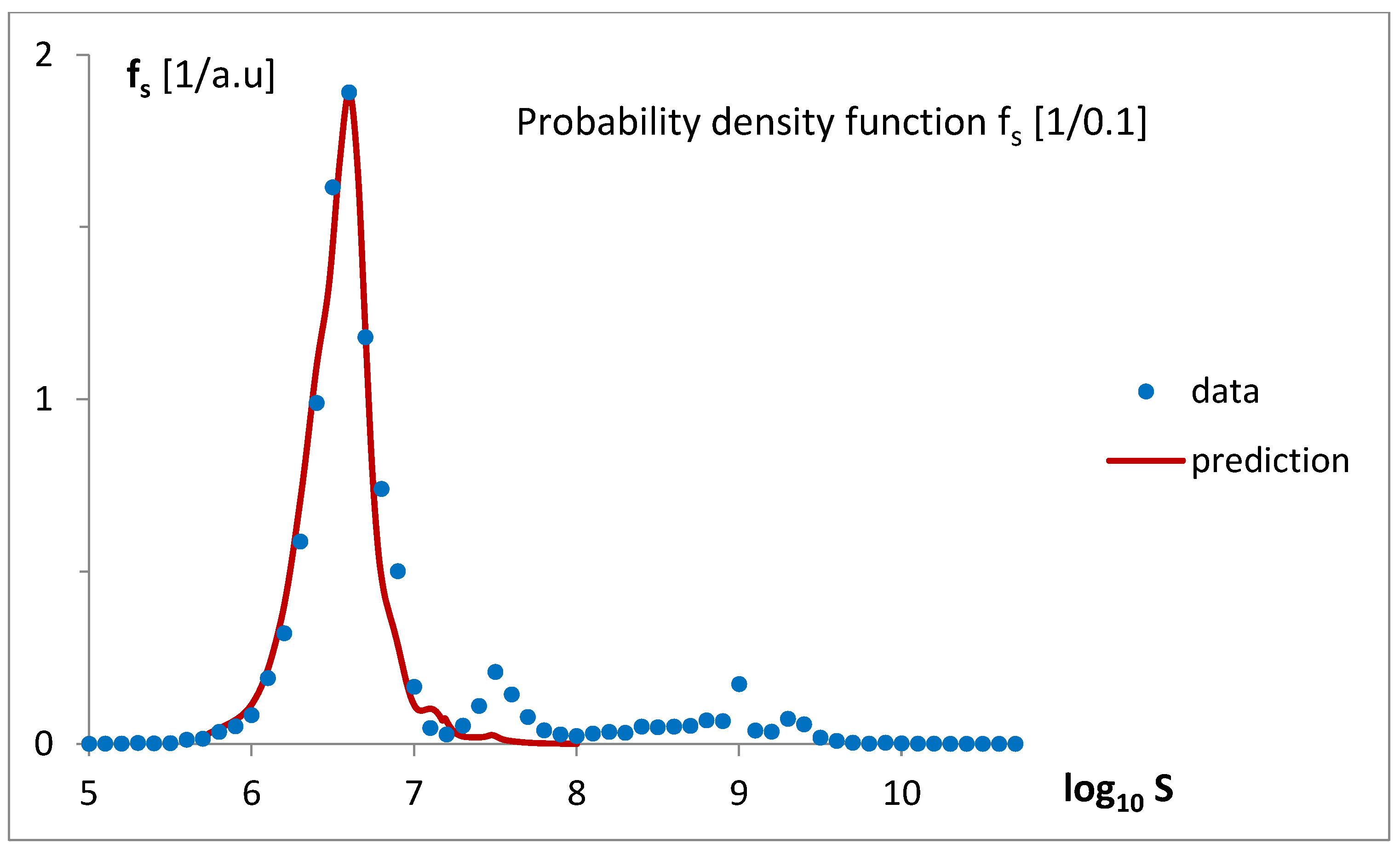
This idea was qualitatively described by the model bg5Ps, which was gradually developed and examined in a series of attempts (
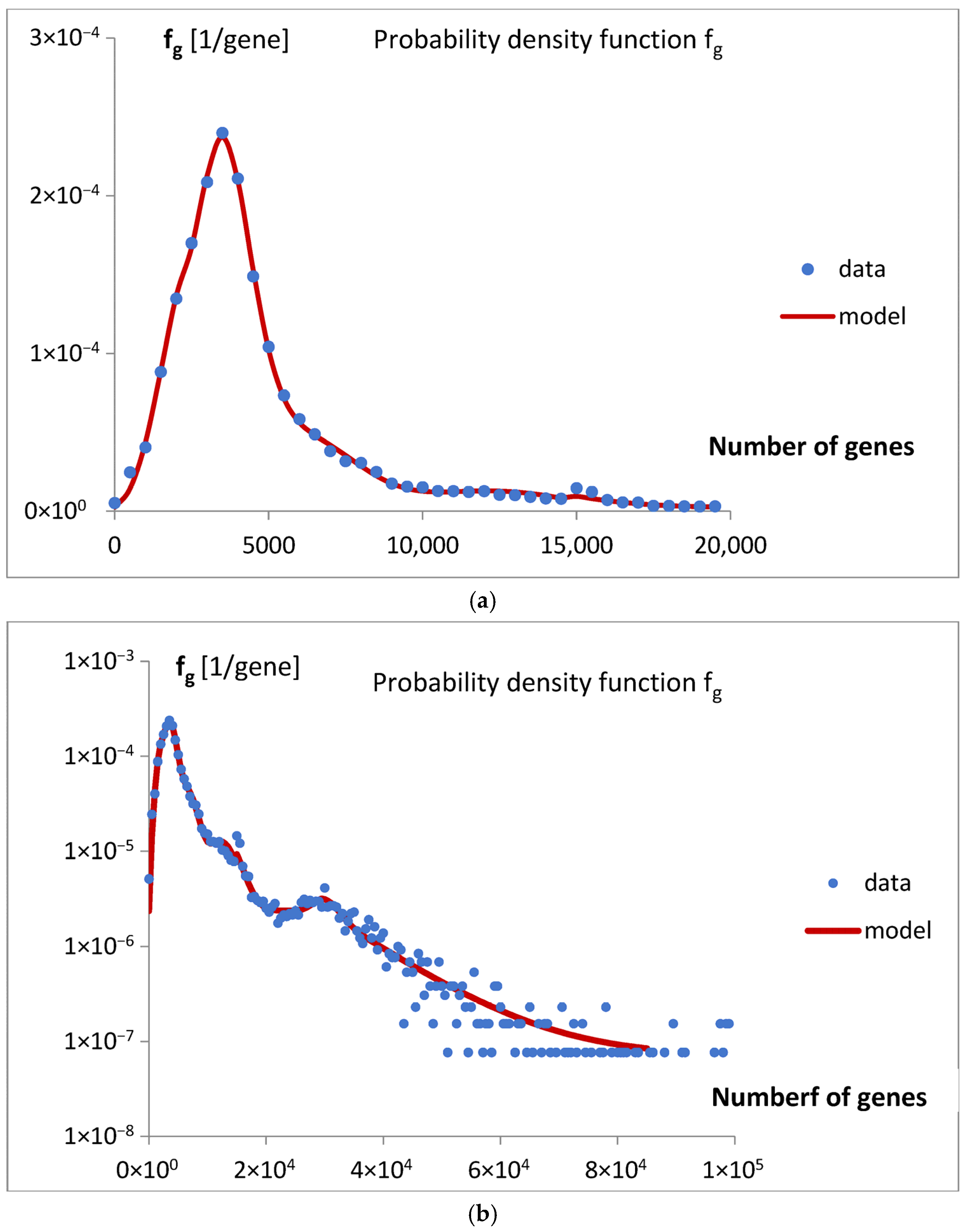
Figure 4). The final fitting of the model to the data for the probability density function of the gene numbers (Equation (2),
Figure 5a,b,
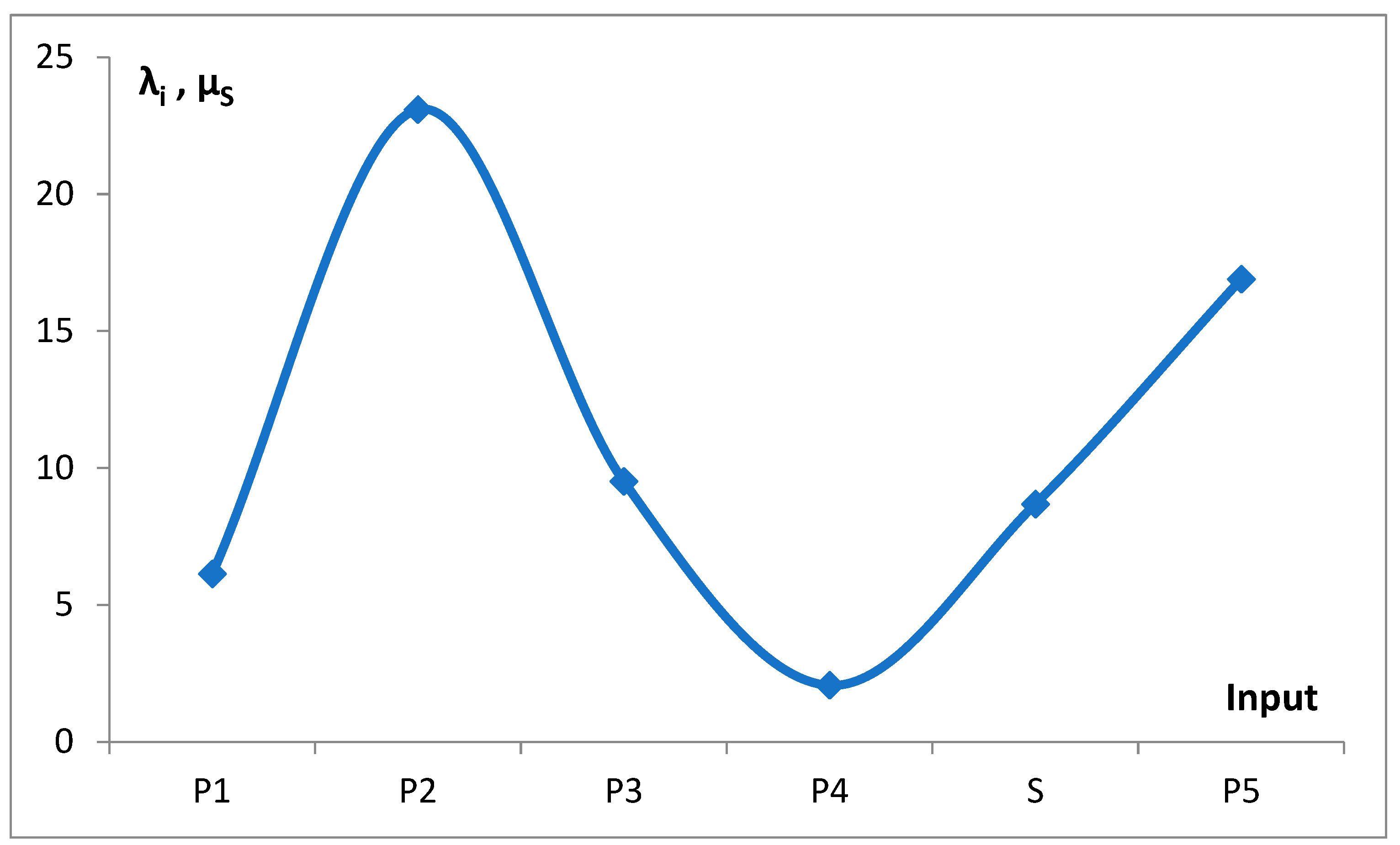
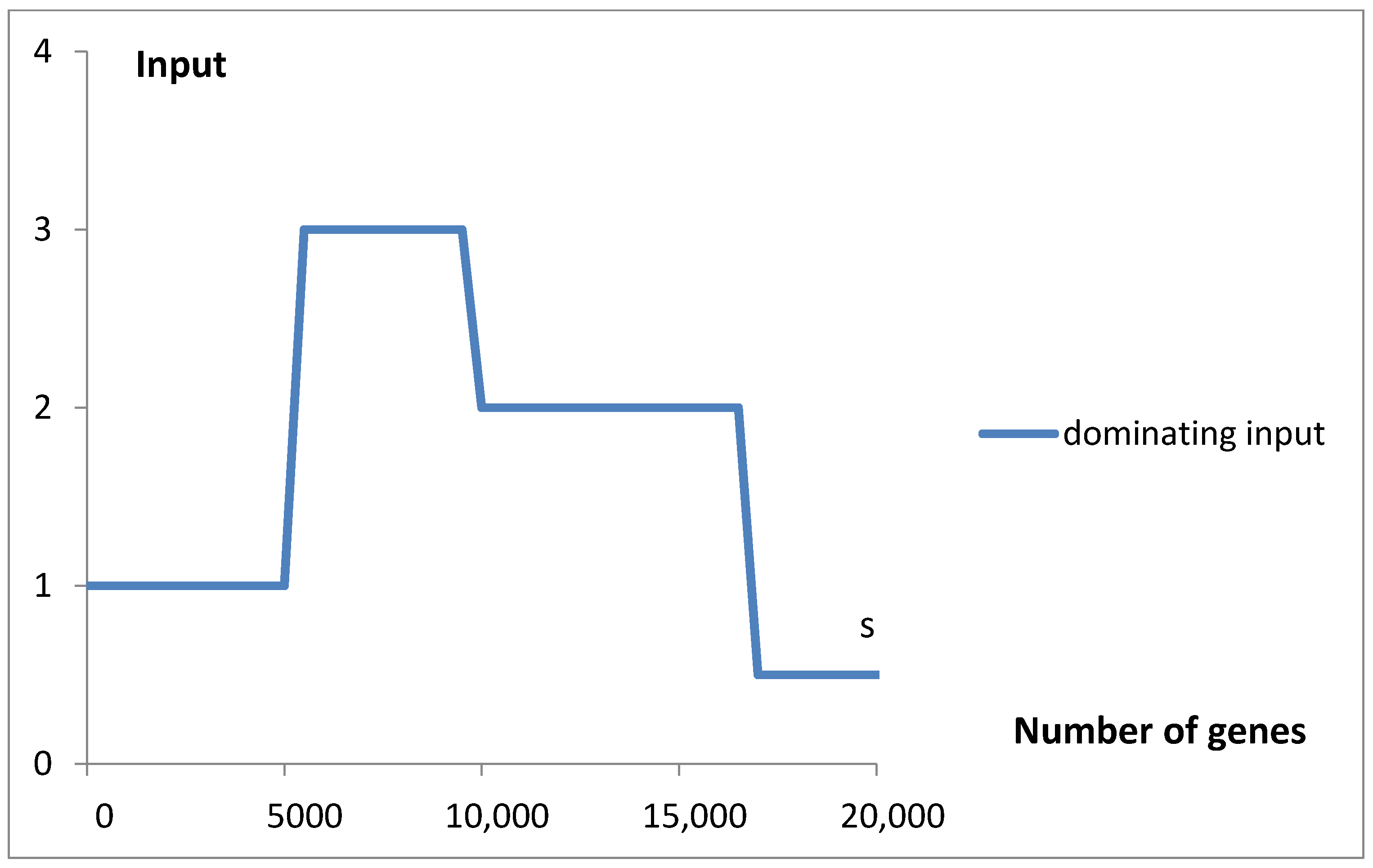
Table 1) and predicted oscillating rates of the considered inputs (
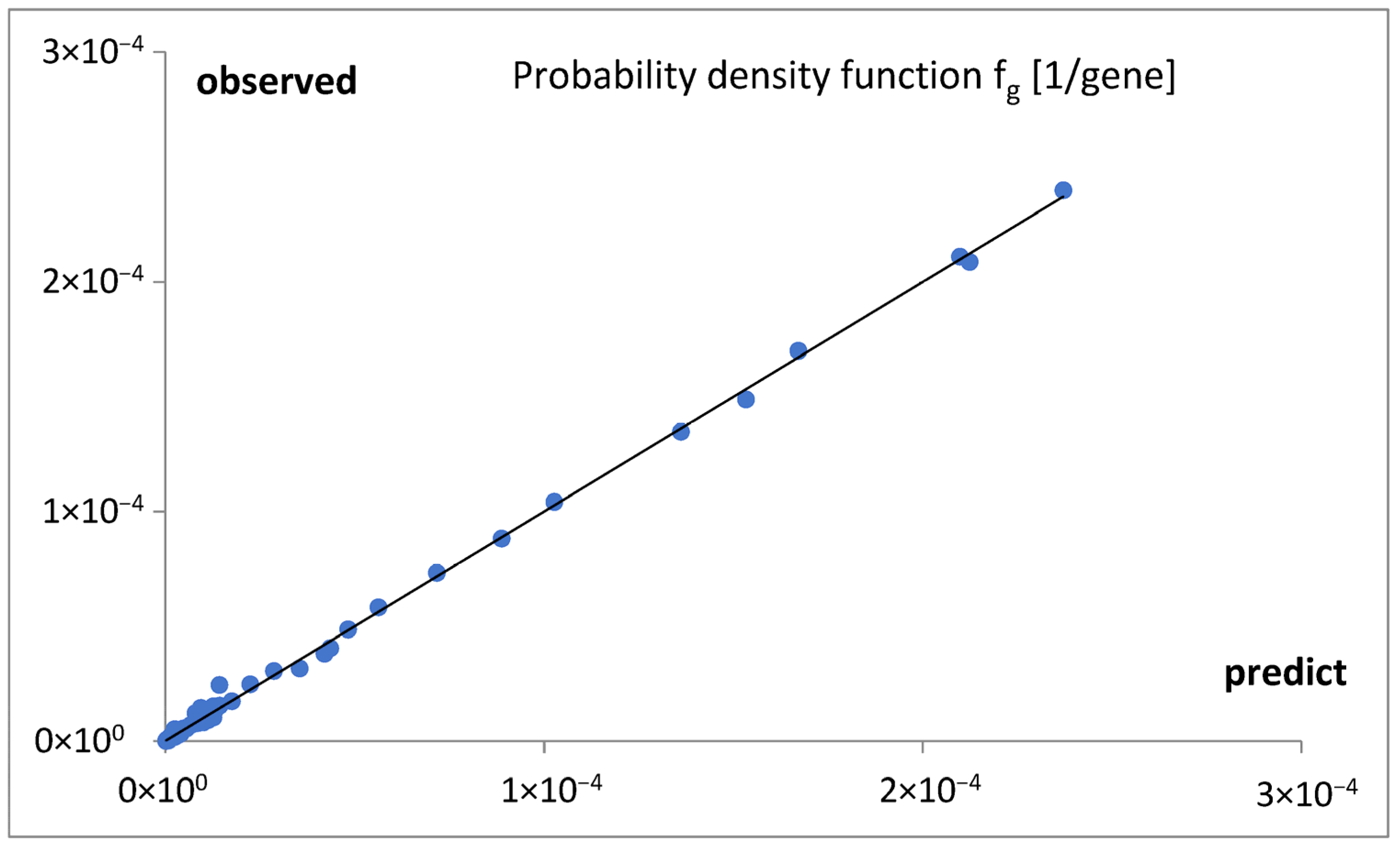
Figure 6), at the obtained quality (
Figure 7), allow us to draw several conclusions.
When decomposing the mathematical model into constitutive components, a landscape of smooth inputs is revealed (
Figure 8a), supplemented by a characteristic decaying short jump in the range of larger gene numbers (
Figure 8b).
Thus, we can identify five evolving Poissonian groups with the different values of N
i and λ
i, starting at different classes (stages of evolution), and a group appearing as step input with an exponential decay. There is also a uniform background of the order of the standard deviation (
Figure 3), which we take for noise of errors. The largest genome content is found in Poissonian group 1, where N
1 represents a fraction a
1 = 0.531584 of the total (
Table 1), accounting for over 53% of the analyzed population, specifically 13,808 (a1 × 25,975) genomes. Despite this, due to evolutionary dispersion of genome gene numbers, it dominates only in the range of the smallest sets of genes (
Figure 9). Genomes with a higher number of genes are predominantly associated with Poissonian group 3. Using
Figure 9, with a low probability of error, one may assign
S. cerevisiae (6477 genes) to the Poissonian group P3 and
H. sapiens (59,715 genes) to the group S resembling step input.
We believe that Poissonian groups can represent genomes progressing through successive phases of evolution, without preserving less distinct intermediate forms or retaining the forms that quickly disappear. In contrast to this, the step-like group represents genomes, which, when moving from class to class, approximately at the same rate, leave behind replicas of their representatives. However, after a certain number of transitions, the progress of this group exponentially vanishes.
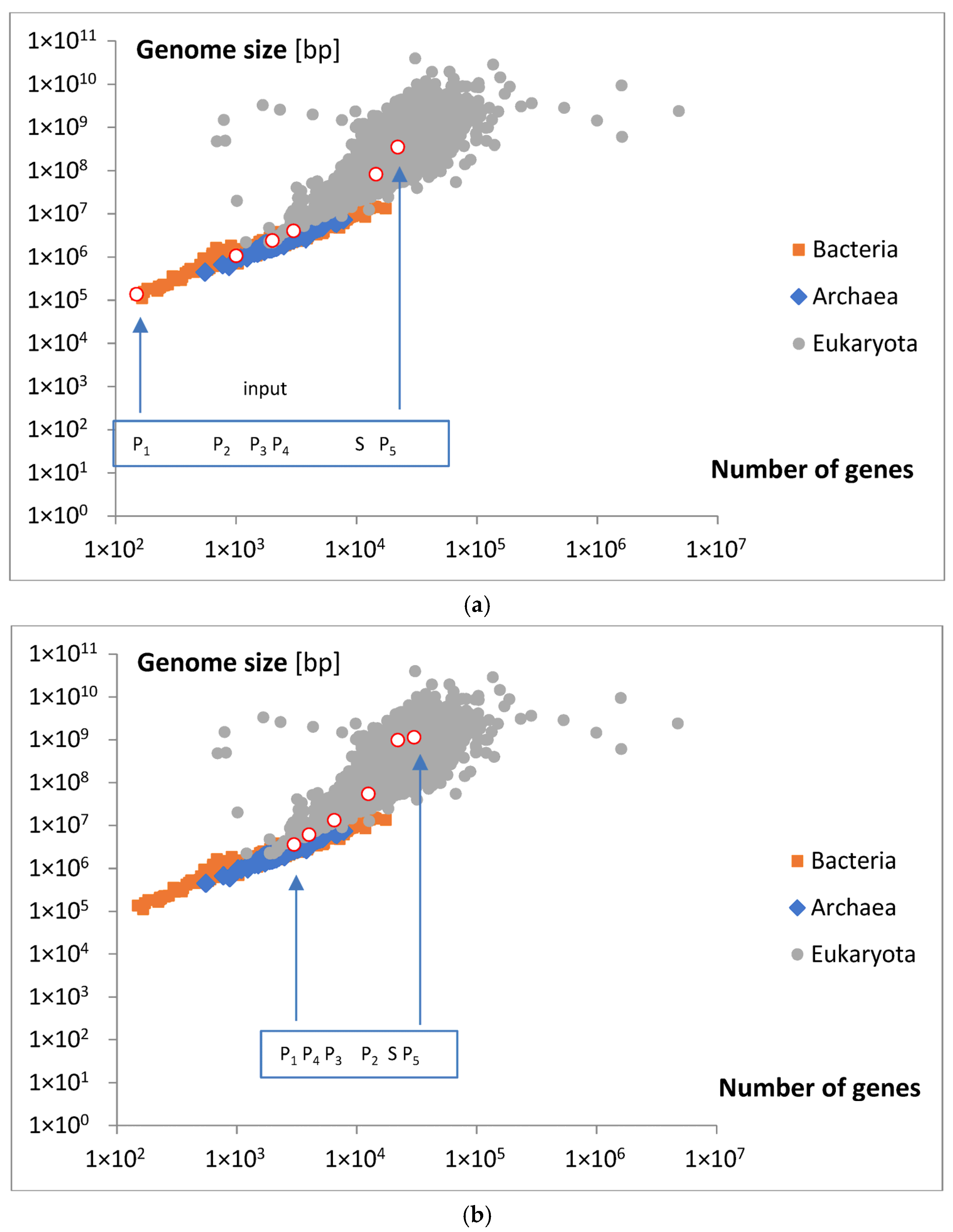
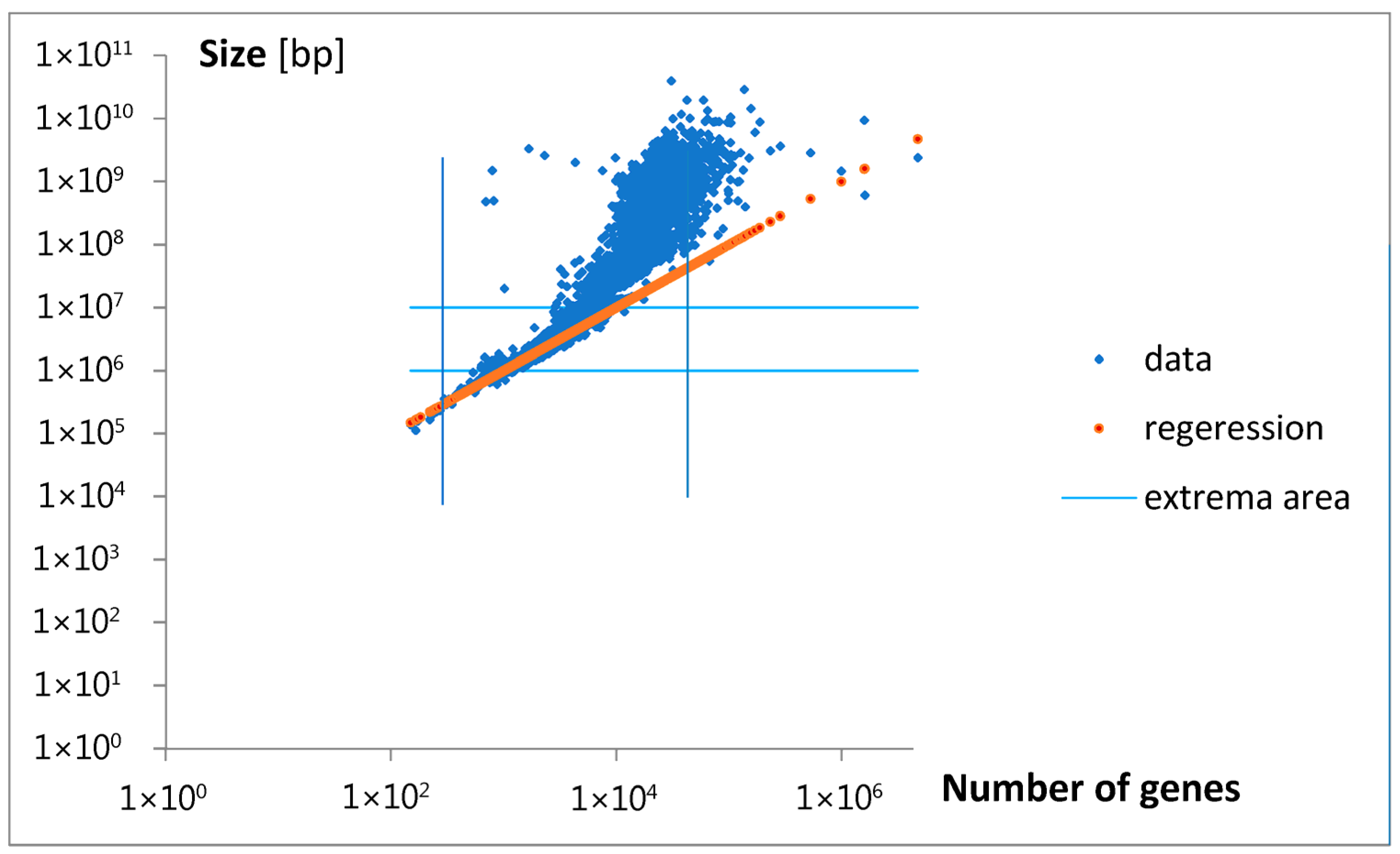
Analyzing the magnitude of the considered inputs, presented in
Figure 8a,b, and locating the appearance of the discussed groups on a map illustrating the relationship between gene numbers and genome size,
Figure 10a,b, we can approximately conclude that only Poissonian group 1 (P
1) may contain the smallest bacteria and archaea (
Figure 10a). Some genes in this area for eukaryota, with bp > 10
7, are probably an error. The P
2 group may contain the smallest and moderate eukaryota. On the other hand, the P
3 and P
4 groups may cover moderate and big bacteria (
Figure 10b). On the contrary, the S group does not cover any bacteria or archaea but includes moderate and big eukaryota. The last, P
5 group, contains only moderate eukaryota. Note that the P
2, S, and P
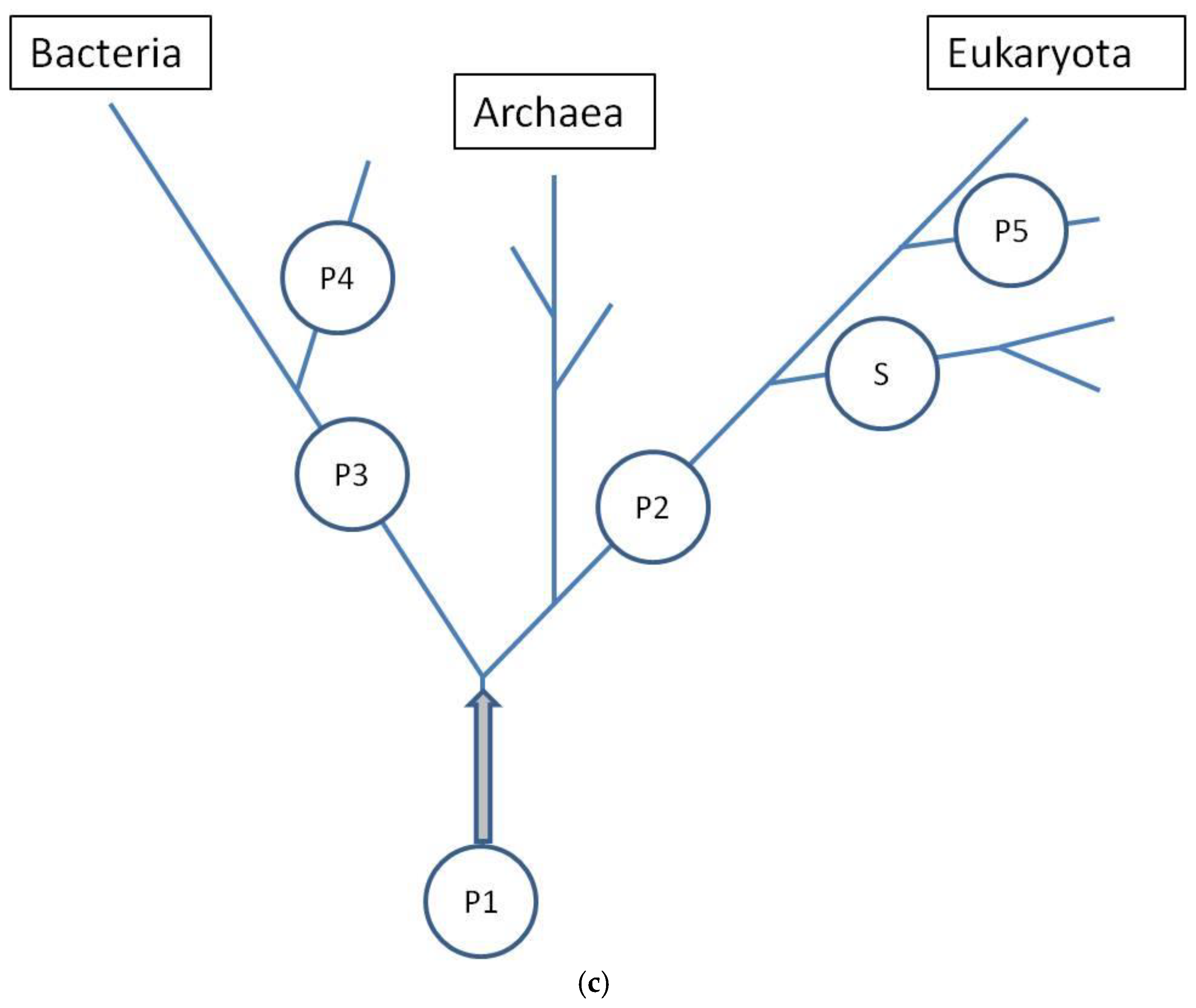
5 inputs increased in size more than was predicted by the emerging initial linear regression. To aid in the visualization of these possibilities, hypothetical points of origin for the discussed inputs along the evolutionary tree are illustrated in the schematic shown in
Figure 10c.
It is important to acknowledge certain limitations of the above conclusions. Due to overlapping contributions and their theoretically infinite range, it is not possible to establish a flawless correspondence between a class of organisms defined by a given gene number and a single characteristic input. Only genomes with fewer than 500 genes can be strictly related to a single input, P1 (neglecting the background error, bg). It is usually a more or less probable relation, so we can only indicate the dominating input among others in a settlement of a certain area of gene numbers. In the opposite direction, search predictability looks better. Only inputs P1 cannot be strictly related to one domain of life. Other inputs can be attributed under the assumption of domain inheritance, whereas P1 lack any domain-specific attribution.
An example below illustrates the above features in practice. Let us consider a histogram bin with gene numbers in the range 5001–5500. It contains genomes that are 94% bacterial, 1% archaeal, and 5% eukaryotic. On the other hand, the inputs contribute to the total number of genomes in this class, as follows: P
1-47%, P
3-23%, and P
4-30% (
Figure 8a). Other inputs may be neglected. One may conclude that P
1 input delivers 6% genomes for archaea and eukaryota and 41% for bacteria. So, 44% of bacterial genomes in this class are P
1 type.
As illustrated by the above example, under certain conditions, the input type of a genome belonging to a specific domain and class can be identified, albeit with a relatively high probability of error. The question arises if additional attributes, e.g., kingdoms, phylogeny, lifestyle, and environment, could reduce this uncertainty up to the level of species. This may be an interesting area for separate statistical study, using machine learning methods, especially classifiers.
Finding a mathematical formula for the probability density function, f
g, regarding gene number, helped in the theoretical determination of the probability density function, f
s, regarding logarithmized genome size. Fitting the transformed f
g (Equation (3a,b)) to the data provides a good estimate of the experimental f
s values (
Figure 11), but only in the gene number range where the density function exhibits a characteristic dominating bell-shaped peak. The linear regression applied during transformation (Equation (3a)), as illustrated in
Figure 12, reveals the limited applicability of this approximation, fortunately confined to the gene number range where genomes occur most frequently, though only among relatively small-sized genomes. The revealed characteristic size per gene (p
1 = 1000) is in the typical reported range for prokaryotic and small or moderate eukaryotic genomes [
27].
In general, the relationship between gene number and genome size is not linear. The first small “acceleration” in the overall genome size may be related to the maximal inputs of the components P
1, P
3, and P
4 (
Figure 10b). The next higher increase falls in the domain of maximal input P
2. The highest increases may be related to maximal inputs S and P
5. The appearance of the discussed inputs may also be associated with a strong divergence in genome size at an approximately constant number of genes.
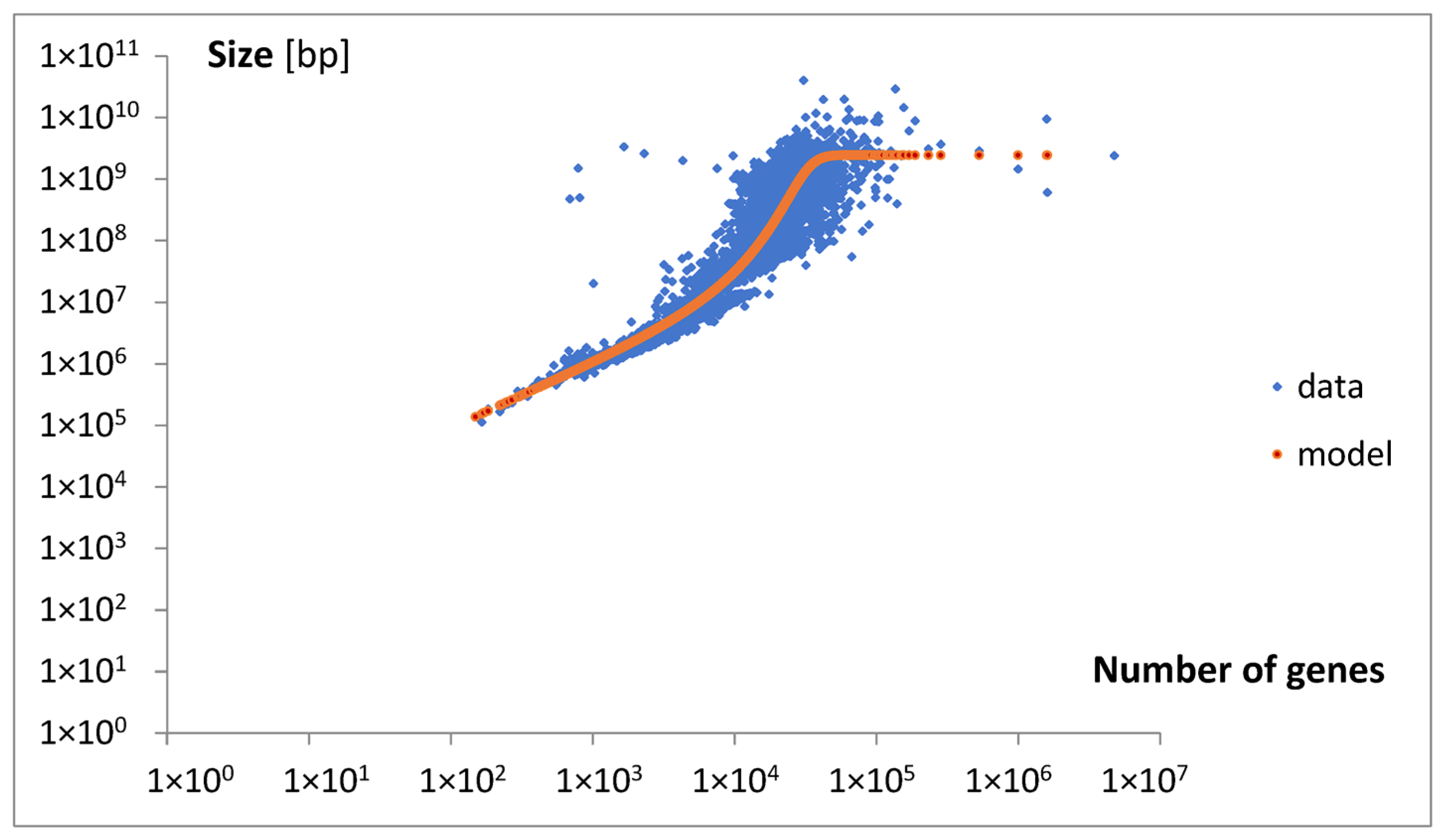
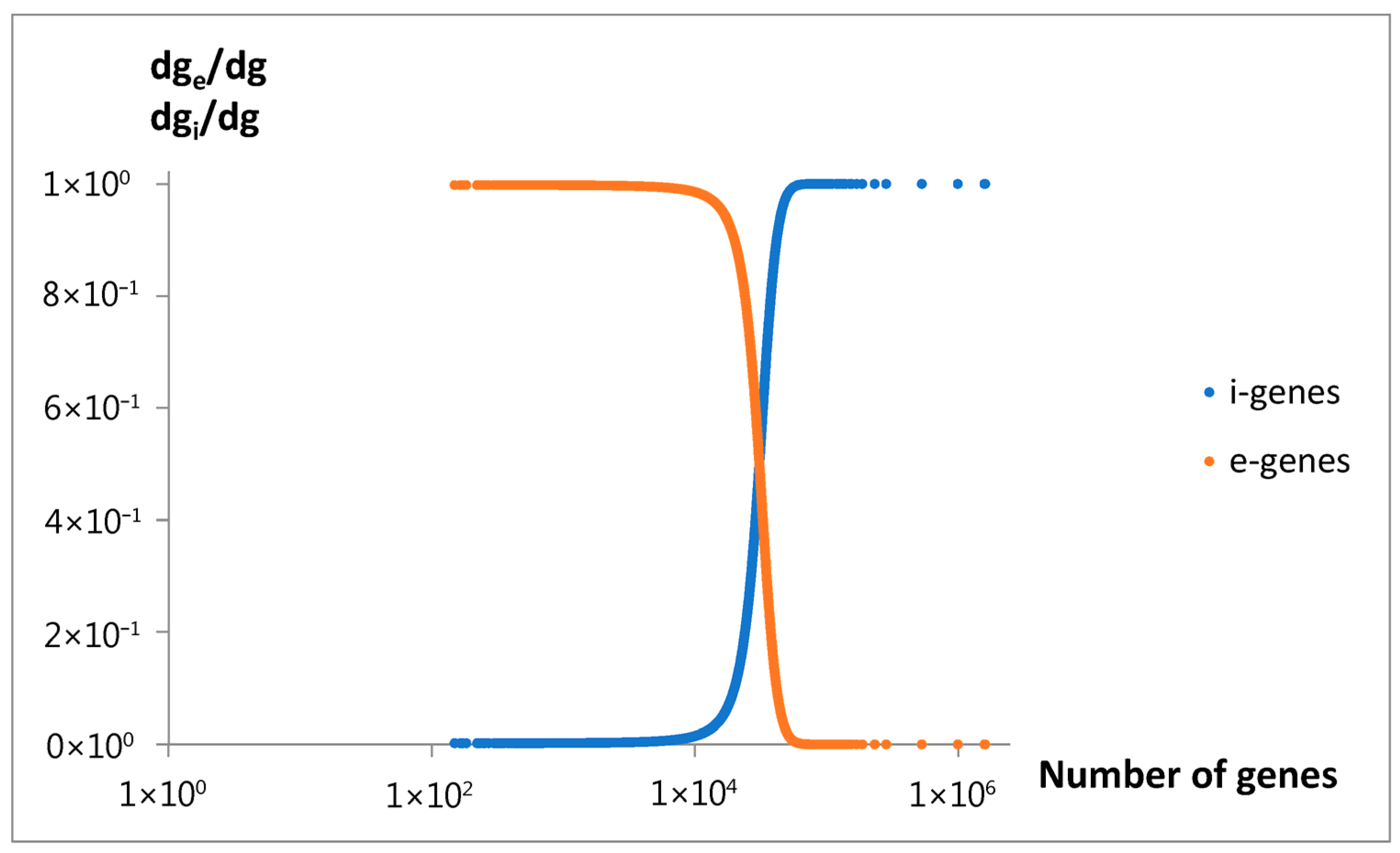
To explain the observed nonlinear behavior, an extended mathematical model of genome size evolution was introduced. The nonlinearity (Equation (13),
Figure 13) is interpreted as resulting from a precise fractioning of new genes into two categories: intensive genes (dg
i), which do not affect genome size, and extensive genes (dg
e), which contribute to its increase. A key variable of the model used in the constitutive equation (Equation (5)), relating the small change in genome size (ds) with the change in dg
e, is the average genome size change per extensive gene, l
e. At the beginning, intensive genes may be related to emerging overlapping genes [
28], and extensive genes may be referred to attached genes from duplication [
29], or horizontal transfer [
30]. According to this model, the fraction of new extensive genes decreases proportionally to the l
e (Equation (6)). This assumption describes the self-limiting attaching of large extensive genes, and the consistently increasing emergence of intensive genes (Equation (8)). A basic source determining such effects may be the evolutionary tendency toward minimization of the size of genome and maximization of the number of its genes. Discussed effects could be especially advantaged in the nucleated (eukaryotic) cells, where the mutations producing overlapping genes prevent enormous increase in the size of big genomes. When applying the discussed model, we also have to assume that the average size of attached new material per extensive gene increases with genome size (Equation (7)).
In summary, according to the model, we may expect that with the increase in genome size, the length of new extensive genes increases, but their fraction decreases. This may lead to a slowdown in genome size growth due to the so-called parabola effect. In the extreme case, when the length le reaches its maximal value lemax, genome size expansion may cease entirely. An accompanying increase in the number of intensive genes could further inhibit growth in the total number of genes, ultimately leading to a complete halt. The predicted maximum length, lemax, is approximately 498,857.
Approximately analyzing the data in
Figure 13 with Equations (6) and (7), we may obtain the following results. For the number of genes around g = 1500 and s = 10
6, the result is l
e = 200, l
e/l
emax = 0.0004, and dg
e = 0.9996dg. For g = 20,000 and s = 2.5 × 10
8, the resulting value is l
e = 50,000, yielding l
e/l
emax = 0.1, and dg
e = 0.9dg. For g = 50,000 and s = 2.5 × 10
9, the result is l
e = 500,000, l
e/l
max = 0.998, and dg
e = 0.002dg.
The discussed model predicts that the minimal length of an extensive gene (le0) is approximately 1009, which is close to the value p1. The values g0 and s0 for the minimal genome used in the model were ultimately set equal to those for the minimal genome in the dataset.
The determined values of the parameters l
emax and a (
Table 2) are effective for modeling the data across the entire range of gene number variability. In the real case, they may differ for the different groups of genomes and could have been slowly modified during evolution to regulate the rate of genome size increase.
As shown in
Figure 6, the dependence of mean values λ
i and µ
s, which characterize the rate of the consecutive inputs to f
g, on the number of genes exhibits an oscillating pattern. The initial upward trend in the rate of emerging new genes with the increase in the gene number, or the genome size, is nothing special. In light of the discussed findings, the superimposed oscillations, e.g., the slowing of the rate in P
3 and P
4, cannot be explained as the result of larger overlapping by an increased number of intensive genes. The mentioned inputs start (
Figure 10a) in the region of unnoticeable changes in the intensive gene income (
Figure 14). As can be seen, the slowdown in P
5 by the same mechanism is also doubtful. It is probably because the slight increase in the fraction of new intensive genes cannot seriously modify the ratio of the total number of these genes to all genes. Thus, the reason may be of a more complex evolutionary nature, modifying the fitness of the genome. In this way, it may also produce a rate increase in inputs P
2 and S. Slow-evolving group P
3 may contain fungal genomes, and faster-evolving groups P
2, S, and P
5 dominate in the gene numbers area of invertebrates, vertebrates, and plants. These kingdoms may have different evolutionary strategies.
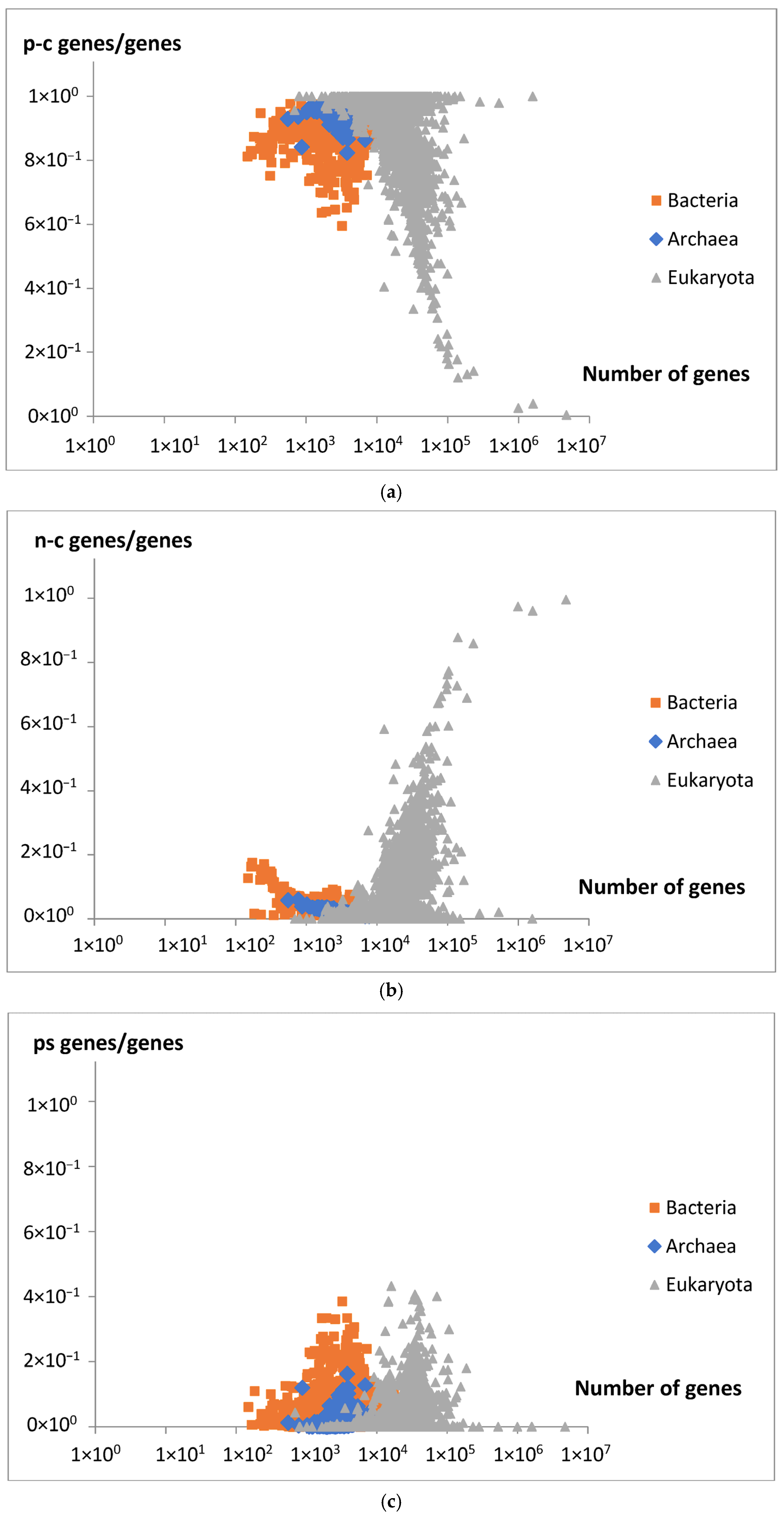
The comparison between the model’s genome size predictions and the experimental data supports the model’s validity in accurately describing the overall relationship between gene number and genome size. Furthermore, as predicted by the model, a decrease in a fraction of new extensive genes (
Figure 14) is consistent with an observed decrease in the fraction of protein-coding genes (p-c genes) in larger genomes (
Figure 15a). It is also consistent with an increasing number of non-coding genes (n-c genes) (
Figure 15b), which can be related to the increasing fraction of new intensive genes (
Figure 14). Pseudogenes (ps genes), like pc-genes, start to vanish around gene number 3 × 10
4 (
Figure 15c). The results in
Figure 15a,b show that the dominant changes concern eukaryota.
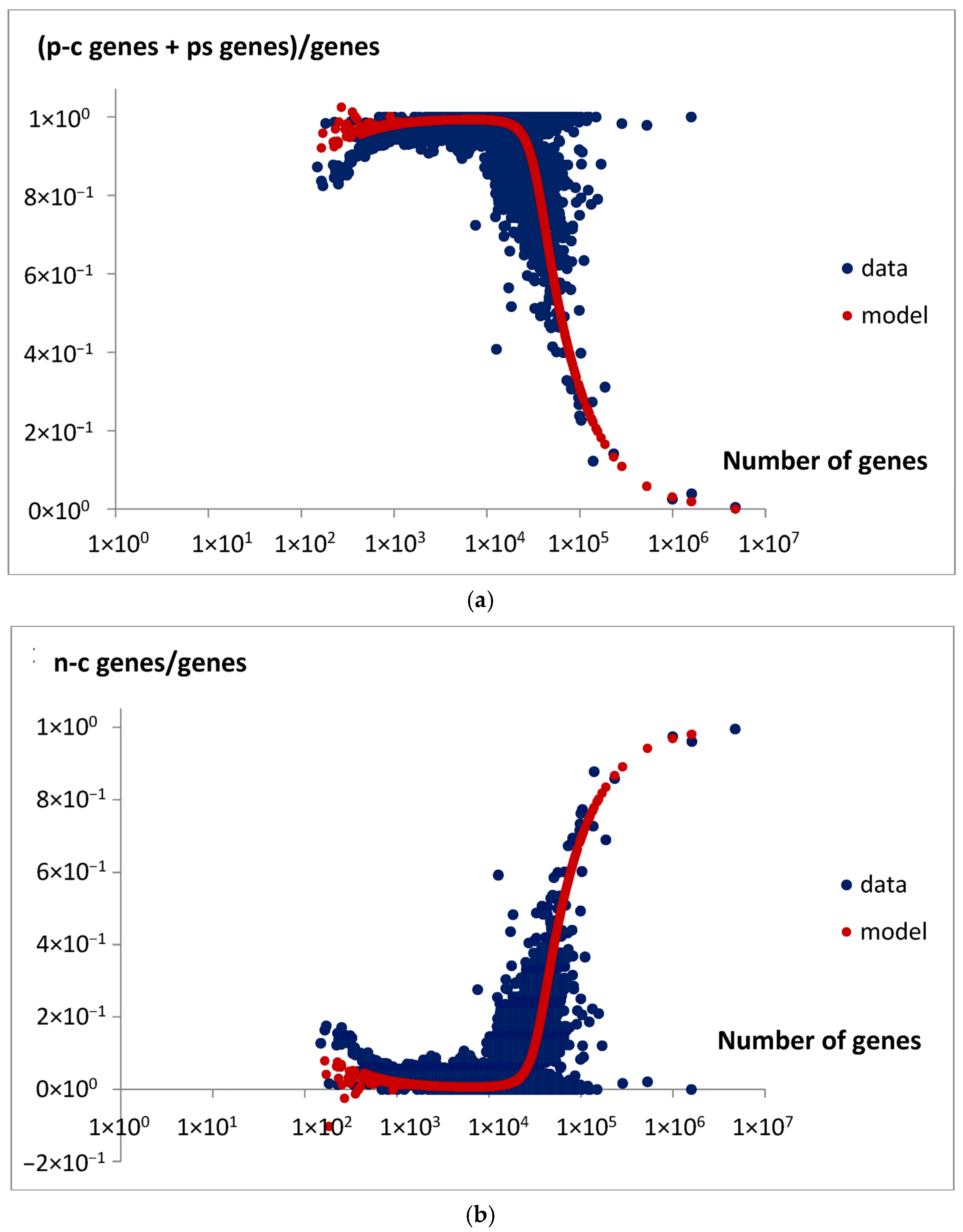
The above observations were verified by the predicted total gene pool in the genome. Specifically, the comparison between the predicted fractions of extensive and intensive genes, ge/g (Equation (16)) and gi/g (Equation (19)), and the derived experimental data support this interpretation. These data include the ratio of the sum of protein-coding genes and pseudogenes to the total number of genes in the genome (
Figure 16a) and the ratio of non-coding genes to total genes (
Figure 16b). This comparison indicates that meaningful relationships between gene types can be established. As suggested, extensive genes that contribute to genome size expansion can be associated with both protein-coding genes and pseudogenes, whereas intensive genes correspond to non-coding genes. Of course, these are not strict rules but rather general observations of dominant trends, which are subject to limitations due to the high dispersion in analyzed data, especially in genomes with large gene counts such as very large eukaryotic genomes. Therefore, they refer to a statistically average situation, and in the specific case, the discrepancy may be particularly large.
Good examples of intensive genes seem to be overlapping genes, especially well-known nested genes. The majority of nested genes are non-coding. For example, in the nematode
C. elegans, over 92% of nested genes are ncRNAs [
31]. Rare examples of coding nested genes are Ins5B and Ins5C in the
E. coli genome [
32], TAR1, NAG1, and CDA12 in
S. cerevisiae [
33], and F8A1 in
H. sapiens [
34].
Extensive genes, on the other hand, can be represented by non-overlapping genes. Approximately 75% of human protein-coding genes were found not to overlap with their neighbors [
35]. Although it was shown that pseudogenes may also be related to extensive genes, in fact a significant number of pseudogenes may overlap with protein-coding genes [
36]. This overlap may be the result of evolutionary progress in sharing a region of initially distinct gene.
The origins of extensive and intensive genes should be sought in an early stage of life, called the “RNA world” [
37,
38], which existed before DNA and proteins became dominant. In this world, RNA performed the functions of both modern DNA (informational), proteins (catalytic), and modern RNA (regulational). The early life proliferation of functional RNA molecules required a relatively large operational space for classic non-overlapping sequences, which could not be sufficiently available within a single-stranded RNA. As a result, both the elongating sequences of self-replicating proto-ribozymes and shorter regulatory elements introducing innovations began to overlap, thereby reusing existing sequence space. Host-nested molecule configuration could be evolutionarily preferred, being a precursor of extensive and intensive genes.
Equations (6) and (8) of the model lead to the equation, which may be presented in the form
describing the balance on a scale with unequal arm lengths (l
emax − l
e) and l
e, where the “weights” correspond to dg
i and dg
e. Such an analogy may inspire the hypothesis of an equilibrium between the emergence of new intensive and extensive genes, a concept that could be explored in future investigations. In our opinion, future development of the model could also describe, in a more detailed way, the dependence of the length l
e on the gene function and a relatively high dispersion of the size of genomes for moderate and high gene numbers.
The authors believe that the aims of this work have been achieved and propose that the analysis of probability density functions, supported by further mathematical modeling, may serve as an effective tool in future bioinformatics research of genomic data, offering valuable insights into the foundations of evolution.