Unraveling the Impact of Dab1 Gene Silencing on the Expression of Autophagy Markers in Lung Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Generation of Dab1 Null Conventional Mutants and Sample Collection
2.3. Immunofluorescence
2.4. Immunofluorescence Signal Quantification
2.5. Statistical Analysis
3. Results
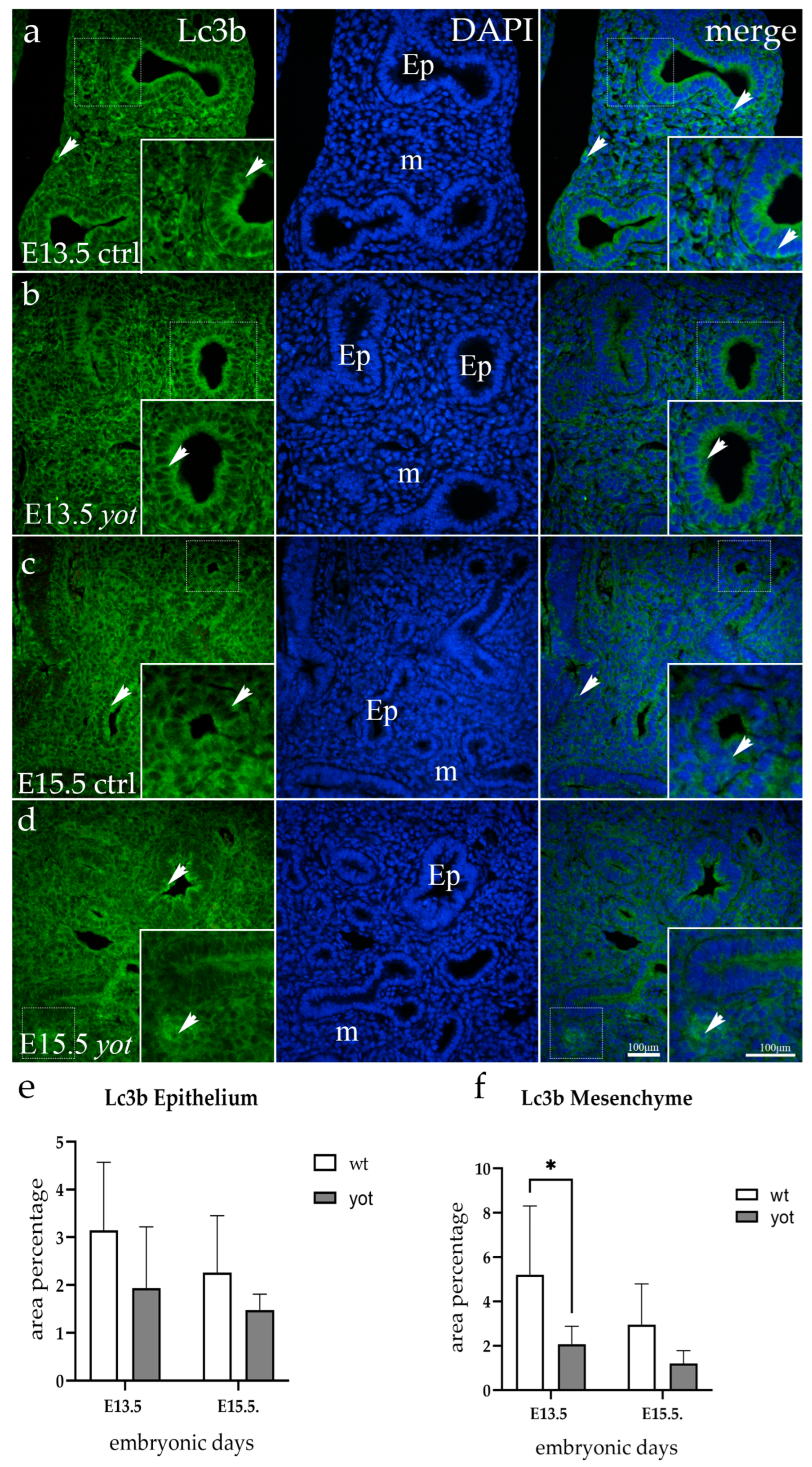
3.1. Lc3b Expression
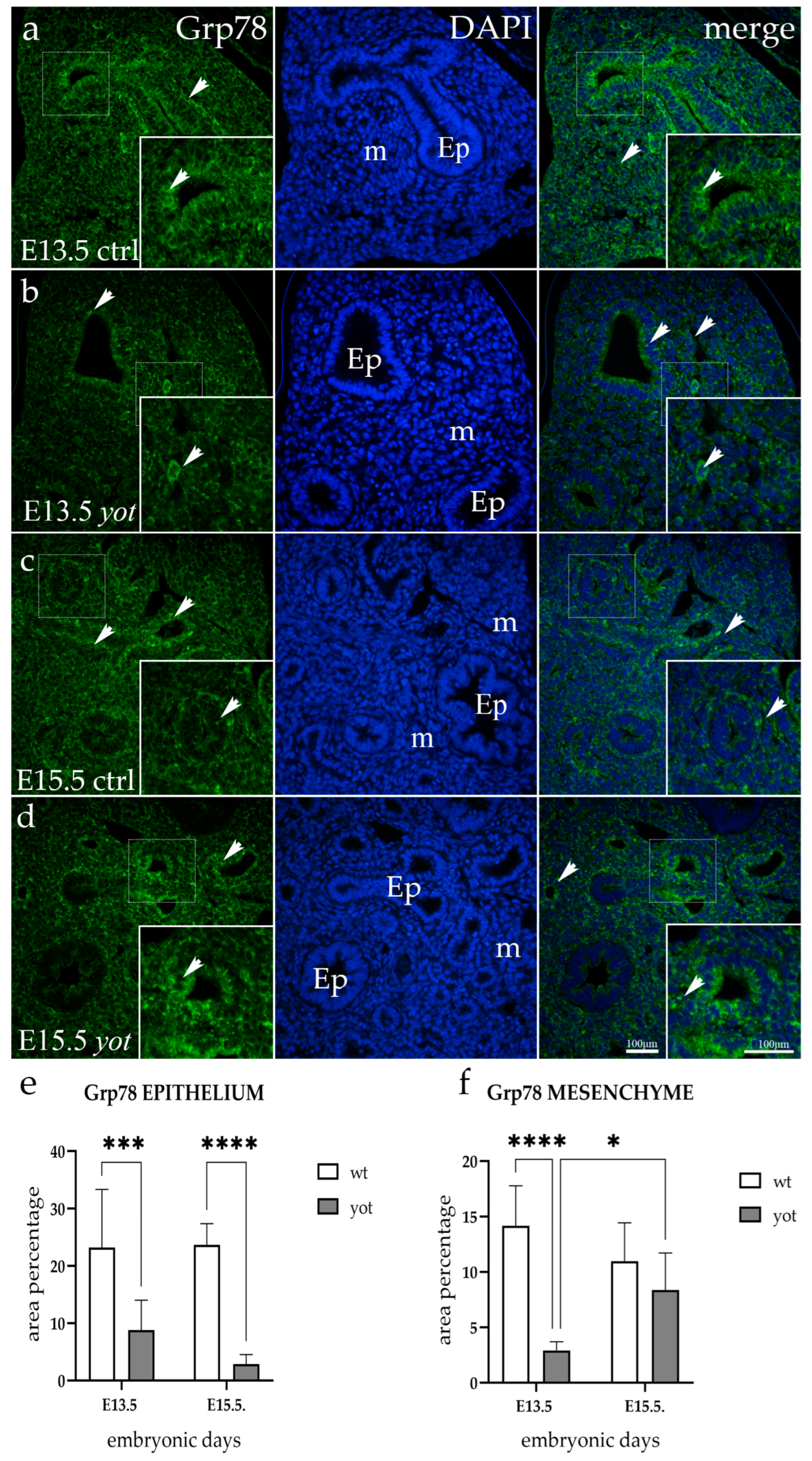
3.2. Grp78 Expression
3.3. Hsc70 Expression
3.4. Lamp2a Expression
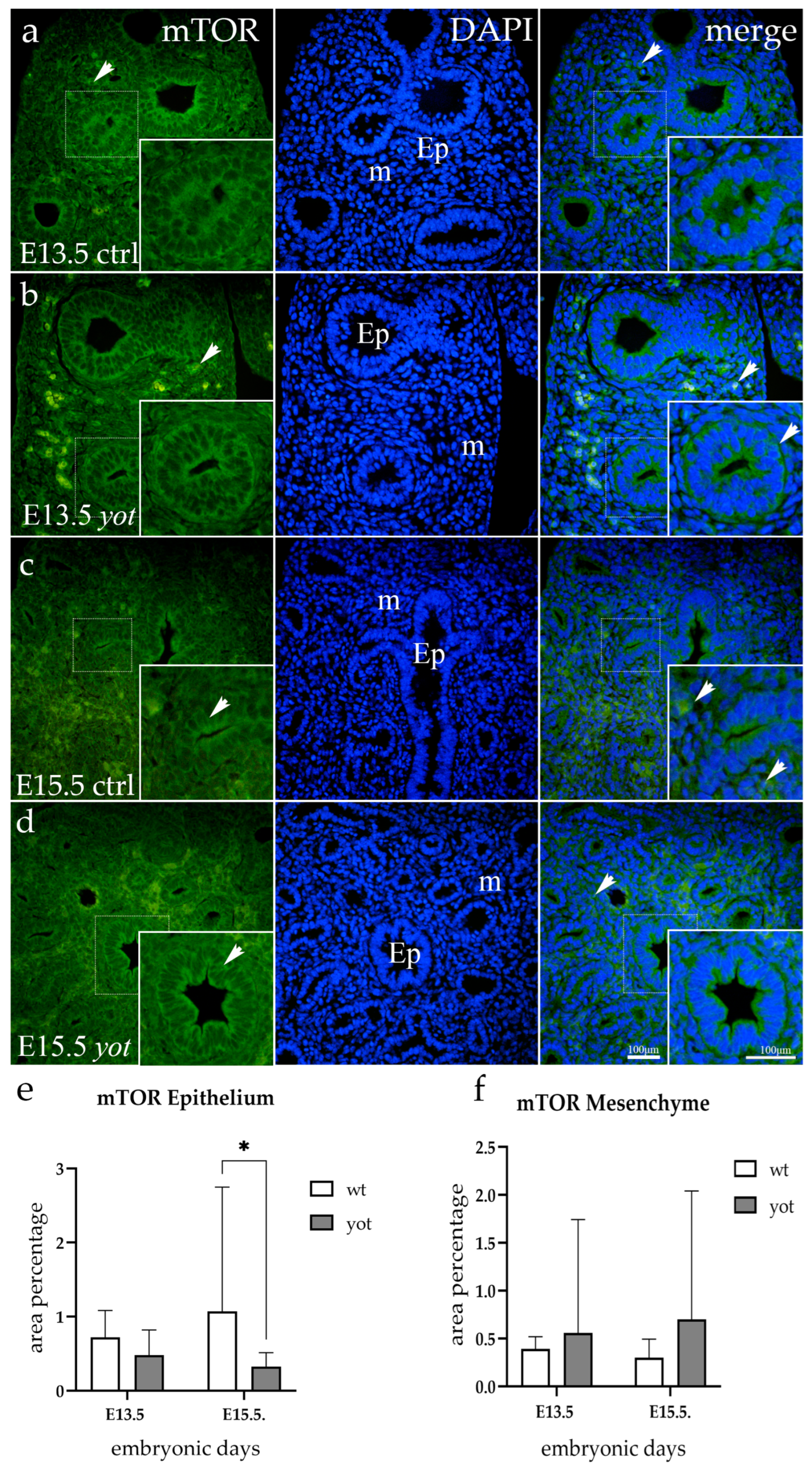
3.5. mTOR Expression
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yoneshima, H.; Nagata, E.; Matsumoto, M.; Yamada, M.; Nakajima, K.; Miyata, T.; Ogawa, M.; Mikoshiba, K. A novel neurological mutant mouse, yotari, which exhibits reeler-like phenotype but expresses CR-50 antigen/reelin. Neurosci. Res. 1997, 29, 217–223. [Google Scholar] [CrossRef]
- Yoshihara, Y.; Setsu, T.; Katsuyama, Y.; Kikkawa, S.; Terashima, T.; Maeda, K. Cortical layer V neurons in the auditory and visual cortices of normal, reeler, and yotari mice. Kobe J. Med. Sci. 2010, 56, E50–E59. [Google Scholar]
- Racetin, A.; Juric, M.; Filipovic, N.; Solic, I.; Kosovic, I.; Glavina Durdov, M.; Kunac, N.; Zekic Tomas, S.; Saraga, M.; Soljic, V.; et al. Expression and localization of DAB1 and Reelin during normal human kidney development. Croat. Med. J. 2019, 60, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Calvier, L.; Demuth, G.; Manouchehri, N.; Wong, C.; Sacharidou, A.; Mineo, C.; Shaul, P.W.; Monson, N.L.; Kounnas, M.Z.; Stuve, O.; et al. Reelin depletion protects against autoimmune encephalomyelitis by decreasing vascular adhesion of leukocytes. Sci. Transl. Med. 2020, 12, eaay7675. [Google Scholar] [CrossRef]
- Ding, Y.; Huang, L.; Xian, X.; Yuhanna, I.S.; Wasser, C.R.; Frotscher, M.; Mineo, C.; Shaul, P.W.; Herz, J. Loss of Reelin protects against atherosclerosis by reducing leukocyte-endothelial cell adhesion and lesion macrophage accumulation. Sci. Signal. 2016, 9, ra29. [Google Scholar] [CrossRef] [PubMed]
- Racanelli, A.C.; Kikkers, S.A.; Choi, A.M.K.; Cloonan, S.M. Autophagy and inflammation in chronic respiratory disease. Autophagy 2018, 14, 221–232. [Google Scholar] [CrossRef]
- Hashimoto, S.; Nakano, H.; Singh, G.; Katyal, S. Expression of Spred and Sprouty in developing rat lung. Mech. Dev. 2002, 119 (Suppl. S1), S303–S309. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.M.; Li, S.; Yang, H.; Morrisey, E.E. Foxp4: A novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech. Dev. 2002, 119 (Suppl. S1), S197–S202. [Google Scholar] [CrossRef]
- Choi, C.H.; Jung, Y.K.; Oh, S.H. Selective induction of catalase-mediated autophagy by dihydrocapsaicin in lung cell lines. Free Radic. Biol. Med. 2010, 49, 245–257. [Google Scholar] [CrossRef]
- Mizumura, K.; Cloonan, S.M.; Haspel, J.A.; Choi, A.M.K. The emerging importance of autophagy in pulmonary diseases. Chest 2012, 142, 1289–1299. [Google Scholar] [CrossRef]
- Deter, R.L.; De Duve, C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J. Cell Biol. 1967, 33, 437–449. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Saftig, P.; Beertsen, W.; Eskelinen, E.L. LAMP-2: A control step for phagosome and autophagosome maturation. Autophagy 2008, 4, 510–512. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 2016, 12, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Klionsky, D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005, 12 (Suppl. S2), 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Perutina, I.; Kelam, N.; Maglica, M.; Racetin, A.; Ogorevc, M.; Filipovic, N.; Katsuyama, Y.; Miskovic, J.; Vukojevic, K. Disturbances in Switching between Canonical and Non-Canonical Wnt Signaling Characterize Developing and Postnatal Kidneys of Dab1−/− (yotari) Mice. Biomedicines 2023, 11, 1321. [Google Scholar] [CrossRef] [PubMed]
- Maglica, M.; Kelam, N.; Haque, E.; Perutina, I.; Racetin, A.; Filipovic, N.; Katsuyama, Y.; Vukojevic, K. Immunoexpression Pattern of Autophagy Markers in Developing and Postnatal Kidneys of Dab1−/− (yotari) Mice. Biomolecules 2023, 13, 402. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef]
- Ohsumi, Y.; Mizushima, N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004, 15, 231–236. [Google Scholar] [CrossRef]
- He, H.; Dang, Y.; Dai, F.; Guo, Z.; Wu, J.; She, X.; Pei, Y.; Chen, Y.; Ling, W.; Wu, C.; et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 2003, 278, 29278–29287. [Google Scholar] [CrossRef]
- Loos, B.; du Toit, A.; Hofmeyr, J.H. Defining and measuring autophagosome flux-concept and reality. Autophagy 2014, 10, 2087–2096. [Google Scholar] [CrossRef]
- Lunding, L.P.; Krause, D.; Stichtenoth, G.; Stamme, C.; Lauterbach, N.; Hegermann, J.; Ochs, M.; Schuster, B.; Sedlacek, R.; Saftig, P.; et al. LAMP3 deficiency affects surfactant homeostasis in mice. PLoS Genet. 2021, 17, e1009619. [Google Scholar] [CrossRef] [PubMed]
- Assaye, M.A.; Gizaw, S.T. Chaperone-Mediated Autophagy and Its Implications for Neurodegeneration and Cancer. Int. J. Gen. Med. 2022, 15, 5635–5649. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Hu, J.; Qiu, X.; Wang, C.; Peng, J.; Zhang, C.; Zhang, M.; Lu, H.; Chen, W. LAMP2A, LAMP2B and LAMP2C: Similar structures, divergent roles. Autophagy 2023, 19, 2837–2852. [Google Scholar] [CrossRef] [PubMed]
- Abdel Fattah, E.; Bhattacharya, A.; Herron, A.; Safdar, Z.; Eissa, N.T. Critical role for IL-18 in spontaneous lung inflammation caused by autophagy deficiency. J. Immunol. 2015, 194, 5407–5416. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T. Mammalian target of rapamycin: A signaling kinase for every aspect of cellular life. In mTOR; Springer: Berlin/Heidelberg, Germany, 2012; Volume 821, pp. 1–14. [Google Scholar] [CrossRef]
- Yoshida, T.; Mett, I.; Bhunia, A.K.; Bowman, J.; Perez, M.; Zhang, L.; Gandjeva, A.; Zhen, L.; Chukwueke, U.; Mao, T.; et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat. Med. 2010, 16, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Tavaria, M.; Gabriele, T.; Kola, I.; Anderson, R.L. A hitchhiker’s guide to the human Hsp70 family. Cell Stress Chaperones 1996, 1, 23–28. [Google Scholar] [CrossRef]
- Behnke, J.; Feige, M.J.; Hendershot, L.M. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J. Mol. Biol. 2015, 427, 1589–1608. [Google Scholar] [CrossRef]
- Jorgensen, E.; Stinson, A.; Shan, L.; Yang, J.; Gietl, D.; Albino, A.P. Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Cancer 2008, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Kim, D.I.; Kang, M.R.; Lee, K.S.; Park, S.Y.; Jeong, J.S.; Lee, Y.C. Endoplasmic reticulum stress influences bronchial asthma pathogenesis by modulating nuclear factor kappaB activation. J. Allergy Clin. Immunol. 2013, 132, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Diehl, J.A.; Yang, W.; Rimerman, R.A.; Xiao, H.; Emili, A. Hsc70 regulates accumulation of cyclin D1 and cyclin D1-dependent protein kinase. Mol. Cell. Biol. 2003, 23, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Agarraberes, F.A.; Terlecky, S.R.; Dice, J.F. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 1997, 137, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Kose, S.; Furuta, M.; Koike, M.; Yoneda, Y.; Imamoto, N. The 70-kD heat shock cognate protein (hsc70) facilitates the nuclear export of the import receptors. J. Cell Biol. 2005, 171, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sellares, J.; Veraldi, K.L.; Thiel, K.J.; Cardenes, N.; Alvarez, D.; Schneider, F.; Pilewski, J.M.; Rojas, M.; Feghali-Bostwick, C.A. Intracellular Heat Shock Protein 70 Deficiency in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2019, 60, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Flodby, P.; Li, C.; Liu, Y.; Wang, H.; Marconett, C.N.; Laird-Offringa, I.A.; Minoo, P.; Lee, A.S.; Zhou, B. The 78-kD Glucose-Regulated Protein Regulates Endoplasmic Reticulum Homeostasis and Distal Epithelial Cell Survival during Lung Development. Am. J. Respir. Cell Mol. Biol. 2016, 55, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Calvier, L.; Xian, X.; Lee, R.G.; Sacharidou, A.; Mineo, C.; Shaul, P.W.; Kounnas, M.Z.; Tsai, S.; Herz, J. Reelin Depletion Protects Against Atherosclerosis by Decreasing Vascular Adhesion of Leukocytes. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 1309–1318. [Google Scholar] [CrossRef]
- Alexander, A.; Herz, J.; Calvier, L. Reelin through the years: From brain development to inflammation. Cell Rep. 2023, 42, 112669. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Song, H.K.; Hwang, D.Y. Use of C57BL/6N mice on the variety of immunological researches. Lab. Anim. Res. 2017, 33, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Shoji, H.; Ogata, M.; Kagawa, Y.; Owada, Y.; Miyakawa, T.; Sakimura, K.; Terashima, T.; Katsuyama, Y. Dorsal Forebrain-Specific Deficiency of Reelin-Dab1 Signal Causes Behavioral Abnormalities Related to Psychiatric Disorders. Cereb. Cortex 2017, 27, 3485–3501. [Google Scholar] [CrossRef] [PubMed]
- Cicchetti, D.V. Guidelines, Criteria, and Rules of Thumb for Evaluating Normed and Standardized Assessment Instrument in Psychology. Psychol. Assess. 1994, 6, 284–290. [Google Scholar] [CrossRef]
- Warburton, D.; El-Hashash, A.; Carraro, G.; Tiozzo, C.; Sala, F.; Rogers, O.; De Langhe, S.; Kemp, P.J.; Riccardi, D.; Torday, J.; et al. Lung organogenesis. Curr. Top. Dev. Biol. 2010, 90, 73–158. [Google Scholar] [CrossRef]
- Soriano, J.B.; Kendrick, P.J.; Paulson, K.R.; Gupta, V.; Abrams, E.M.; Adedoyin, R.A.; Adhikari, T.B.; Advani, S.M.; Agrawal, A.; Ahmadian, E.; et al. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Chen, Z.H.; Lam, H.C.; Jin, Y.; Kim, H.P.; Cao, J.; Lee, S.J.; Ifedigbo, E.; Parameswaran, H.; Ryter, S.W.; Choi, A.M. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. USA 2010, 107, 18880–18885. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Smith, A.; Guo, L.; Alastalo, T.P.; Li, M.; Sawada, H.; Liu, X.; Chen, Z.H.; Ifedigbo, E.; Jin, Y.; et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2011, 183, 649–658. [Google Scholar] [CrossRef]
- Ayaub, E.A.; Kolb, P.S.; Mohammed-Ali, Z.; Tat, V.; Murphy, J.; Bellaye, P.S.; Shimbori, C.; Boivin, F.J.; Lai, R.; Lynn, E.G.; et al. GRP78 and CHOP modulate macrophage apoptosis and the development of bleomycin-induced pulmonary fibrosis. J. Pathol. 2016, 239, 411–425. [Google Scholar] [CrossRef]
- Lam, T.Y.W.; Nguyen, N.; Peh, H.Y.; Shanmugasundaram, M.; Chandna, R.; Tee, J.H.; Ong, C.B.; Hossain, M.Z.; Venugopal, S.; Zhang, T.; et al. ISM1 protects lung homeostasis via cell-surface GRP78-mediated alveolar macrophage apoptosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2019161119. [Google Scholar] [CrossRef]
- Malhotra, J.D.; Kaufman, R.J. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 2007, 18, 716–731. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Mao, C.; Lee, B.; Lee, A.S. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol. Cell. Biol. 2006, 26, 5688–5697. [Google Scholar] [CrossRef]
- Aschkenasy, G.; Bromberg, Z.; Raj, N.; Deutschman, C.S.; Weiss, Y.G. Enhanced Hsp70 expression protects against acute lung injury by modulating apoptotic pathways. PLoS ONE 2011, 6, e26956. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; He, S.; Huang, L.; Ren, D.; Nie, T.; Tao, K.; Xia, L.; Lu, F.; Mao, Z.; Yang, Q. Chaperone-mediated autophagy degrades Keap1 and promotes Nrf2-mediated antioxidative response. Aging Cell 2022, 21, e13616. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, K.; Kato, Y.; Ikami, Y.; Sakamoto, K.; Ohtake, K.; Kusano, S.; Tomabechi, Y.; Kukimoto-Niino, M.; Shirouzu, M.; Guan, J.L.; et al. Direct homophilic interaction of LAMP2A with the two-domain architecture revealed by site-directed photo-crosslinks and steric hindrances in mammalian cells. Autophagy 2021, 17, 4286–4304. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, Y.; Araya, J.; Fujita, Y.; Kadota, T.; Tsubouchi, K.; Yoshida, M.; Minagawa, S.; Hara, H.; Kawamoto, H.; Watanabe, N.; et al. Chaperone-Mediated Autophagy Suppresses Apoptosis via Regulation of the Unfolded Protein Response during Chronic Obstructive Pulmonary Disease Pathogenesis. J. Immunol. 2020, 205, 1256–1267. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Li, Z.Y.; Dong, L.L.; Li, W.J.; Wu, Y.P.; Wang, J.; Chen, H.P.; Liu, H.W.; Li, M.; Jin, C.L.; et al. Inactivation of MTOR promotes autophagy-mediated epithelial injury in particulate matter-induced airway inflammation. Autophagy 2020, 16, 435–450. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, H.W. Role of mTOR in the Development of Asthma in Mice with Cigarette Smoke-Induced Cellular Senescence. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2022, 77, 433–442. [Google Scholar] [CrossRef]
- Ikeda, H.; Shiojima, I.; Oka, T.; Yoshida, M.; Maemura, K.; Walsh, K.; Igarashi, T.; Komuro, I. Increased Akt-mTOR signaling in lung epithelium is associated with respiratory distress syndrome in mice. Mol. Cell. Biol. 2011, 31, 1054–1065. [Google Scholar] [CrossRef][Green Version]
- Russell, R.C.; Yuan, H.X.; Guan, K.L. Autophagy regulation by nutrient signaling. Cell Res. 2014, 24, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Houssaini, A.; Breau, M.; Kebe, K.; Abid, S.; Marcos, E.; Lipskaia, L.; Rideau, D.; Parpaleix, A.; Huang, J.; Amsellem, V.; et al. mTOR pathway activation drives lung cell senescence and emphysema. JCI Insight 2018, 3, e93203. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]


| Antibodies | Host | Dilution | Source | |
|---|---|---|---|---|
| Primary | Anti-Lc3b/ab48394 | Rabbit | 1:100 | Abcam (Cambridge, UK) |
| Anti-Grp78/PA5-19503 | Rabbit | 1:300 | Thermo Fisher Scientific (Waltham, MA, USA) | |
| Ani-Hsc70/ab51052 | Rabbit | 1:50 | Abcam (Cambridge, UK) | |
| Anti-Lamp2a/ab18528 | Rabbit | 1:100 | Abcam (Cambridge, UK) | |
| Anti-mTOR/ab32028 | Rabbit | 1:100 | Abcam (Cambridge, UK) | |
| Secondary | Anti-Rabbit IgG, Alexa Fluor® 488, 711-545-152 | Donkey | 1:300 | Jackson Immuno Research Laboratories, Inc. (Baltimore, PA, USA) |
| Embryonic Day (E) | Animal | Structure | Antibody | ||||
|---|---|---|---|---|---|---|---|
| Lc3b | Grp78 | Hsc70 | mTOR | Lamp2a | |||
| E13.5 | wild-type | Ep | + | ++ | − | + | + |
| m | ++ | ++ | + | −/+ | −/+ | ||
| yotari | Ep | −/+ | −/+ | − | −/+ | + | |
| m | −/+ | −/+ | + | + | − | ||
| E15.5 | wild-type | Ep | −/+ | ++ | −/+ | ++ | −/+ |
| m | + | + | + | −/+ | −/+ | ||
| yotari | Ep | −/+ | −/+ | − | −/+ | −/+ | |
| m | −/+ | + | + | + | −/+ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizikalo, A.; Maglica, M.; Kelam, N.; Perutina, I.; Ogorevc, M.; Racetin, A.; Filipović, N.; Katsuyama, Y.; Zovko, Z.; Mišković, J.; et al. Unraveling the Impact of Dab1 Gene Silencing on the Expression of Autophagy Markers in Lung Development. Life 2024, 14, 316. https://doi.org/10.3390/life14030316
Rizikalo A, Maglica M, Kelam N, Perutina I, Ogorevc M, Racetin A, Filipović N, Katsuyama Y, Zovko Z, Mišković J, et al. Unraveling the Impact of Dab1 Gene Silencing on the Expression of Autophagy Markers in Lung Development. Life. 2024; 14(3):316. https://doi.org/10.3390/life14030316
Chicago/Turabian StyleRizikalo, Azer, Mirko Maglica, Nela Kelam, Ilija Perutina, Marin Ogorevc, Anita Racetin, Natalija Filipović, Yu Katsuyama, Zdenka Zovko, Josip Mišković, and et al. 2024. "Unraveling the Impact of Dab1 Gene Silencing on the Expression of Autophagy Markers in Lung Development" Life 14, no. 3: 316. https://doi.org/10.3390/life14030316
APA StyleRizikalo, A., Maglica, M., Kelam, N., Perutina, I., Ogorevc, M., Racetin, A., Filipović, N., Katsuyama, Y., Zovko, Z., Mišković, J., & Vukojević, K. (2024). Unraveling the Impact of Dab1 Gene Silencing on the Expression of Autophagy Markers in Lung Development. Life, 14(3), 316. https://doi.org/10.3390/life14030316









