Potato Wart Isolates from Europe and North America Form Distinct Clusters of Genetic Variation
Abstract
1. Introduction
- To develop and test an extended set of nuclear SSR markers for the analysis of genetic diversity in Synchytrium endobioticum;
- To analyze genetic differentiation in a set of 51 isolates from diverse geographic origins, mostly from Europe, with high reliability using a combination of the new and published markers and various computational methods;
- To search for potential diagnostic markers characteristic of the distinct clusters, useful for the characterization of new undefined field samples.
2. Materials and Methods
2.1. Potato Wart Isolates
2.2. Simple Sequence Repeat Markers
2.3. Data Analysis
3. Results
3.1. Development of New SSR and SCAR Markers
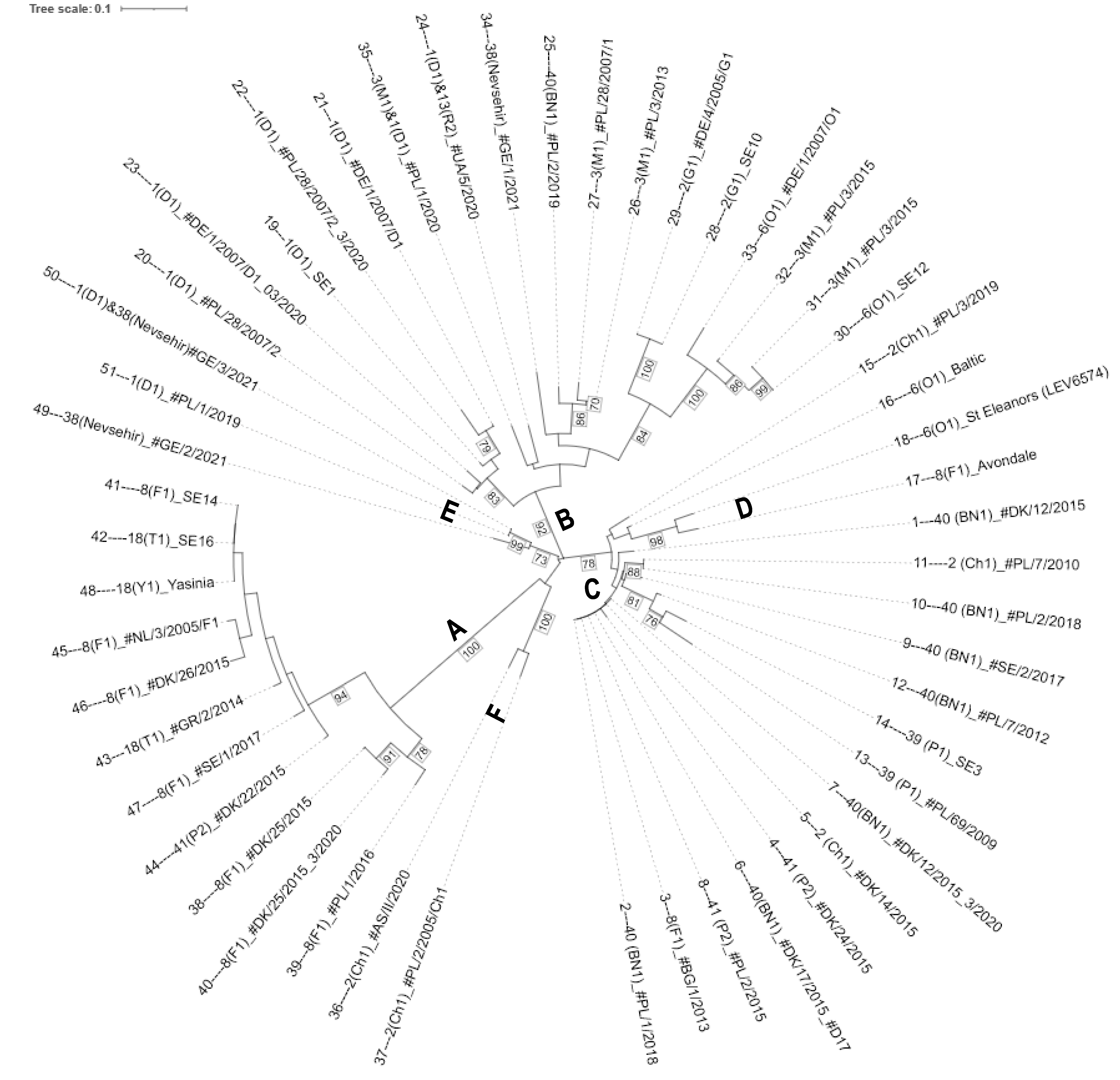
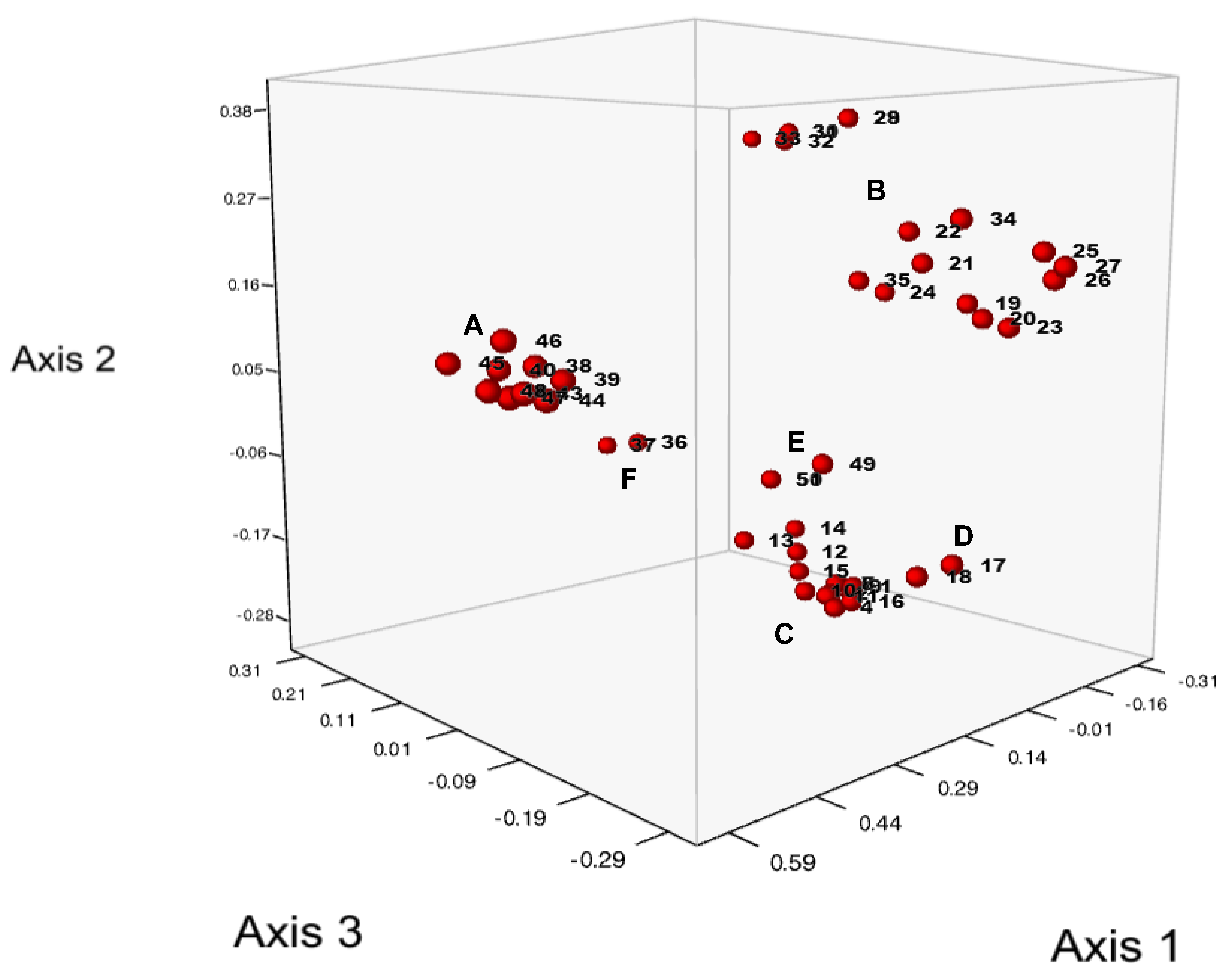
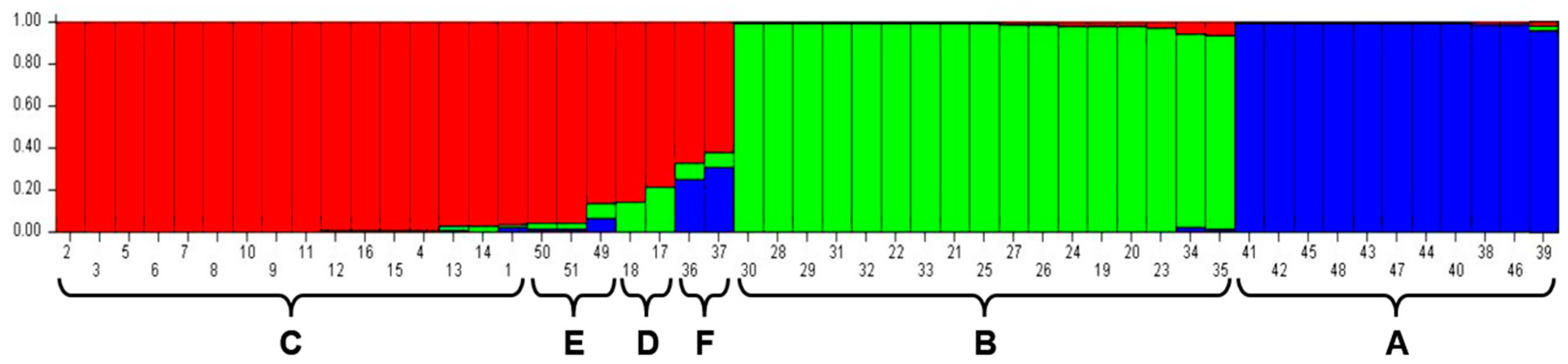
3.2. Analysis of Genetic Diversity of Wart Isolates
3.3. Markers Specific to Subclusters A and B
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Obidiegwu, J.E.; Flath, K.; Gebhardt, C. Managing potato wart: A review of present research status and future perspective. Theor. Appl. Genet. 2014, 127, 763–780. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.L.H.; Prodhomme, C.; Vossen, J.H.; van der Lee, T.A.J. Synchytrium endobioticum, the potato wart disease pathogen. Mol. Plant Pathol. 2022, 23, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Prodhomme, C.; van Arkel, G.; Plich, J.; Tammes, J.E.; Rijk, J.; van Eck, H.J.; Visser, R.G.F.; Vossen, J.H. A Hitchhiker’s guide to the potato wart disease resistance galaxy. Theor. Appl. Genet. 2020, 133, 3419–3439. [Google Scholar] [CrossRef] [PubMed]
- Schleusner, Y.; Gossmann, M.; Bandte, M.; Buttner, C. Inactivation of plant pathogens during anaerobic digestion based on case studies. Julius-Kuhn-Archiv 2012, 438, 61. [Google Scholar] [CrossRef]
- van Leeuwen, G.; Heungens, K.K.; Przetakiewicz, J.; Boerma, M.; Dimitrova, L.; Flath, K. A standardised set of differential potato cultivars to identify pathotypes in Synchytrium endobioticum. Phytopathol. Off. J. Am. Phytopathol. Soc. 2018, 108, 1. [Google Scholar]
- Luderer, R.; Joosten, M.H. Avirulence proteins of plant pathogens: Determinants of victory and defeat. Mol. Plant Pathol. 2001, 2, 355–364. [Google Scholar] [CrossRef]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal effectors, the double edge sword of phytopathogens. Curr. Genet. 2021, 67, 27–40. [Google Scholar] [CrossRef]
- Erper, I.; Ozer, G.; Kalendar, R.; Avci, S.; Yildirim, E.; Alkan, M.; Turkkan, M. Genetic Diversity and Pathogenicity of Rhizoctonia spp. Isolates Associated with Red Cabbage in Samsun (Turkey). J. Fungi 2021, 7, 234. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; McDonald, B.A. The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 2008, 46, 75–100. [Google Scholar] [CrossRef]
- Bonants, P.J.M.; van Gent-Pelzer, M.P.E.; van Leeuwen, G.C.M.; van der Lee, T.A.J. A real-time TaqMan PCR assay to discriminate between pathotype 1 (D1) and non-pathotype 1 (D1) isolates of Synchytrium endobioticum. Eur. J. Plant Pathol. 2015, 143, 495–506. [Google Scholar] [CrossRef]
- Smith, D.S.; Rocheleau, H.; Chapados, J.T.; Abbott, C.; Ribero, S.; Redhead, S.A.; Lévesque, C.A.; de Boer, S.H. Phylogeny of the genus Synchytrium and the development of TaqMan PCR assay for sensitive detection of Synchytrium endobioticum in soil. Phytopathology 2014, 104, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.S.; Singh, U. A molecular method for determining the viability of Synchytrium endobioticum. Can. J. Plant Pathol. 2017, 39, 108. [Google Scholar]
- van de Vossenberg, B.T.L.H.; te Braak, N.; Warbroek, T.; Meffert, J.P.; Rosendahl-Peters, K.H.M.; van Gent-Pelzer, M.P.E.; van Rijswick, P.C.J. Comparison of real-time PCR tests for the detection of Synchytrium endobioticum resting spores in soil and their potential application in descheduling of previously infested plots. EPPO Bull. 2022, 52, 154–163. [Google Scholar] [CrossRef]
- van den Boogert, P.H.J.F.; van Gent-Pelzer, M.P.E.; Bonants, P.J.M.; de Boer, S.H.; Wander, J.G.N.; Lévesque, C.A.; van Leeuwen, G.C.M.; Baayen, R.P. Development of PCR-based Detection Methods for the Quarantine Phytopathogen Synchytrium endobioticum, Causal Agent of Potato Wart Disease. Eur. J. Plant Pathol. 2005, 113, 47–57. [Google Scholar] [CrossRef]
- Busse, F.; Bartkiewicz, A.; Terefe-Ayana, D.; Niepold, F.; Schleusner, Y.; Flath, K.; Sommerfeldt-Impe, N.; Lübeck, J.; Strahwald, J.; Tacke, E.; et al. Genomic and Transcriptomic Resources for Marker Development in Synchytrium endobioticum, an Elusive but Severe Potato Pathogen. Phytopathology 2017, 107, 322–328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gagnon, M.-C.; van der Lee, T.A.J.; Bonants, P.J.M.; Smith, D.S.; Li, X.; Lévesque, C.A.; Bilodeau, G.J. Development of Polymorphic Microsatellite Loci for Potato Wart from Next-Generation Sequence Data. Phytopathology 2016, 106, 636–644. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.L.H.; Warris, S.; Nguyen, H.D.T.; van Gent-Pelzer, M.P.E.; Joly, D.L.; van de Geest, H.C.; Bonants, P.J.M.; Smith, D.S.; Lévesque, C.A.; van der Lee, T.A.J. Comparative genomics of chytrid fungi reveal insights into the obligate biotrophic and pathogenic lifestyle of Synchytrium endobioticum. Sci. Rep. 2019, 9, 8672. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.L.H.; Brankovics, B.; Nguyen, H.D.T.; van Gent-Pelzer, M.P.E.; Smith, D.; Dadej, K.; Przetakiewicz, J.; Kreuze, J.F.; Boerma, M.; van Leeuwen, G.C.M.; et al. The linear mitochondrial genome of the quarantine chytrid Synchytrium endobioticum; insights into the evolution and recent history of an obligate biotrophic plant pathogen. BMC Evol. Biol. 2018, 18, 136. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.; van Gent, M.; Meffert, J.P.; Nguyen, H.D.; Smith, D.; van Kempen, T.; Helderman, C.M.; Rosendahl-Peters, K.C.; Braak, N.t.; Flath, K.; et al. Molecular characterization and comparisons of potato wart (Synchytrium endobioticum) in historic collections to recent findings in Canada and the Netherlands. J. Plant Pathol. 2023, 1–13. [Google Scholar] [CrossRef]
- Przetakiewicz, J. Sampling, maintenance and pathotype identification of Synchytrium endobioticum (Schilb.) perc. Plant Breed. Seed Sci. 2017, 76, 29–36. [Google Scholar] [CrossRef]
- Przetakiewicz, J. The Viability of Winter Sporangia of Synchytrium endobioticum (Schilb.) Perc. from Poland. Am. J. Potato Res. 2015, 92, 704–708. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: https://darwin.cirad.fr/ (accessed on 6 July 2023).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- van de Vossenberg, B.T.L.H.; Prodhomme, C.; van Arkel, G.; van Gent-Pelzer, M.P.E.; Bergervoet, M.; Brankovics, B.; Przetakiewicz, J.; Visser, R.G.F.; van der Lee, T.A.J.; Vossen, J.H. The Synchytrium endobioticum AvrSen1 triggers a hypersensitive response in Sen1 potatoes while natural variants evade detection. Mol. Plant-Microbe Interact. 2019, 32, 1536–1546. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure. A simulation study. Mol.Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- McDonald, B.A. How can research on pathogen population biology suggest disease management strategies? The example of barley scald (Rhynchosporium commune). Plant Pathology 2015, 64, 1005–1013. [Google Scholar] [CrossRef]
- MCDonald, B.A.; Stukenbrock, E.H. Rapid emergence of pathogens in agro-ecosystems: Global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. London, Ser. B-Biol. Sci. 2016, 371, 20160026. [Google Scholar] [CrossRef]
- MCDonald, B.A.; Mundt, C.C. How Knowledge of Pathogen Population Biology Informs Management of Septoria Tritici Blotch. Phytopathology 2016, 106, 948–955. [Google Scholar] [CrossRef]
- Hampson, M.C.; Wood, S.L.; Coombes, J.W. Detection of resting spores of Synchytrium endobioticum in soil from vehicles at Port-aux-Basques, Newfoundland. Can. J. Plant Pathol. 1996, 18, 59–63. [Google Scholar] [CrossRef]
- Baayen, R.P.; Cochius, G.; Hendriks, H.; Meffert, J.P.; Bakker, J.; Bekker, M.; van den Boogert, P.H.J.F.; Stachewicz, H.; van Leeuwen, G.C.M. History of potato wart disease in Europe—A proposal for harmonisation in defining pathotypes. Eur. J. Plant. Pathol. 2006, 116, 21–31. [Google Scholar] [CrossRef]
- Bojnansky, V. Potato wart pathotypes in Europe from the ecological point of view. EPPO Bull. 1984, 14, 141–146. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röhrs, I.; Linde, M.; Przetakiewicz, J.; Zelya, A.; Zelya, G.; Pucher, A.; Tlapák, H.; Debener, T. Potato Wart Isolates from Europe and North America Form Distinct Clusters of Genetic Variation. Life 2023, 13, 1883. https://doi.org/10.3390/life13091883
Röhrs I, Linde M, Przetakiewicz J, Zelya A, Zelya G, Pucher A, Tlapák H, Debener T. Potato Wart Isolates from Europe and North America Form Distinct Clusters of Genetic Variation. Life. 2023; 13(9):1883. https://doi.org/10.3390/life13091883
Chicago/Turabian StyleRöhrs, Ina, Marcus Linde, Jaroslaw Przetakiewicz, Avrelia Zelya, George Zelya, Anna Pucher, Hana Tlapák, and Thomas Debener. 2023. "Potato Wart Isolates from Europe and North America Form Distinct Clusters of Genetic Variation" Life 13, no. 9: 1883. https://doi.org/10.3390/life13091883
APA StyleRöhrs, I., Linde, M., Przetakiewicz, J., Zelya, A., Zelya, G., Pucher, A., Tlapák, H., & Debener, T. (2023). Potato Wart Isolates from Europe and North America Form Distinct Clusters of Genetic Variation. Life, 13(9), 1883. https://doi.org/10.3390/life13091883




