Evidence of Heritability in Prebiotically Realistic Membrane-Bound Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Vesicle and Solution Preparation
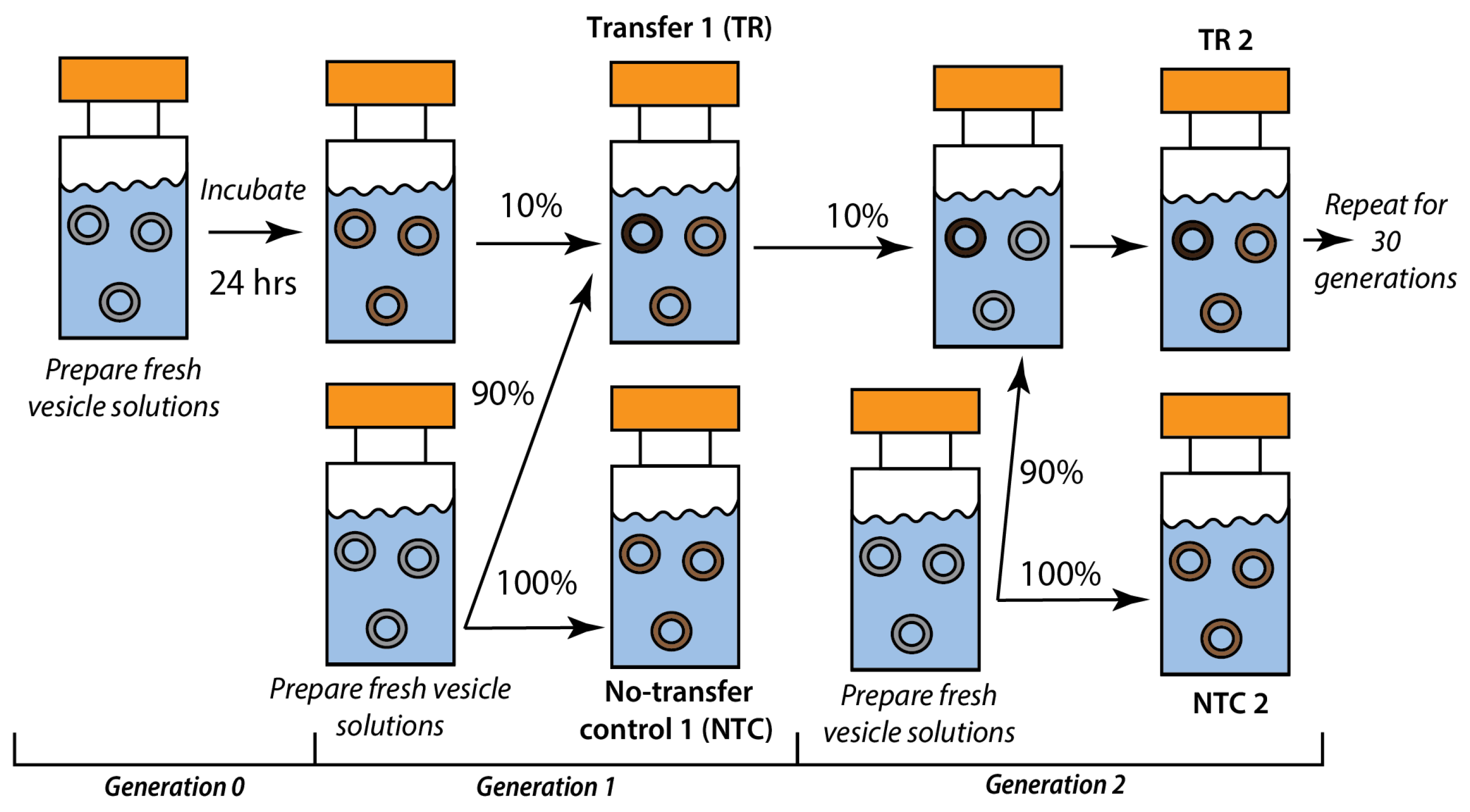
2.2. Recursive Seeding Design
2.3. Nile Red Fluorescence Measurements
2.4. Dynamic Light Scattering
2.5. LC/MS Analyses
3. Results
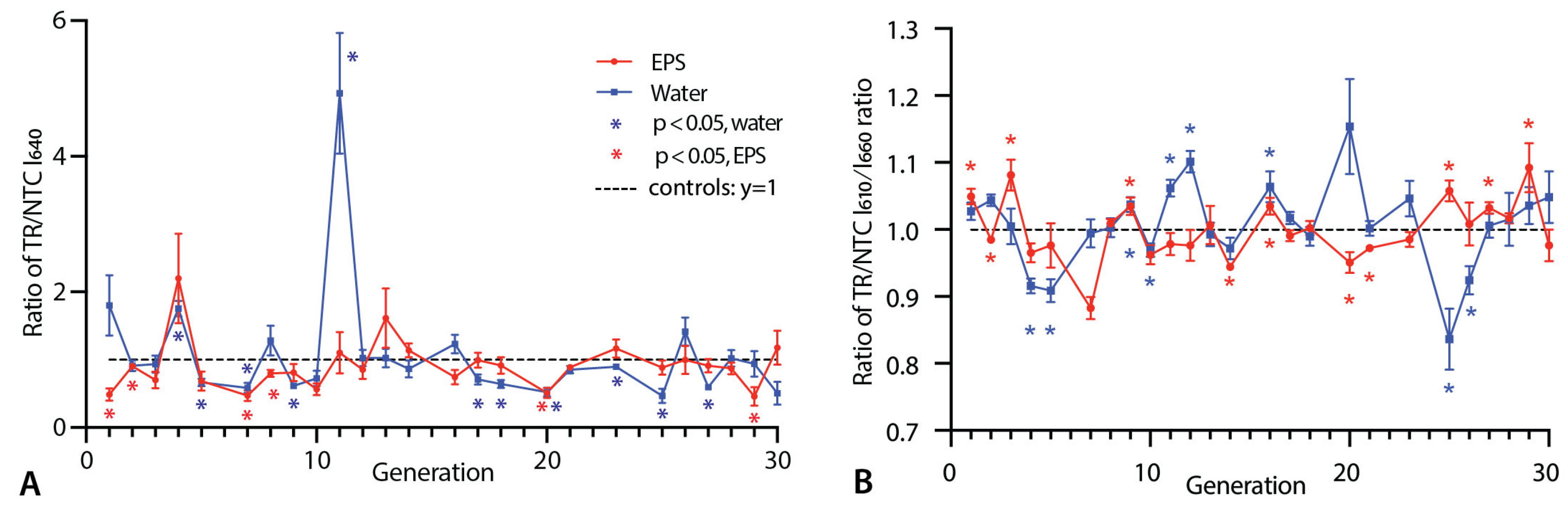
3.1. Nile Red Fluorescence Measurements
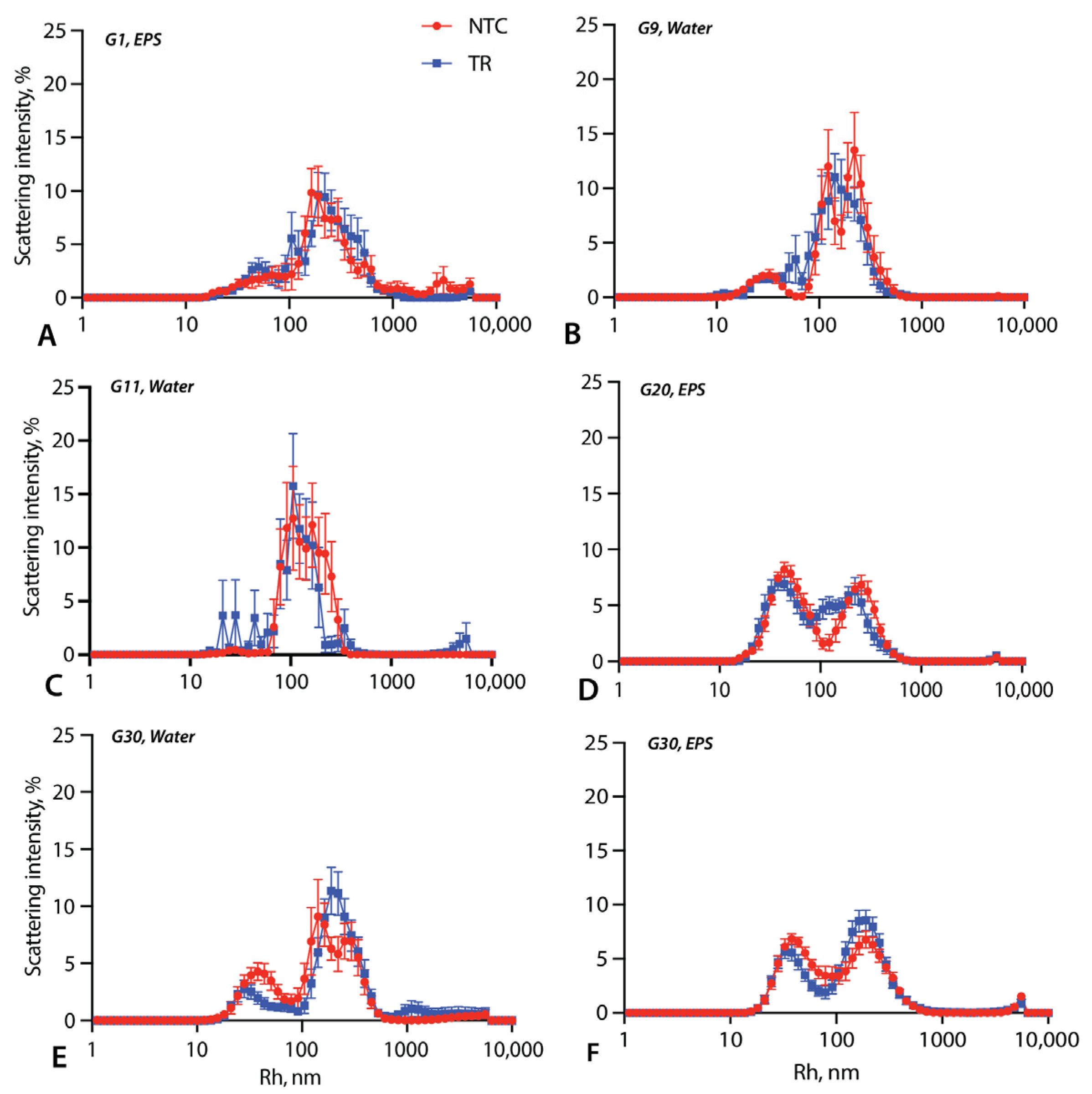
3.2. Vesicle Size Measurements
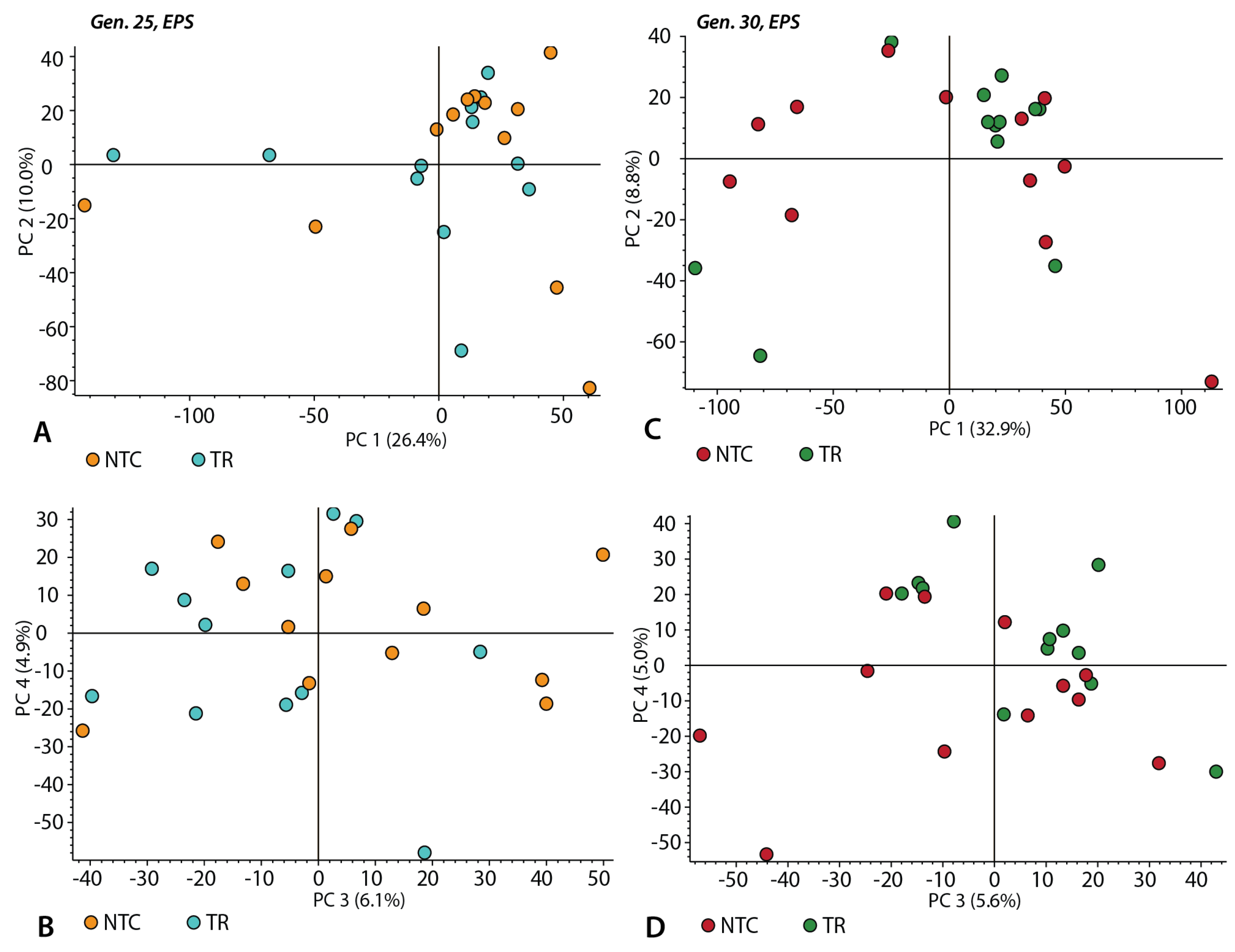
3.3. Chemical Composition of the EPS Samples without Triton
4. Discussion
4.1. Limitations of the Study
4.2. Explanations of the Observed Patterns
4.3. Heritability in Vesicle Populations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical roots of biological evolution: The origins of life as a process of development of autonomous functional systems. Open Biol. 2017, 7, 170050. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.A.; Walde, P. Current ideas about prebiological compartmentalization. Life 2015, 5, 1239–1263. [Google Scholar] [CrossRef] [PubMed]
- Mizuuchi, R.; Ichihashi, N. Primitive compartmentalization for the sustainable replication of genetic molecules. Life 2021, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Camprubi, E.; Jordan, S.F.; Vasiliadou, R.; Lane, N. Iron catalysis at the origin of life. IUBMB Life 2017, 69, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Pyrite formation, the first energy source for life: A hypothesis. Syst. Appl. Microbiol. 1988, 10, 207–210. [Google Scholar] [CrossRef]
- Baum, D.A. The origin and early evolution of life in chemical composition space. J. Theor. Biol. 2018, 456, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Tuck, A. The role of atmospheric aerosols in the origin of life. Surv. Geophys. 2002, 23, 379–409. [Google Scholar] [CrossRef]
- Ghosh, B.; Bose, R.; Tang, T.D. Can coacervation unify disparate hypotheses in the origin of cellular life? Curr. Opin. Colloid Interface Sci. 2021, 52, 101415. [Google Scholar] [CrossRef]
- Martin, N.; Douliez, J.P. Fatty acid vesicles and coacervates as model prebiotic protocells. ChemSystemsChem 2021, 3, e2100024. [Google Scholar] [CrossRef]
- Kahana, A.; Lancet, D. Self-reproducing catalytic micelles as nanoscopic protocell precursors. Nat. Rev. Chem. 2021, 5, 870–878. [Google Scholar] [CrossRef]
- Deamer, D.W. Prebiotic amphiphilic compounds. In Origins—Genesis, Evolution and Diversity of Life; Springer: Berlin/Heidelberg, Germany, 2004; pp. 75–89. [Google Scholar]
- Fiore, M.; Strazewski, P. Prebiotic lipidic amphiphiles and condensing agents on the early Earth. Life 2016, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Z.R.; Todd, Z.R.; Wogan, N.; Black, R.A.; Keller, S.L.; Catling, D.C. Plausible sources of membrane-forming fatty acids, on the early Earth: A review of the literature and an estimation of amounts. ACS Earth Space Chem. 2022, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- McCollom, T.M.; Ritter, G.; Simoneit, B.R. Lipid synthesis under hydrothermal conditions by Fischer-Tropsch-type reactions. Orig. Life Evol. Biosph. 1999, 29, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.H.; Nagy, B. Distribution and enantiomeric composition of amino acids in the Murchison meteorite. Nature 1982, 296, 837–840. [Google Scholar] [CrossRef]
- Vincent, L.; Colón-Santos, S.; Cleaves, H.J.; Baum, D.A.; Maurer, S.E. The Prebiotic Kitchen: A Guide to Composing Prebiotic Soup Recipes to Test Origins of Life Hypotheses. Life 2021, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, J.P.; Deamer, D.W.; Sandford, S.A.; Allamandola, L.J. Self-assembling amphiphilic molecules: Synthesis in simulated interstellar/precometary ices. Proc. Natl. Acad. Sci. USA 2001, 98, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Szostak, J.W. Replicating vesicles as models of primitive cell growth and division. Curr. Opin. Chem. Biol. 2004, 8, 660–664. [Google Scholar] [CrossRef]
- Blöchliger, E.; Blocher, M.; Walde, P.; Luisi, P.L. Matrix effect in the size distribution of fatty acid vesicles. J. Phys. Chem. B 1998, 102, 10383–10390. [Google Scholar] [CrossRef]
- Luisi, P.L.; Souza, T.P.D.; Stano, P. Vesicle behavior: In search of explanations. J. Phys. Chem. B 2008, 112, 14655–14664. [Google Scholar] [CrossRef]
- Markvoort, A.J.; Pfleger, N.; Staffhorst, R.; Hilbers, P.A.; Van Santen, R.A.; Killian, J.A.; De Kruijff, B. Self-reproduction of fatty acid vesicles: A combined experimental and simulation study. Biophys. J. 2010, 99, 1520–1528. [Google Scholar] [CrossRef]
- Segrè, D.; Ben-Eli, D.; Deamer, D.W.; Lancet, D. The lipid world. Orig. Life Evol. Biosph. 2001, 31, 119–145. [Google Scholar] [CrossRef] [PubMed]
- Segrè, D.; Ben-Eli, D.; Lancet, D. Compositional genomes: Prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl. Acad. Sci. USA 2000, 97, 4112–4117. [Google Scholar] [CrossRef]
- Vasas, V.; Szathmáry, E.; Santos, M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc. Natl. Acad. Sci. USA 2010, 107, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Baum, D.A.; Vetsigian, K. An experimental framework for generating evolvable chemical systems in the laboratory. Orig. Life Evol. Biosph. 2017, 47, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution 1988, 42, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Kahana, A.; Segev, L.; Lancet, D. Attractor dynamics drives self-reproduction in protobiological catalytic networks. Cell Rep. Phys. Sci. 2023, 4, 101384. [Google Scholar] [CrossRef]
- Maurer, S.E.; Tølbøl Sørensen, K.; Iqbal, Z.; Nicholas, J.; Quirion, K.; Gioia, M.; Monnard, P.A.; Hanczyc, M.M. Vesicle self-assembly of monoalkyl amphiphiles under the effects of high ionic strength, extreme pH, and high temperature environments. Langmuir 2018, 34, 15560–15568. [Google Scholar] [CrossRef]
- Vincent, L.; Berg, M.; Krismer, M.; Saghafi, S.T.; Cosby, J.; Sankari, T.; Vetsigian, K.; Cleaves, H.J.; Baum, D.A. Chemical ecosystem selection on mineral surfaces reveals long-term dynamics consistent with the spontaneous emergence of mutual catalysis. Life 2019, 9, 80. [Google Scholar] [CrossRef]
- Noguchi, H. Structure formation in binary mixtures of lipids and detergents: Self-assembly and vesicle division. J. Chem. Phys. 2013, 138, 01B611. [Google Scholar] [CrossRef]
- Lin, C.; Zhao, J.; Jiang, R. Nile red probing for the micelle-to-vesicle transition of AOT in aqueous solution. Chem. Phys. Lett. 2008, 464, 77–81. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 57. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.Y.; Dowling, Q.M.; Sivananthan, S.J.; Kramer, R.M. Particle sizing of nanoparticle adjuvant formulations by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA). In Vaccine Adjuvants: Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 239–252. [Google Scholar]
- Xu, H.; Du, N.; Song, Y.; Song, S.; Hou, W. Spontaneous vesicle formation and vesicle-to-micelle transition of sodium 2-ketooctanate in water. J. Colloid Interface Sci. 2018, 509, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Dagar, S.; Rajamani, S. Influence of Wet–Dry Cycling on the Self-Assembly and Physicochemical Properties of Model Protocellular Membrane Systems. ChemSystemsChem 2021, 3, e2100014. [Google Scholar] [CrossRef]
- Cohen, Z.R.; Todd, Z.R.; Catling, D.C.; Black, R.A.; Keller, S.L. Prebiotic Vesicles Retain Solutes and Grow by Micelle Addition after Brief Cooling below the Membrane Melting Temperature. Langmuir 2022, 38, 13407–13413. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.; Das, K.; Nath, D.N.; Bhattacharyya, K. Twisted charge transfer processes of nile red in homogeneous solutions and in faujasite zeolite. Langmuir 1994, 10, 326–329. [Google Scholar] [CrossRef]
- Teo, W.; Caprariello, A.V.; Morgan, M.L.; Luchicchi, A.; Schenk, G.J.; Joseph, J.T.; Geurts, J.J.; Stys, P.K. Nile Red fluorescence spectroscopy reports early physicochemical changes in myelin with high sensitivity. Proc. Natl. Acad. Sci. USA 2021, 118, e2016897118. [Google Scholar] [CrossRef] [PubMed]
- Ikari, K.; Sakuma, Y.; Jimbo, T.; Kodama, A.; Imai, M.; Monnard, P.A.; Rasmussen, S. Dynamics of fatty acid vesicles in response to pH stimuli. Soft Matter 2015, 11, 6327–6334. [Google Scholar] [CrossRef] [PubMed]
- Hitz, T.; Luisi, P.L. Liposome-assisted selective polycondensation of α-amino acids and peptides. Pept. Sci. 2000, 55, 381–390. [Google Scholar] [CrossRef]
- Zepik, H.H.; Rajamani, S.; Maurel, M.C.; Deamer, D. Oligomerization of thioglutamic acid: Encapsulated reactions and lipid catalysis. Orig. Life Evol. Biosph. 2007, 37, 495–505. [Google Scholar] [CrossRef]
- Joshi, M.P.; Sawant, A.A.; Rajamani, S. Spontaneous emergence of membrane-forming protoamphiphiles from a lipid–amino acid mixture under wet–dry cycles. Chem. Sci. 2021, 12, 2970–2978. [Google Scholar] [CrossRef]
- Berclaz, N.; Müller, M.; Walde, P.; Luisi, P.L. Growth and transformation of vesicles studied by ferritin labeling and cryotransmission electron microscopy. J. Phys. Chem. B 2001, 105, 1056–1064. [Google Scholar] [CrossRef]
- Cornell, C.E.; Black, R.A.; Xue, M.; Litz, H.E.; Ramsay, A.; Gordon, M.; Mileant, A.; Cohen, Z.R.; Williams, J.A.; Lee, K.K.; et al. Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc. Natl. Acad. Sci. USA 2019, 116, 17239–17244. [Google Scholar] [CrossRef]
- Todd, Z.R.; Cohen, Z.R.; Catling, D.C.; Keller, S.L.; Black, R.A. Growth of prebiotically plausible fatty acid vesicles proceeds in the presence of prebiotic amino acids, dipeptides, sugars, and nucleic acid components. Langmuir 2022, 38, 15106–15112. [Google Scholar] [CrossRef]

| Generations | Water [NTC] | Water [TR] | EPS [NTC] | EPS [TR] |
|---|---|---|---|---|
| 1–2 | −0.18 | −0.62 * | 0.43 | −0.09 |
| 2–3 | 0.12 | 0.01 | 0.24 | 0.65 * |
| 3–4 | 0.00 | −0.08 | −0.38 | −0.05 |
| 4–5 | −0.25 | −0.41 | 0.71 * | 0.03 |
| 5–7 | −0.11 | 0.02 | −0.51 | -0.29 |
| 7–8 | 0.50 | −0.20 | 0.04 | 0.68 * |
| 8–9 | −0.55 | −0.77 * | 0.36 | −0.02 |
| 9–10 | −0.50 | −0.52 | 0.17 | −0.19 |
| 10–11 | 0.19 | 0.42 | 0.30 | 0.58 * |
| 11–12 | 0.04 | 0.18 | −0.01 | 0.34 |
| 12–13 | −0.14 | 0.27 | 0.16 | −0.07 |
| 13–14 | 0.01 | 0.22 | 0.18 | 0.17 |
| 14–16 | 0.39 | −0.06 | 0.28 | 0.47 |
| 16–17 | 0.29 | −0.18 | 0.06 | 0.28 |
| 17–18 | 0.43 | −0.43 | 0.31 | 0.37 |
| 18–20 | 0.13 | 0.11 | −0.19 | −0.09 |
| 20–21 | 0.04 | −0.57 | −0.27 | 0.12 |
| 21–23 | 0.59 * | 0.13 | 0.09 | 0.31 |
| 23–25 | 0.43 | 0.21 | −0.22 | −0.23 |
| 25–26 | −0.58 * | −0.04 | −0.52 | 0.32 |
| 26–27 | 0.14 | −0.23 | 0.50 | 0.64 * |
| 27–28 | −0.15 | −0.23 | −0.15 | −0.35 |
| 28–29 | 0.24 | −0.25 | −0.09 | 0.07 |
| 29–30 | −0.21 | 0.76 * | 0.09 | −0.04 |
| Mean r | 0.04 | −0.09 | 0.07 | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolskyi, T.; Ganju, P.; Montgomery-Taylor, R.; Baum, D.A. Evidence of Heritability in Prebiotically Realistic Membrane-Bound Systems. Life 2024, 14, 284. https://doi.org/10.3390/life14030284
Sokolskyi T, Ganju P, Montgomery-Taylor R, Baum DA. Evidence of Heritability in Prebiotically Realistic Membrane-Bound Systems. Life. 2024; 14(3):284. https://doi.org/10.3390/life14030284
Chicago/Turabian StyleSokolskyi, Tymofii, Pavani Ganju, Ronan Montgomery-Taylor, and David A. Baum. 2024. "Evidence of Heritability in Prebiotically Realistic Membrane-Bound Systems" Life 14, no. 3: 284. https://doi.org/10.3390/life14030284
APA StyleSokolskyi, T., Ganju, P., Montgomery-Taylor, R., & Baum, D. A. (2024). Evidence of Heritability in Prebiotically Realistic Membrane-Bound Systems. Life, 14(3), 284. https://doi.org/10.3390/life14030284







