Chondrocyte Homeostasis and Differentiation: Transcriptional Control and Signaling in Healthy and Osteoarthritic Conditions
Abstract
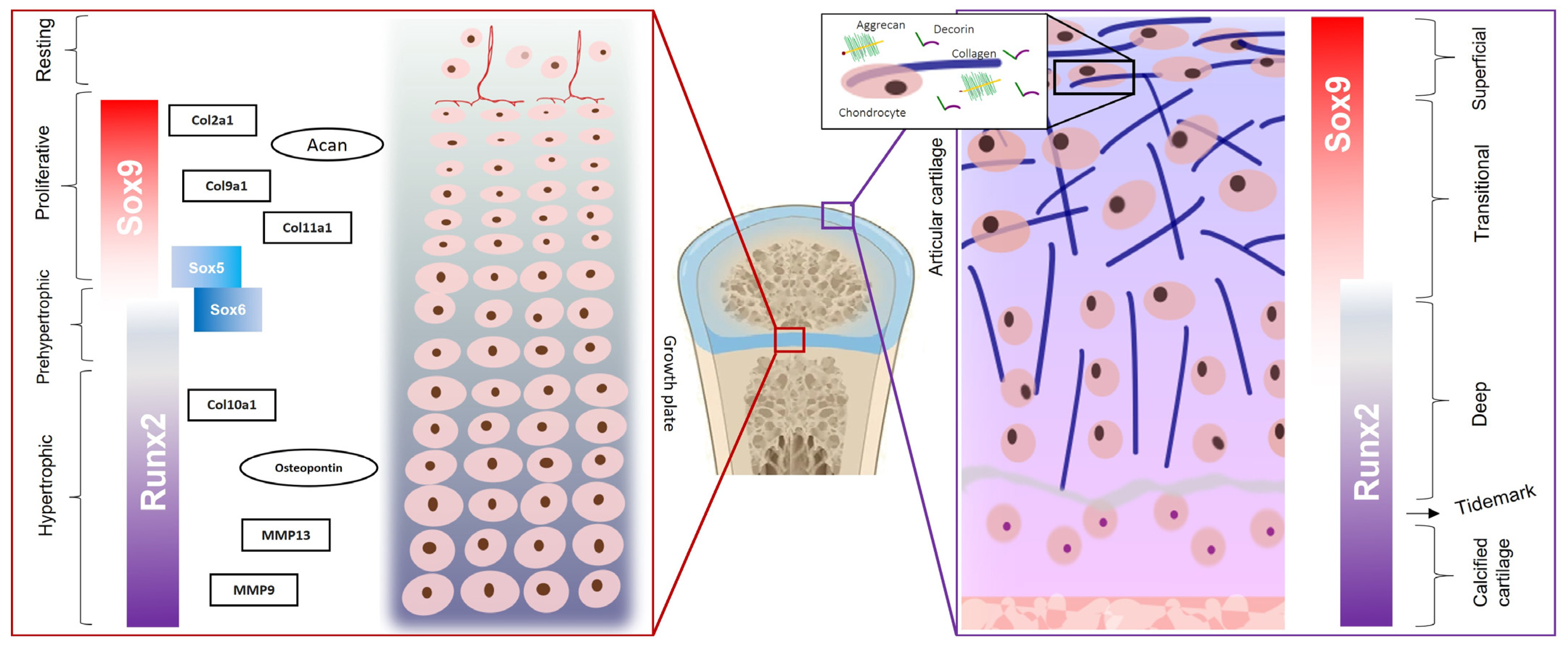
1. Cartilage Organization: Chondrocytes and Extracellular Matrix
2. Chondrocyte Differentiation
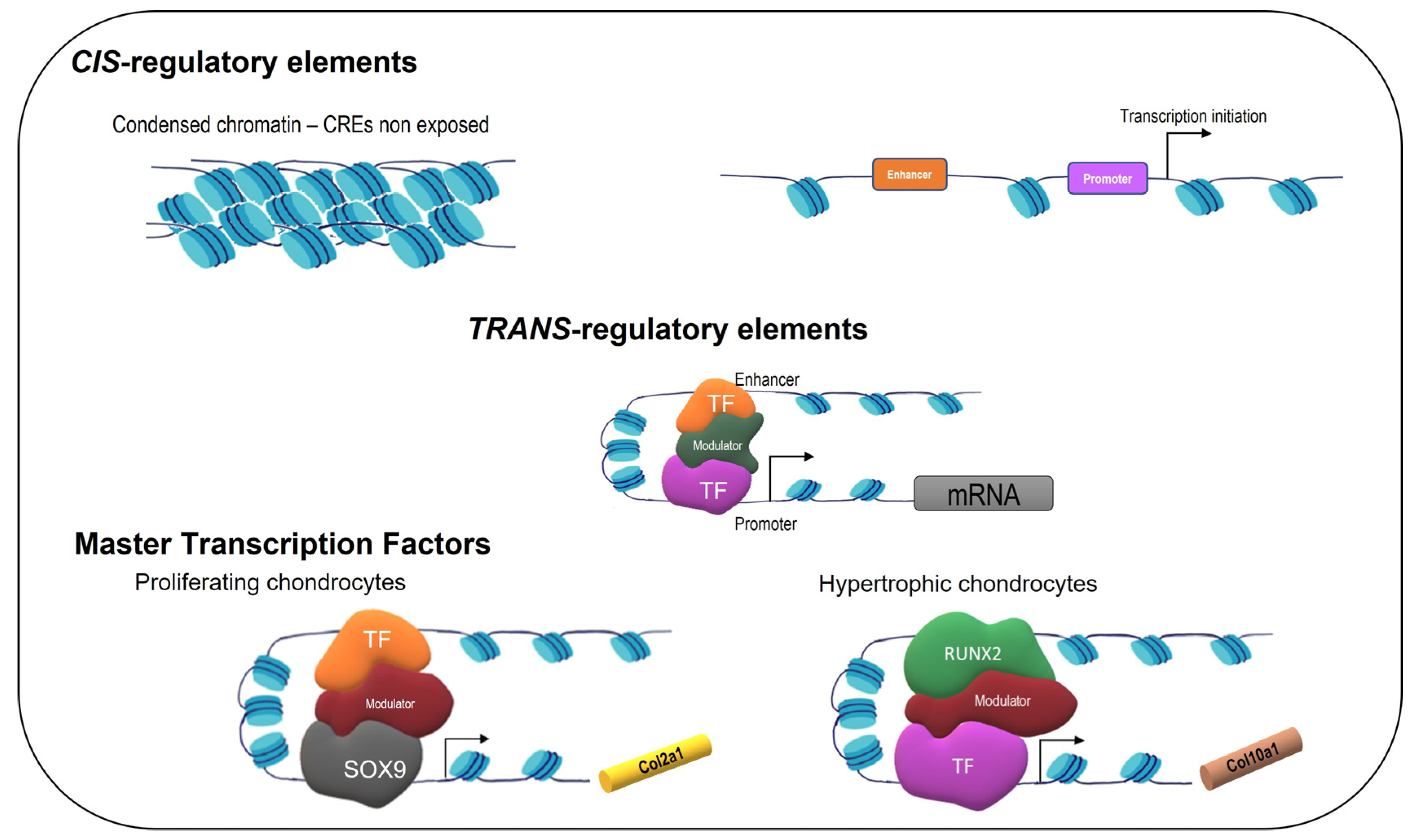
2.1. Control of Gene Expression
2.2. SOX9, SOX5, and SOX6
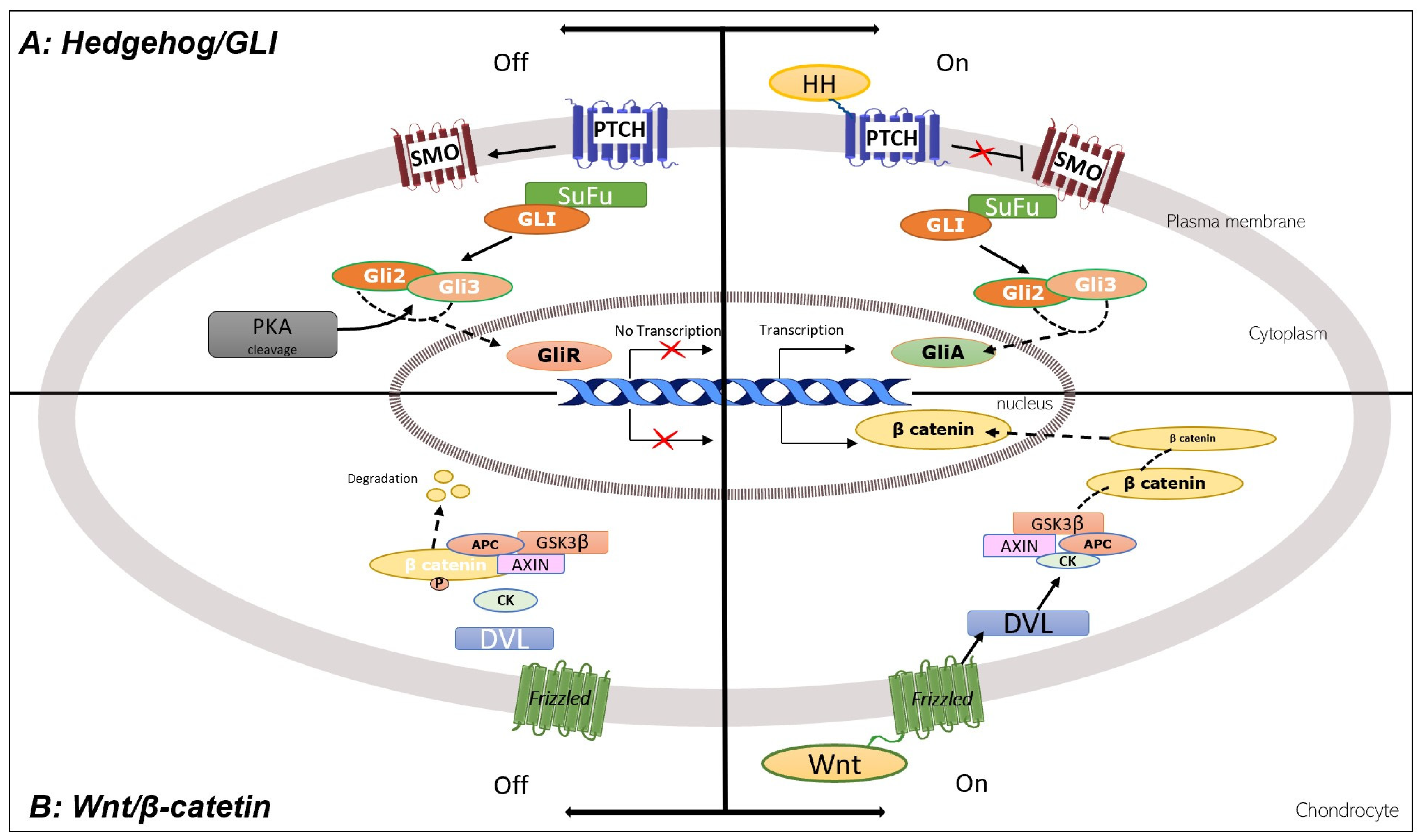
2.3. GLI1, GLI2, and GLI3
2.4. RUNX2
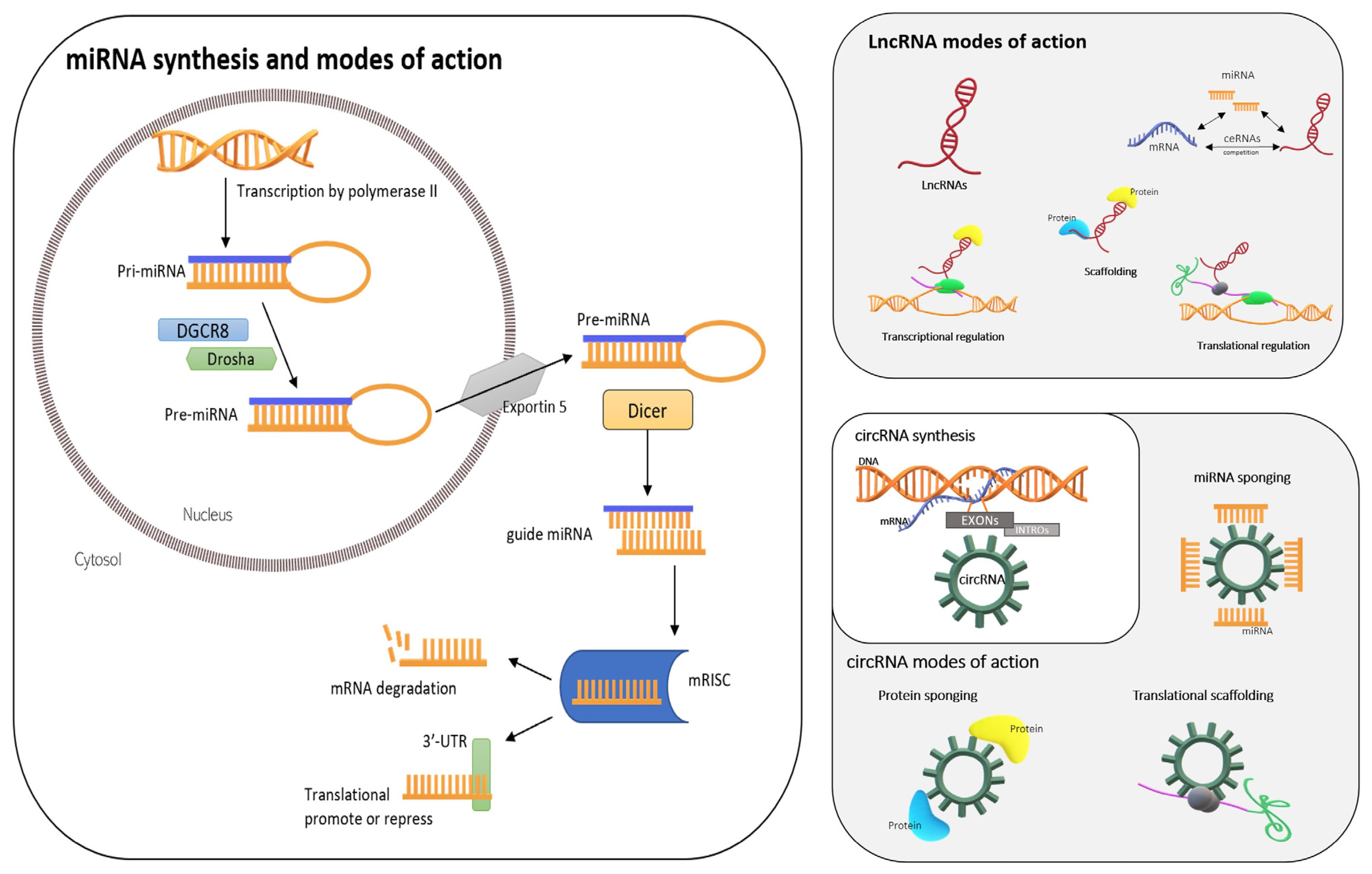
2.5. ncRNAs: miRNAs, lncRNAs, and circRNA
2.5.1. miRNAs
2.5.2. lncRNAs
2.5.3. circRNAs
3. Signaling in Chondrocyte
3.1. Hedgehog, GLI, TRPSI, and Wnt/Catenin
3.2. TGFβ, BMP, and SMAD4
3.3. FGF
3.4. Notch
3.5. NFkB, TLR, TNF-α, and IL-1
4. Chondrocyte Death
4.1. Necrosis, Apoptosis, Chondroptosis, and Autophagy
4.2. Oxidative Stress-Induced Senescence
5. Protection of Chondrocytes against Inflammation
5.1. Chondroitin Sulfate and Glucosamine
5.2. Other Agents
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, Y.; Sinkeviciute, D.; He, Y.; Karsdal, M.; Henrotin, Y.; Mobasheri, A.; Önnerfjord, P.; Bay-Jensen, A. The minor collagens in articular cartilage. Protein Cell 2017, 8, 560–572. [Google Scholar] [CrossRef]
- Pfaff, M.; Aumailley, M.; Specks, U.; Knolle, J.; Zerwes, H.; Timpl, R. Integrin and Arg-Gly-Asp dependence of cell adhesion to the native and unfolded triple helix of collagen type VI. Exp. Cell Res. 1993, 206, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.; van der Rest, M.; Bruckner, P.; Rodriguez, E.; Winterhalter, K.; Vaughan, L. Identification of the type IX collagen polypeptide chains. The alpha 2(IX) polypeptide carries the chondroitin sulfate chain(s). J. Biol. Chem. 1986, 261, 5965–5968. [Google Scholar] [CrossRef]
- Gannon, J.; Walker, G.; Fischer, M.; Carpenter, R.; Thompson, R.; Oegema, T. Localization of type X collagen in canine growth plate and adult canine articular cartilage. J. Orthop. Res. 1991, 9, 485–494. [Google Scholar] [CrossRef]
- Iozzo, R. Matrix proteoglycans: From molecular design to cellular function. Annu. Rev. Biochem. 1998, 67, 609–652. [Google Scholar] [CrossRef]
- Iozzo, R.; Murdoch, A. Proteoglycans of the extracellular environment: Clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996, 10, 598–614. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.; Cohen, I.; Grässel, S.; Murdoch, A. The biology of perlecan: The multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem. J. 1994, 302, 625–639. [Google Scholar] [CrossRef]
- Roughley, P. The structure and function of cartilage proteoglycans. Eur. Cells Mater. 2006, 12, 92–101. [Google Scholar] [CrossRef]
- Rucci, N.; Rufo, A.; Alamanou, M.; Capulli, M.; Del Fattore, A.; Ahrman, E.; Capece, D.; Iansante, V.; Zazzeroni, F.; Alesse, E.; et al. The glycosaminoglycan-binding domain of PRELP acts as a cell type-specific NF-kappaB inhibitor that impairs osteoclastogenesis. J. Cell Biol. 2009, 187, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Wiberg, C.; Klatt, A.; Wagener, R.; Paulsson, M.; Bateman, J.; Heinegård, D.; Morgelin, M. Complexes of matrilin-1 and biglycan or decorin connect collagen VI microfibrils to both collagen II and aggrecan. J. Biol. Chem. 2003, 278, 37698–37704. [Google Scholar] [CrossRef]
- Baccarin, R.; Seidel, S.; Michelacci, Y.; Tokawa, P.; Oliveira, T. Osteoarthritis: A common disease that should be avoided in the athletic horse’s life. Anim. Front. 2022, 12, 25–36. [Google Scholar] [CrossRef]
- Poole, A. Cartilage in health and disease. In Arthritis and Allied Conditions: A Textbook of Rheumatology; Koopman, W.S., Ed.; Lippincott, Williams, and Wilkins: Philadelphia, PA, USA, 2005; Volume 15, pp. 223–269. [Google Scholar]
- Tomkoria, S.; Patel, R.; Ma, J. Heterogeneous nanomechanical properties of superficial and zonal regions of articular cartilage of the rabbit proximal radius condyle by atomic force microscopy. Med. Eng. Phys. 2004, 26, 815–822. [Google Scholar] [CrossRef]
- Lippiello, L.; Hall, D.; Mankin, H. Collagen synthesis in normal and osteoarthritic human cartilage. J. Clin. Investig. 1977, 59, 593–600. [Google Scholar] [CrossRef] [PubMed]
- de Crombrugghe, B.; Lefebvre, V.; Behringer, R.; Bi, W.; Murakami, S.; Huang, W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000, 19, 389–394. [Google Scholar] [CrossRef]
- Leung, V.; Gao, B.; Leung, K.; Melhado, I.; Wynn, S.; Au, T.; Dung, N.; Lau, J.; Mak, A.; Chan, D.; et al. SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 2011, 7, e1002356. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.; Bedi, A.; Rodeo, S. The basic science of articular cartilage. Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar]
- Hojo, H. Emerging RUNX2-mediated gene regulatory mechanisms consisting of multi-layered regulatory networks in skeletal development. Int. J. Mol. Sci. 2023, 24, 2979. [Google Scholar] [CrossRef]
- Davie, J.; Moniwa, M. Control of chromatin remodeling. Crit. Rev. Eukaryot. Gene Expr. 2000, 10, 303–325. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Rasheed, Z.; Ramamurthy, S.; Anbazhagan, A.; Voss, F.; Haqqi, T. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010, 62, 1361–1371. [Google Scholar] [CrossRef]
- Lafont, J.; Moustaghfir, S.; Durand, A.; Mallein-Gerin, F. The epigenetic players and the chromatin marks involved in the articular cartilage during osteoarthritis. Front. Physiol. 2023, 14, 1070241. [Google Scholar] [CrossRef]
- Lefebvre, V.; Zhou, G.; Mukhopadhyay, K.; Smith, C.; Zhang, Z.; Eberspaecher, H.; Zhou, X.; Sinha, S.; Maity, S.; de Crombrugghe, B. An 18-base-pair sequence in the mouse proalpha1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell. Biol. 1996, 16, 4512–4523. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Keller, B.; Zhou, G.; Napierala, D.; Chen, Y.; Zabel, B.; Parker, A.; Lee, B. Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J. Bone Miner. Res. 2009, 24, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lu, Y.; Ding, M.; Napierala, D.; Abbassi, S.; Chen, Y.; Duan, X.; Wang, S.; Lee, B.; Zheng, Q. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J. Bone Miner. Res. 2011, 26, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Lories, R.; Luyten, F. The bone-cartilage unit in osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Michelacci, Y.; Mourão, P.; Laredo, J.; Dietrich, C. Chondroitin sulfates and proteoglycans from normal and arthrosic human cartilage. Connect. Tissue Res. 1979, 7, 29–36. [Google Scholar] [CrossRef]
- Burr, D.; Gallant, M. Bone remodelling in osteoarthritis. Nat. Rev. Rheumatol. 2012, 8, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.; Goldring, S. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann. N. Y. Acad. Sci. 2010, 1192, 230–237. [Google Scholar] [CrossRef]
- Grafe, I.; Alexander, S.; Peterson, J.; Snider, T.; Levi, B.; Lee, B.; Mishina, Y. TGF-β Family Signaling in Mesenchymal Differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10, a022202. [Google Scholar] [CrossRef]
- Bi, W.; Deng, J.; Zhang, Z.; Behringer, R.; de Crombrugghe, B. Sox9 is required for cartilage formation. Nat. Genet. 1999, 22, 85–89. [Google Scholar] [CrossRef]
- Henry, S.; Liang, S.; Akdemir, K.; de Crombrugghe, B. The postnatal role of Sox9 in cartilage. J. Bone Miner. Res. 2012, 27, 2511–2525. [Google Scholar] [CrossRef]
- Dy, P.; Wang, W.; Bhattaram, P.W.Q.W.L.; Ballock, R.; Lefebvre, V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell 2012, 22, 597–609. [Google Scholar] [CrossRef]
- Tew, S.; Li, Y.; Pothacharoen, P.; Tweats, L.; Hawkins, R.; Hardingham, T. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthr. Cartil. 2005, 13, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V. The SoxD transcription factors–Sox5, Sox6, and Sox13–are key cell fate modulators. Int. J. Biochem. Cell Biol. 2010, 42, 429–432. [Google Scholar] [CrossRef]
- Akiyama, H.; Lefebvre, V. Unraveling the transcriptional regulatory machinery in chondrogenesis. J. Bone Miner. Metab. 2011, 29, 390–395. [Google Scholar] [CrossRef]
- Ng, L.; Wheatley, S.; Muscat, G.; Conway-Campbell, J.; Bowles, J.; Wright, E.; Bell, D.; Tam, P.; Cheah, K.; Koopman, P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol. 1997, 183, 108–121. [Google Scholar] [CrossRef]
- Dy, P.; Smits, P.; Silvester, A.; Penzo-Mendez, A.; Dumitriu, B.; Han, Y.; de la Motte, C.; Kingsley, D.; Lefebvre, V. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev. Biol. 2010, 341, 346–359. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.; McMahon, A. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef]
- Komori, T. Whole aspect of Runx2 functions in skeletal development. Int. J. Mol. Sci. 2022, 23, 5776. [Google Scholar] [CrossRef] [PubMed]
- Stricker, S.; Fundele, R.; Vortkamp, A.; Mundlos, S. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 2002, 245, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Guo, Y.; Wang, C.; Chen, F.; Di, C.; Dong, S.; Cao, Q.; Guo, J.; Rong, Y.; Yao, S.; et al. Lineage-specific rearrangement of chromatin loops and epigenomic features during adipocytes and osteoblasts commitment. Cell Death Differ. 2022, 29, 2503–2518. [Google Scholar] [CrossRef]
- Nagata, K.; Hojo, H.; Chang, S.; Okada, H.; Yano, F.; Chijimatsu, R.; Omata, Y.; Mori, D.; Makii, Y.; Kawata, M.; et al. Runx2 and Runx3 differentially regulate articular chondrocytes during surgically induced osteoarthritis development. Nat. Commun. 2022, 13, 6187. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, N.; De Guire, V. miRNAs: Their discovery, biogenesis and mechanism of action. Clin. Biochem. 2013, 46, 842–845. [Google Scholar] [CrossRef]
- Bartel, D. microRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Marcu, K.; Otero, M.; Olivotto, E.; Borzi, R.; Goldring, M. NF-kappaB signaling: Multiple angles to target OA. Curr. Drug Targets 2010, 11, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Meng, H.; Yuan, X.; Wang, Y.; Guo, Q.; Peng, J.; Wang, A.; Lu, S. MicroRNAs’ involvement in osteoarthritis and the prospects for treatments. Evid.-Based Complement. Altern. Med. 2015, 2015, 236179. [Google Scholar] [CrossRef]
- Endisha, H.; Rockel, J.; Jurisica, I.; Kapoor, M. The complex landscape of microRNAs in articular cartilage: Biology, pathology, and therapeutic targets. JCI Insight 2018, 3, e121630. [Google Scholar] [CrossRef]
- Miyaki, S.; Nakasa, T.; Otsuki, S.; Grogan, S.; Higashiyama, R.; Inoue, A.; Kato, Y.; Sato, T.; Lotz, M.; Asahara, H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009, 60, 2723–2730. [Google Scholar] [CrossRef]
- Miyaki, S.; Sato, T.; Inoue, A.; Otsuki, S.; Ito, Y.; Yokoyama, S.; Kato, Y.; Takemoto, F.; Nakasa, T.; Yamashita, S.; et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010, 24, 1173–1185. [Google Scholar] [CrossRef]
- Qin, H.; Wang, C.; He, Y.; Lu, A.; Li, T.; Zhang, B.; Shen, J. Silencing miR-146a-5p protects against injury-induced osteoarthritis in mice. Biomolecules 2023, 13, 123. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.J. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Sun, H.; Peng, G.; Ning, X.; Wang, J.; Yang, H.; Deng, J. Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis. Am. J. Transl. Res. 2019, 11, 16–30. [Google Scholar] [PubMed]
- Patty, B.; Hainer, S. Non-Coding RNAs and nucleosome remodeling complexes: An intricate regulatory relationship. Biology 2020, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Carlson, H.; Quinn, J.; Yang, Y.; Thornburg, C.; Chang, H.; Stadler, H. LncRNA-HIT Functions as an Epigenetic Regulator of Chondrogenesis through Its Recruitment of p100/CBP Complexes. PLoS Genet. 2015, 11, e1005680. [Google Scholar] [CrossRef] [PubMed]
- Barter, M.; Gomez, R.; Hyatt, S.; Cheung, K.; Skelton, A.; Xu, Y.; Clark, I.; Young, D. The long non-coding RNA ROCR contributes to SOX9 expression and chondrogenic differentiation of human mesenchymal stem cells. Development 2017, 144, 4510–4521. [Google Scholar]
- Zhu, J.; Yu, W.; Wang, Y.; Xia, K.; Huang, Y.; Xu, A.; Chen, Q.; Liu, B.; Tao, H.; Li, F.; et al. lncRNAs: Function and mechanism in cartilage development, degeneration, and regeneration. Stem Cell Res. Ther. 2019, 10, 344. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Zhang, Y.; Lu, X.; Chen, L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Busa, V.; Leung, A. Thrown for a (stem) loop: How RNA structure impacts circular RNA regulation and function. Methods 2021, 196, 56–67. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, L.; Liu, K.; Wang, M.; Su, X.; Wang, J. Regulatory mechanism of circular RNA involvement in osteoarthritis. Front. Surg. 2023, 9, 1049513. [Google Scholar] [CrossRef]
- Usami, Y.; Gunawardena, A.; Iwamoto, M.; Enomoto-Iwamoto, M. Wnt signaling in cartilage development and diseases: Lessons from animal studies. Lab. Investig. 2016, 96, 186–196. [Google Scholar] [CrossRef]
- Chung, U.; Schipani, E.; McMahon, A.; Kronenberg, H. Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J. Clin. Investig. 2001, 107, 295–304. [Google Scholar] [CrossRef]
- Lim, J.; Tu, X.; Choi, K.; Akiyama, H.; Mishina, Y.; Long, F. BMP-Smad4 signaling is required for pre-cartilaginous mesenchymal condensation independent of Sox9 in the mouse. Dev. Biol. 2015, 400, 132–138. [Google Scholar] [CrossRef]
- Ellman, M.B.; Yan, D.; Ahmadinia, K.; Chen, D.; An, H.S.; Im, H.J. Fibroblast growth factor control of cartilage homeostasis. J. Cell. Biochem. 2013, 114, 735–742. [Google Scholar] [CrossRef]
- Zieba, J.; Chen, Y.; Lee, B.; Bae, Y. Notch signaling in skeletal development, homeostasis and pathogenesis. Biomolecules 2020, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Therond, P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 14751–14756. [Google Scholar] [CrossRef]
- Gai, Z.; Gui, T.; Muragaki, Y. The function of TRPS1 in the development and differentiation of bone, kidney, and hair follicles. Histol. Histopathol. 2011, 26, 915–921. [Google Scholar] [PubMed]
- Sassi, N.; Laadhar, L.; Allouche, M.; Achek, A.; Kallel-Sellami, M.; Makni, S.; Sellami, S. WNT signaling and chondrocytes: From cell fate determination to osteoarthritis physiopathology. J. Recept. Signal Transduct. 2014, 34, 73–80. [Google Scholar] [CrossRef]
- Chun, J.; Oh, H.; Yang, S.; Park, M. Wnt signaling in cartilage development and degeneration. BMB Rep. 2008, 41, 485–494. [Google Scholar] [CrossRef]
- Wuelling, M.; Schneider, S.; Schröther, V.; Waterkamp, C.; Hoffmann, D.; Vortkamp, A. Wnt5a is a transcriptional target of Gli3 and Trps1 at the onset of chondrocyte hypertrophy. Dev. Biol. 2020, 457, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, C.; Li, Y. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef]
- Wu, M.; Chen, G.; Li, Y. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.; Itoh, N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef]
- Wing, L.; Chen, H.; Chuang, P.; Wu, M.; Tsai, S. The mammalian target of rapamycin p70 ribosomal S6 kinase but not phosphatidylinositol 3 kinase akt signaling is responsible for fibroblast growth factor 9 induced cell proliferation. J. Biol. Chem. 2005, 280, 19937. [Google Scholar] [CrossRef]
- Sahni, M.; Ambrosetti, D.; Mansukhani, A.; Gertner, R.; Levy, D.; Basilico, C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT 1 pathway. Genes Dev. 1999, 13, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- El Seoudi, A.; Abd El Kader, T.; Nishida, T.; Eguchi, T.; Aoyama, E.; Takigawa, M.; Kubota, S. Catabolic effects of FGF 1 on chondrocytes and its possible role in osteoarthritis. J. Cell Commun. Signal. 2017, 11, 255–263. [Google Scholar] [CrossRef]
- Yan, D.; Chen, D.; Cool, S.; van Wijnen, A.; Mikecz, K.; Murphy, G.; Im, H. Fibroblast growth factor receptor 1 is principally responsible for fibroblast growth factor 2 induced catabolic activities in human articular chondrocytes. Arthritis Res. Ther. 2011, 13, R130. [Google Scholar] [CrossRef]
- Nummenmaa, E.; Hämäläinen, M.; Moilanen, T.; Vuolteenaho, K.; Moilanen, E. Effects of FGF 2 and FGF receptor antagonists on MMP enzymes, aggrecan, and type II collagen in primary human OA chondrocytes. Scand. J. Rheumatol. 2015, 44, 321–330. [Google Scholar] [CrossRef]
- Uchii, M.; Tamura, T.; Suda, T.; Kakuni, M.; Tanaka, A.; Miki, I. Role of fibroblast growth factor 8 (FGF8) in animal models of osteoarthritis. Arthritis Res. Ther. 2008, 10, R9. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Z.; Tang, J.; Li, W.; Huang, J.; Xu, W.; Luo, F.; Xu, M.; Wang, J.; Wen, X.; et al. Exogenous fibroblast growth factor 9 attenuates cartilage degradation and aggravates osteophyte formation in post-traumatic osteoarthritis. Osteoarthr. Cartil. 2016, 24, 2181–2192. [Google Scholar] [CrossRef]
- Ellsworth, J.; Berry, J.; Bukowski, T.; Claus, J.; Feldhaus, A.; Holderman, S.; Holdren, M.; Lum, K.; Moore, E.R.F.; Ren, H.S.P.S.C.; et al. Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthr. Cartil. 2002, 10, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.; Bendele, A.; Thompson, D.; Littau, A.; Waggie, K.; Reardon, B.; Ellsworth, J. Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthr. Cartil. 2005, 13, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Saito, T.C.S.; Kobayashi, H.; Ladel, C.; Guehring, H.; Chung, U.; Kawaguchi, H. Identification of fibroblast growth factor-18 as a molecule to protect adult articular cartilage by gene expression profiling. J. Biol. Chem. 2014, 289, 10192–10200. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Li, Y.; Shang, C.; Shang, G.; Kou, H.; Li, J.; Chen, S.; Liu, H. Sprifermin: Effects on cartilage homeostasis and therapeutic prospects in cartilage-related diseases. Front. Cell Dev. Biol. 2021, 9, 786546. [Google Scholar] [CrossRef]
- Hosaka, Y.; Saito, T.; Sugita, S.; Hikata, T.; Kobayashi, H.; Fukai, A.; Taniguchi, Y.; Hirata, M.; Akiyama, H.; Chung, U.; et al. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc. Natl. Acad. Sci. USA 2013, 110, 1875–1880. [Google Scholar] [CrossRef]
- Saito, T.; Tanaka, S. Molecular mechanisms underlying osteoarthritis development: Notch and NF-Κb. Arthritis Res. Ther. 2017, 19, 94. [Google Scholar] [CrossRef]
- Salucci, S.; Falcieri, E.; Battistelli, M. Chondrocyte death involvement in osteoarthritis. Cell Tissue Res. 2022, 389, 159–170. [Google Scholar] [CrossRef]
- Jørgensen, A.; Kjær, A.; Heinemeier, K. The effect of aging and mechanical loading on the metabolism of articular cartilage. J. Rheumatol. 2017, 44, 410–417. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, X.; Shang, W.; Xu, J.; Wang, X.; Hu, X.; Ao, Y.; Cheng, H. Proinflammatory cytokines stimulate mitochondrial superoxide flashes in articular chondrocytes in vitro and in situ. PLoS ONE 2013, 8, e66444. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Y.; Zhang, D.; Zhang, M.; Wang, M.; Zhang, Y.; Ma, T.; Chen, J. Death of chondrocytes in Kashin-Beck disease: Apoptosis, necrosis or necroptosis? Int. J. Exp. Pathol. 2018, 99, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.; Gupta, K.; Franco, S.; Liu, B. Necroptosis in the pathophysiology of disease. Am. J. Pathol. 2020, 190, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Xiang, S.; Wang, Y.; Dong, Y.; Li, Z.; Xiang, Y.; Bian, Y.; Feng, B.; Yang, B.; Weng, X. NR4A1 promotes TNF-α-induced chondrocyte death and migration injury via activating the AMPK/Drp1/mitochondrial fission pathway. Int. J. Mol. Med. 2020, 45, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Stolberg, J.; Sambale, M.; Hansen, U.; Schäfer, A.; Raschke, M.; Bertrand, J.; Pap, T.; Sherwood, J. Cartilage trauma induces necroptotic chondrocyte death and expulsion of cellular contents. Int. J. Mol. Sci. 2020, 21, 4204. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Hwang, H.; Kim, H. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef]
- Charlier, E.; Relic, B.; Deroyer, C.; Malaise, O.; Neuville, S.; Collée, J.; Malaise, M.; De Seny, D. Insights on molecular mechanisms of chondrocytes death in osteoarthritis. Int. J. Mol. Sci. 2016, 17, 2146. [Google Scholar] [CrossRef]
- Roach, H.; Aigner, T.; Kouri, J. Chondroptosis: A variant of apoptotic cell death in chondrocytes? Apoptosis 2004, 9, 265–277. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Huang, J.; Lam, G.; Brumell, J. Autophagy signaling through reactive oxygen species. Antioxid. Redox Signal. 2011, 14, 2215–2231. [Google Scholar] [CrossRef]
- Sasaki, H.; Takayama, K.; Matsushita, T.; Ishida, K.; Kubo, S.; Matsumoto, T.; Fujita, N.; Oka, S.; Kurosaka, M.; Kuroda, R. Autophagy modulates osteoarthritis-related gene expression in human chondrocytes. Arthritis Rheum. 2012, 64, 1920–1928. [Google Scholar] [CrossRef]
- Maiuri, M.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef]
- Caramés, B.; Taniguchi, N.; Otsuki, S.; Blanco, F.; Lotz, M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010, 62, 791–801. [Google Scholar] [CrossRef]
- Gao, T.; Guo, W.; Chen, M.; Huang, J.; Yuan, Z.; Zhang, Y.; Wang, M.; Li, P.; Peng, J.; Wang, A.; et al. Extracellular Vesicles and Autophagy in Osteoarthritis. Biomed. Res. Int. 2016, 2016, 2428915. [Google Scholar] [CrossRef] [PubMed]
- Miyaki, S.; Lotz, M. Extracellular vesicles in cartilage homeostasis and osteoarthritis. Curr. Opin. Rheumatol. 2018, 30, 129–135. [Google Scholar] [CrossRef]
- Bovellan, M.; Fritzsche, M.; Stevens, C.; Charras, G. Death associated protein kinase (DAPK) and signal transduction: Blebbing in programmed cell death. FEBS J. 2010, 277, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Jaovisidha, K.; Hung, J.; Ning, G.; Ryan, L.; Derfus, B. Comparative calcification of native articular cartilage matrix vesicles and nitroprusside-generated vesicles. Osteoarthr. Cartil. 2002, 10, 646–652. [Google Scholar] [CrossRef]
- Kirsch, T.; Wang, W.; Pfander, D. Functional differences between growth plate apoptotic bodies and matrix vesicles. J. Bone Miner. Res. 2003, 18, 1872–1881. [Google Scholar] [CrossRef]
- Wang, A.; Lukas, T.; Yuan, M.; Du, N.; Tso, M.; Neufeld, A. Autophagy and exosomes in the aged retinal pigment epithelium: Possible relevance to drusen formation and age-related macular degeneration. PLoS ONE 2009, 4, e4160. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, A.; Gohr, C.; Mitton-Fitzgerald, E.; Grewal, R.; Ninomiya, J.; Coyne, C.; Jackson, W. Autophagy modulates articular cartilage vesicle formation in primary articular chondrocytes. J. Biol. Chem. 2015, 290, 13028–13038. [Google Scholar] [CrossRef]
- Gao, S.; Zeng, C.; Li, L.; Luo, W.; Zhang, F.; Tian, J.; Cheng, C.; Tu, M.; Xiong, Y.; Jiang, W.; et al. Correlation between senescence-associated beta-galactosidase expression in articular cartilage and disease severity of patients with knee osteoarthritis. Int. J. Rheum. Dis. 2016, 19, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Z.; Li, T.; Xu, H.; Zhang, H. Senescence in osteoarthritis: From mechanism to potential treatment. Arthritis Res. Ther. 2022, 24, 174. [Google Scholar] [CrossRef]
- Wu, C.; Liu, R.; Huan, S.; Tang, W.; Zeng, Y.; Zhang, J.; Yang, J.; Li, Z.; Zhou, Y.; Zha, Z.; et al. Senescent skeletal cells cross-talk with synovial cells plays a key role in the pathogenesis of osteoarthritis. Arthritis Res. Ther. 2022, 24, 59. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of cellular senescence: Cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol. 2021, 29, 645593. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lipa, K.; Alexander, P.; Clark, K.; Lin, H. Potential methods of targeting cellular aging hallmarks to reverse osteoarthritic phenotype of chondrocytes. Biology 2022, 11, 996. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, X.; Zhou, J.; Wei, L. The age-related changes in cartilage and osteoarthritis. Biomed. Res. Int. 2013, 2013, 916530. [Google Scholar] [CrossRef]
- Varesi, A.; Chirumbolo, S.; Campagnoli, L.; Pierella, E.; Piccini, G.; Carrara, A.; Ricevuti, G.; Scassellati, C.; Bonvicini, C.; Pascale, A. The role of antioxidants in the interplay between oxidative stress and senescence. Antioxidants 2022, 11, 1224. [Google Scholar] [CrossRef]
- Lettieri-Barbato, D.; Aquilano, K.; Punziano, C.; Minopoli, G.; Faraonio, R. MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in the redox control of cell senescence. Antioxidants 2022, 11, 480. [Google Scholar] [CrossRef]
- Loeser, R. Aging and osteoarthritis: The role of chondrocyte senescence and aging changes in the cartilage. Osteoarthr. Cartil. 2009, 17, 971–979. [Google Scholar] [CrossRef]
- Henrotin, Y.; Bruckner, P.; Pujol, J. The role of reactive oxygen species in homeostasis and degradation of cartilage. Osteoarthr. Cartil. 2003, 11, 747–755. [Google Scholar] [CrossRef]
- Vinatier, C.; Domínguez, E.; Guicheux, J.; Caramés, B. Role of the inflammation-autophagy-senescence integrative network in osteoarthritis. Front. Physiol. 2018, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Suh, N. MicroRNA controls of cellular senescence. BMB Rep. 2018, 51, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 71–478. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.; Fahmi, H. Role of pro-inflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Baccarin, B.; Machado, T.; Lopes-Moraes, A.; Vieira, F.; Michelacci, Y. Urinary glycosaminoglycans in horse osteoarthritis. Effects of chondroitin sulfate and glucosamine. Res. Vet. Sci. 2012, 93, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Mikuls, T.; O’dell, J.; Moore, G.; Palmer, W. Dietary supplement use by rheumatology and internal medicine clinic patients results of a survey questionnaire. J. Clin. Rheumatol. 1999, 5, 255–258. [Google Scholar] [CrossRef]
- Miller, K.; Clegg, D. Glucosamine and chondroitin sulfate. Rheum. Dis. Clin. N. Am. 2011, 37, 103–118. [Google Scholar] [CrossRef]
- da Cunha, A.; de Oliveira, L.; Maia, L.; de Oliveira, L.; Michelacci, Y.; de Aguiar, J. Pharmaceutical grade chondroitin sulfate: Structural analysis and identification of contaminants in different commercial preparations. Carbohydr. Polym. 2015, 134, 300–308. [Google Scholar] [CrossRef]
- Ruiz-Romero, V.; Toledano-Serrabona, J.; Gay-Escoda, C. Efficacy of the use of chondroitin sulphate and glucosamine for the treatment of temporomandibular joint dysfunction: A systematic review and meta-analysis. CRANIO 2022, 19, 1–10. [Google Scholar] [CrossRef]
- Wandel, S.; Jüni, P.; Tendal, B.; Nüesch, E.; Villiger, P.M.; Welton, N.J.; Reichenbach, S.; Trelle, S. Effects of glucosamine, chondroitin, or placebo in patients with osteoarthritis of hip or knee: Network meta-analysis. BMJ 2010, 341, c4675. [Google Scholar] [CrossRef]
- Lippiello, L. Glucosamine and chondroitin sulfate: Biological response modifiers of chondrocytes under simulated conditions of joint stress. Osteoarthr. Cartil. 2003, 11, 335–342. [Google Scholar] [CrossRef] [PubMed]
- da Cunha, A.; Aguiar, J.; Correa da Silva, F.; Michelacci, Y. Do chondroitin sulfates with different structures have different activities on chondrocytes and macrophages? Int. J. Biol. Macromol. 2017, 103, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Therkildsen, M.; Aluko, R.; Lametsch, R. Exploration of collagen recovered from animal by-products as a precursor of bioactive peptides: Successes and challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 2011–2027. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Puig, D.; Costa-Larrión, E.; Rubio-Rodríguez, N.; Gálvez-Martín, P. Collagen supplementation for joint health: The link between composition and scientific knowledge. Nutrients 2023, 15, 1332. [Google Scholar] [CrossRef]
- Colletti, A.; Cicero, A. Nutraceutical approach to chronic osteoarthritis: From molecular research to clinical evidence. Int. J. Mol. Sci. 2021, 22, 12920. [Google Scholar] [CrossRef]
- Itano, N.; Sawai, T.; Yoshida, M.; Lenas, P.; Yamada, Y.; Imagawa, M.; Shinomura, T.; Hamaguchi, M.; Yoshida, Y.; Ohnuki, Y.; et al. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J. Biol. Chem. 1999, 274, 25085–25092. [Google Scholar] [CrossRef]
- Gupta, R.; Lall, R.; Srivastava, A.; Sinha, A. Hyaluronic acid: Molecular mechanisms and therapeutic trajectory. Front. Vet. Sci. 2019, 25, 192. [Google Scholar] [CrossRef]
- Neuenschwander, H.; Moreira, J.; Vendruscolo, C.; Fülber, J.; Seidel, S.; Michelacci, Y.; Baccarin, R. Hyaluronic acid has chondroprotective and joint-preserving effects on LPS-induced synovitis in horses. J. Vet. Sci. 2019, 20, e67. [Google Scholar] [CrossRef]
- Marinho, A.; Nunes, C.; Reis, S. Hyaluronic acid: A key ingredient in the therapy of inflammation. Biomolecules 2021, 11, 1518. [Google Scholar] [CrossRef]
- Peng, Y.; Ao, M.; Dong, B.; Jiang, Y.; Yu, L.; Chen, Z.; Hu, C.; Xu, R. Anti-Inflammatory effects of curcumin in the inflammatory diseases: Status, limitations and countermeasures. Drug Des. Devel. Ther. 2021, 15, 4503–4525. [Google Scholar] [CrossRef]
- Yang, S.; Sun, M.; Zhang, X. Protective effect of resveratrol on knee osteoarthritis and its molecular mechanisms: A recent review in preclinical and clinical trials. Front. Pharmacol. 2022, 13, 921003. [Google Scholar] [CrossRef] [PubMed]
- Cholet, J.; Decombat, C.; Delort, L.; Gainche, M.; Berry, A.; Ogeron, C.; Ripoche, I.; Vareille-Delarbre, M.; Vermerie, M.; Fraisse, D.; et al. Potential anti-inflammatory and chondroprotective effect of Luzula sylvatica. Int. J. Mol. Sci. 2022, 24, 127. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michelacci, Y.M.; Baccarin, R.Y.A.; Rodrigues, N.N.P. Chondrocyte Homeostasis and Differentiation: Transcriptional Control and Signaling in Healthy and Osteoarthritic Conditions. Life 2023, 13, 1460. https://doi.org/10.3390/life13071460
Michelacci YM, Baccarin RYA, Rodrigues NNP. Chondrocyte Homeostasis and Differentiation: Transcriptional Control and Signaling in Healthy and Osteoarthritic Conditions. Life. 2023; 13(7):1460. https://doi.org/10.3390/life13071460
Chicago/Turabian StyleMichelacci, Yara M., Raquel Y. A. Baccarin, and Nubia N. P. Rodrigues. 2023. "Chondrocyte Homeostasis and Differentiation: Transcriptional Control and Signaling in Healthy and Osteoarthritic Conditions" Life 13, no. 7: 1460. https://doi.org/10.3390/life13071460
APA StyleMichelacci, Y. M., Baccarin, R. Y. A., & Rodrigues, N. N. P. (2023). Chondrocyte Homeostasis and Differentiation: Transcriptional Control and Signaling in Healthy and Osteoarthritic Conditions. Life, 13(7), 1460. https://doi.org/10.3390/life13071460





