An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections
Abstract
1. Introduction
2. Material and Methods
2.1. Materials
2.2. GC-MS Analysis
2.3. Pharmacokinetics and Physicochemical Characteristics of WGO Compounds
2.4. Gene Interaction Network with Inflammatory Pathways and Genes
2.5. Enrichment Analysis for Anti-Inflammatory Action
2.6. Heatmap Cluster Analysis
2.7. Molecular Docking Analysis
3. Results
3.1. GC-MS Analysis
3.2. SwissADME Analysis
3.3. Heat Map Analysis
3.4. GeneMANIA Analysis
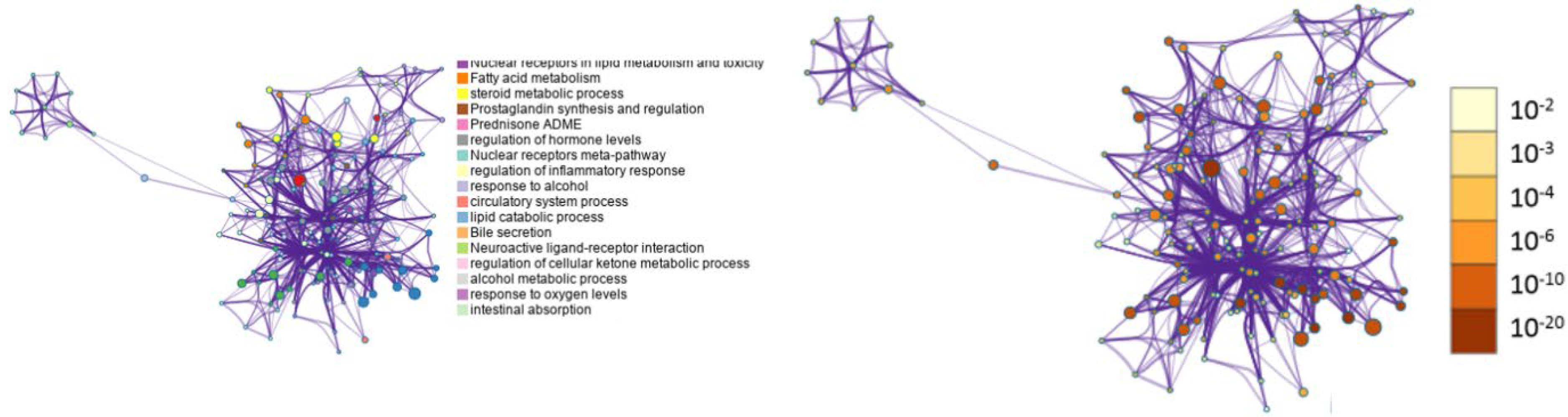
3.5. Metascape Analysis
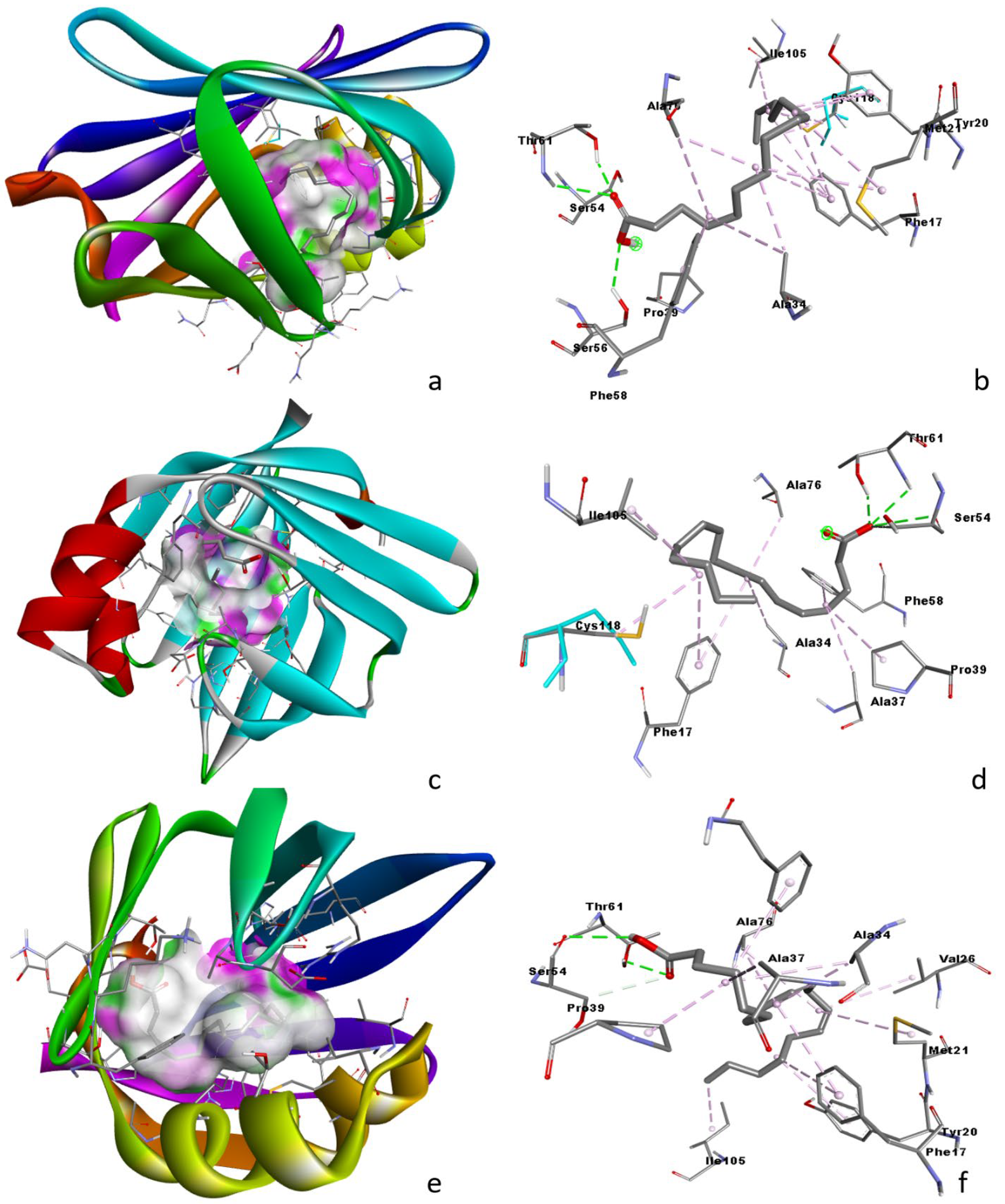
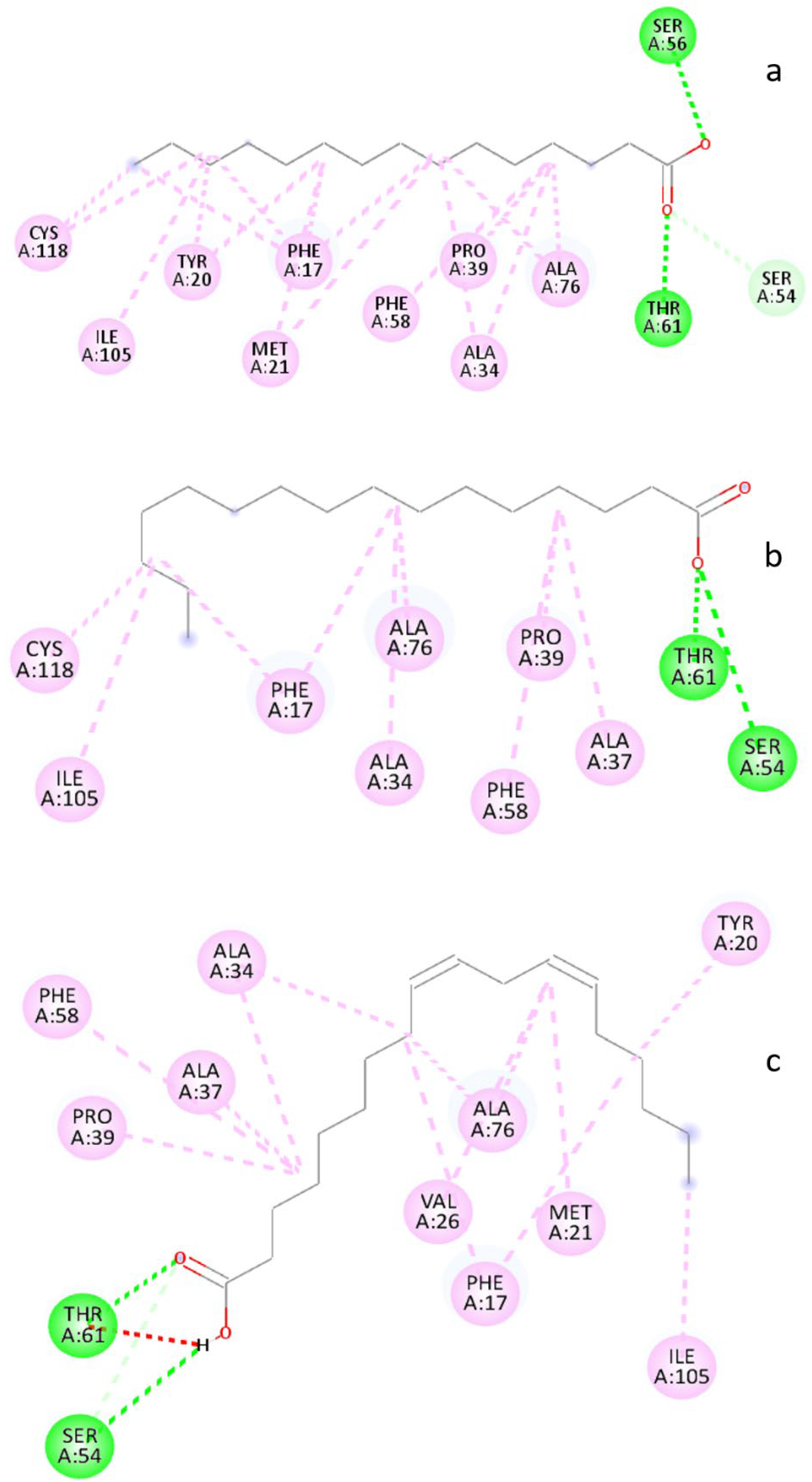
3.6. Molecular Docking Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slavin, J. Whole grains and human health. Nutr. Res. Rev. 2004, 17, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Vaher, M.; Matso, K.; Levandi, T.; Helmja, K.; Kaljurand, M. Phenolic compounds and the antioxidant activity of the bran, flour and whole grain of different wheat varieties. Procedia Chem. 2010, 2, 76–82. [Google Scholar] [CrossRef]
- Zhu, K.-X.; Lian, C.-X.; Guo, X.-N.; Peng, W.; Zhou, H.-M. Antioxidant activities and total phenolic contents of various extracts from defatted wheat germ. Food Chem. 2011, 126, 1122–1126. [Google Scholar] [CrossRef]
- Mahmoud, A.A.; Mohdaly, A.A.; Elneairy, N.A. Wheat germ: An overview on nutritional value, antioxidant potential and antibacterial characteristics. Food Nutr. Sci. 2015, 6, 265. [Google Scholar] [CrossRef]
- Weng, Z.; Chen, Y.; Liang, T.; Lin, Y.; Cao, H.; Song, H.; Xiong, L.; Wang, F.; Shen, X.; Xiao, J. A review on processing methods and functions of wheat germ-derived bioactive peptides. Crit. Rev. Food Sci. Nutr. 2021, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Meriles, S.P.; Steffolani, M.E.; León, A.E.; Penci, M.C.; Ribotta, P.D. Physico-chemical characterization of protein fraction from stabilized wheat germ. Food Sci. Biotechnol. 2019, 28, 1327–1335. [Google Scholar] [CrossRef]
- Budhwar, S.; Chakraborty, M.; Sethi, K.; Chatterjee, A. Antidiabetic properties of rice and wheat bran—A review. J. Food Biochem. 2020, 44, e13424. [Google Scholar] [CrossRef]
- Siraj, N. Wheat germ oil: A comprehensive review. Food Sci. Technol. 2022, 42. [Google Scholar] [CrossRef]
- Liaqat, H.; Jeong, E.; Kim, K.J.; Kim, J.Y. Effect of wheat germ on metabolic markers: A systematic review and meta-analysis of randomized controlled trials. Food Sci. Biotechnol. 2020, 29, 739–749. [Google Scholar] [CrossRef]
- Mazzocchi, A.; De Cosmi, V.; Risé, P.; Milani, G.P.; Turolo, S.; Syrén, M.-L.; Sala, A.; Agostoni, C. Bioactive compounds in edible oils and their role in oxidative stress and inflammation. Front. Physiol. 2021, 12, 659551. [Google Scholar] [CrossRef]
- Fajka-Boja, R.; Hidvegi, M.; Shoenfeld, Y.; Ion, G.; Demydenko, D.; Tomoskozi-Farkas, R.; Vizler, C.; Telekes, A.; Resetar, A.; Monostori, E. Fermented wheat germ extract induces apoptosis and downregulation of major histocompatibility complex class I proteins in tumor T and B cell lines. Int. J. Oncol. 2002, 20, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Heimbach, J.T.; Sebestyen, G.; Semjen, G.; Kennepohl, E. Safety studies regarding a standardized extract of fermented wheat germ. Int. J. Toxicol. 2007, 26, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lapis, K.; Szende, B. Effect of Avemar and Avemar+ Vitamin C on tumor growth and metastasis in experimental animals. Anticancer Res. 1998, 1000, 2353–2358. [Google Scholar]
- Farkas, E. Fermented wheat germ extract in the supportive therapy of colorectal cancer. Orv. Hetil. 2005, 146, 1925–1931. [Google Scholar]
- Koh, E.M.; Lee, E.K.; Song, J.; Kim, S.J.; Song, C.H.; Seo, Y.; Chae, C.H.; Jung, K.J. Anticancer activity and mechanism of action of fermented wheat germ extract against ovarian cancer. J. Food Biochem. 2018, 42, e12688. [Google Scholar] [CrossRef]
- Rezq, A.A.; Mahmoud, M.Y. Preventive effect of wheat germ on hypercholesteremic and atherosclerosis in rats fed cholesterol-containing diet. Pak J Nutr 2011, 10, 424–432. [Google Scholar] [CrossRef]
- Chadha, R.; Ram, H.; Purohit, A. Hypolipidemic effect of wheat germ oil in cholesterol fed Rabbits. Med. Drug Res. 2015, 3, 15–20. [Google Scholar]
- Akool, E.-S. Molecular mechanisms of the protective role of wheat germ oil against cyclosporin A-induced hepatotoxicity in rats. Pharm. Biol. 2015, 53, 1311–1317. [Google Scholar] [CrossRef]
- Iyer, A.; Brown, L. Fermented wheat germ extract (Avemar) in the treatment of cardiac remodeling and metabolic symptoms in rats. Evid.-Based Complement. Altern. Med. 2011, 2011, 508957. [Google Scholar] [CrossRef]
- Ojo, B.; Simenson, A.J.; O’Hara, C.; Wu, L.; Gou, X.; Peterson, S.K.; Lin, D.; Smith, B.J.; Lucas, E.A. Wheat germ supplementation alleviates insulin resistance and cardiac mitochondrial dysfunction in an animal model of diet-induced obesity. Br. J. Nutr. 2017, 118, 241–249. [Google Scholar] [CrossRef]
- Shahidi, F.; Danielski, R.; Rhein, S.O.; Meisel, L.A.; Fuentes, J.; Speisky, H.; Schwember, A.R.; de Camargo, A.C. Wheat and Rice beyond Phenolic Acids: Genetics, Identification Database, Antioxidant Properties, and Potential Health Effects. Plants 2022, 11, 3283. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.-Y.; Choi, Y.-S.; Lee, J.-K.; Lee, B.-J.; Kim, W.-K.; Kang, H. Anti-inflammatory activity of citric acid-treated wheat germ extract in lipopolysaccharide-stimulated macrophages. Nutrients 2017, 9, 730. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, H.O.; Kim, G.-N.; Song, J.-H. Anti-oxidant and anti-adipogenic effects of ethanol extracts from wheat germ and wheat germ fermented with Aspergillus oryzae. Prev. Nutr. Food Sci. 2015, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Alamery, S.; Zargar, S.; Yaseen, F.; Wani, T.A.; Siyal, A. Evaluation of the Effect of Wheat Germ Oil and Olmutinib on the Thioacetamide-Induced Liver and Kidney Toxicity in Mice. Life 2022, 12, 900. [Google Scholar] [CrossRef] [PubMed]
- Fărcaș, A.C.; Socaci, S.A.; Nemeș, S.A.; Pop, O.L.; Coldea, T.E.; Fogarasi, M.; Biriș-Dorhoi, E.S. An update regarding the bioactive compound of cereal by-products: Health benefits and potential applications. Nutrients 2022, 14, 3470. [Google Scholar] [CrossRef]
- Hanhoff, T.; Lücke, C.; Spener, F. Insights into binding of fatty acids by fatty acid binding proteins. In Cellular Lipid Binding Proteins; Springer: Berlin/Heidelberg, Germany, 2002; pp. 45–54. [Google Scholar]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Calder, P.C. Functional roles of fatty acids and their effects on human health. J. Parenter. Enter. Nutr. 2015, 39, 18S–32S. [Google Scholar] [CrossRef]
- Sokoła-Wysoczańska, E.; Wysoczański, T.; Wagner, J.; Czyż, K.; Bodkowski, R.; Lochyński, S.; Patkowska-Sokoła, B. Polyunsaturated fatty acids and their potential therapeutic role in cardiovascular system disorders—A review. Nutrients 2018, 10, 1561. [Google Scholar] [CrossRef]
- Domenichiello, A.F.; Kitson, A.P.; Bazinet, R.P. Is docosahexaenoic acid synthesis from α-linolenic acid sufficient to supply the adult brain? Prog. Lipid Res. 2015, 59, 54–66. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network pharmacology databases for traditional Chinese medicine: Review and assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef]
- Jia, C.-Y.; Li, J.-Y.; Hao, G.-F.; Yang, G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 2020, 25, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. Application of the SwissDrugDesign online resources in virtual screening. Int. J. Mol. Sci. 2019, 20, 4612. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Jo, W.H.; Hoang, N.H.M.; Kim, M.-S. Curcumin-attenuated TREM-1/DAP12/NLRP3/Caspase-1/IL1B, TLR4/NF-κB pathways, and tau hyperphosphorylation induced by 1, 2-diacetyl benzene: An in vitro and in silico study. Neurotox. Res. 2022, 40, 1272–1291. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.D.; Oh, H.; Kim, M.-S. The effects of chemical mixtures on lipid profiles in the Korean adult population: Threshold and molecular mechanisms for dyslipidemia involved. Environ. Sci. Pollut. Res. 2022, 29, 39182–39208. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, S.C. A Coding Basis and Three-in-One Integrated Data Visualization Method ‘Ana’for the Rapid Analysis of Multidimensional Omics Dataset. Life 2022, 12, 1864. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Balsinde, J. Anti-inflammatory Mechanisms of 7-cis-Hexadecenoic acid. In Proceedings of the CIBERDEM Annual Meeting, Cerdanyola del Valles, Spain, 17–19 May 2017. [Google Scholar]
- Bassaganya-Riera, J.; Hontecillas, R.; Beitz, D. Colonic anti-inflammatory mechanisms of conjugated linoleic acid. Clin. Nutr. 2002, 21, 451–459. [Google Scholar] [CrossRef]
- Venn-Watson, S.K.; Butterworth, C.N. Broader and safer clinically-relevant activities of pentadecanoic acid compared to omega-3: Evaluation of an emerging essential fatty acid across twelve primary human cell-based disease systems. PLoS ONE 2022, 17, e0268778. [Google Scholar] [CrossRef] [PubMed]
- Abdulkhaleq, L.; Assi, M.; Abdullah, R.; Zamri-Saad, M.; Taufiq-Yap, Y.; Hezmee, M. The crucial roles of inflammatory mediators in inflammation: A review. Vet. World 2018, 11, 627. [Google Scholar] [CrossRef] [PubMed]
- Izano, M.; Wei, E.K.; Tai, C.; Swede, H.; Gregorich, S.; Harris, T.B.; Klepin, H.; Satterfield, S.; Murphy, R.; Newman, A.B. Chronic inflammation and risk of colorectal and other obesity-related cancers: The health, aging and body composition study. Int. J. Cancer 2016, 138, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Perretti, M.; Leroy, X.; Bland, E.J.; Montero-Melendez, T. Resolution pharmacology: Opportunities for therapeutic innovation in inflammation. Trends Pharmacol. Sci. 2015, 36, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Laine, L. Gastrointestinal effects of NSAIDs and coxibs. J. Pain Symptom Manage. 2003, 25, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.; Knaus, E.E. Evolution of nonsteroidal anti-inflammatory drugs (NSAIDs): Cyclooxygenase (COX) inhibition and beyond. J. Pharm. Pharm. Sci. 2008, 11, 81s–110s. [Google Scholar] [CrossRef]
- Boros, L.G.; Lapis, K.; Szende, B.; Tömösközi-Farkas, R.; Balogh, Á.; Boren, J.; Marin, S.; Cascante, M.; Hidvégi, M. Wheat germ extract decreases glucose uptake and RNA ribose formation but increases fatty acid synthesis in MIA pancreatic adenocarcinoma cells. Pancreas 2001, 23, 141–147. [Google Scholar] [CrossRef]
- Anwar, M.; Mohamed, N. Amelioration of liver and kidney functions disorders induced by sodium nitrate in rats using wheat germ oil. J. Radiat. Res. Appl. Sci. 2015, 8, 77–83. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Ritchie, T.J.; Macdonald, S.J.; Peace, S.; Pickett, S.D.; Luscombe, C.N. Increasing small molecule drug developability in sub-optimal chemical space. MedChemComm 2013, 4, 673–680. [Google Scholar] [CrossRef]
- Li, N.; Kulkarni, P.; Badrinarayanan, A.; Kefelegn, A.; Manoukian, R.; Li, X.; Prasad, B.; Karasu, M.; McCarty, W.J.; Knutson, C.G. P-glycoprotein substrate assessment in drug discovery: Application of modeling to bridge differential protein expression across in vitro tools. J. Pharm. Sci. 2021, 110, 325–337. [Google Scholar] [CrossRef]
- Mao, H.; Han, B.; Li, H.; Tao, Y.; Wu, W. FABP4 knockdown suppresses inflammation, apoptosis and extracellular matrix degradation in IL-1β-induced chondrocytes by activating PPARγ to regulate the NF-κB signaling pathway. Mol. Med. Rep. 2021, 24, 855. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Vanden Berghe, W.; Vermeulen, L.; Delerive, P.; De Bosscher, K.; Staels, B.; Haegeman, G. A paradigm for gene regulation: Inflammation, NF-κB and PPAR. In Peroxisomal Disorders and Regulation of Genes; Springer: Berlin/Heidelberg, Germany, 2003; pp. 181–196. [Google Scholar]
- Tsuchida, A.; Yamauchi, T.; Takekawa, S.; Hada, Y.; Ito, Y.; Maki, T.; Kadowaki, T. Peroxisome proliferator–activated receptor (PPAR) α activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: Comparison of activation of PPARα, PPARγ, and their combination. Diabetes 2005, 54, 3358–3370. [Google Scholar] [CrossRef]
- Liu, N.; Liu, J.-T.; Ji, Y.-Y.; Lu, P.-P. C-reactive protein triggers inflammatory responses partly via TLR4/IRF3/NF-κB signaling pathway in rat vascular smooth muscle cells. Life Sci. 2010, 87, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L. Lipoprotein lipase: New roles for an “old” enzyme. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 111. [Google Scholar] [CrossRef] [PubMed]
- Di Filippo, L.; De Lorenzo, R.; Sciorati, C.; Capobianco, A.; Lorè, N.I.; Giustina, A.; Manfredi, A.A.; Rovere-Querini, P.; Conte, C. Adiponectin to leptin ratio reflects inflammatory burden and survival in COVID-19. Diabetes Metab. 2021, 47, 101268. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.; Zhang, L.; Halasz, N.A.; Hunter, T.; Chojkier, M. Nuclear export of phosphorylated C/EBPβ mediates the inhibition of albumin expression by TNF-α. EMBO J. 2001, 20, 6712–6723. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Romigh, T.; Arab, K.; Teresi, R.E.; Tada, Y.; Eng, C.; Plass, C. Activator protein 2 alpha (AP2α) suppresses 42 kDa C/CAAT enhancer binding protein α (p42C/EBPα) in head and neck squamous cell carcinoma. Int. J. Cancer 2009, 124, 1285–1292. [Google Scholar] [CrossRef]
- Wani, T.A.; Bakheit, A.H.; Al-Majed, A.A.; Altwaijry, N.; Baquaysh, A.; Aljuraisy, A.; Zargar, S. Binding and drug displacement study of colchicine and bovine serum albumin in presence of azithromycin using multispectroscopic techniques and molecular dynamic simulation. J. Mol. Liq. 2021, 333, 115934. [Google Scholar] [CrossRef]
- Wani, T.A.; Bakheit, A.H.; Zargar, S.; Alamery, S. Mechanistic competitive binding interaction study between olmutinib and colchicine with model transport protein using spectroscopic and computer simulation approaches. J. Photochem. Photobiol. A Chem. 2022, 426, 113794. [Google Scholar] [CrossRef]
- Zargar, S.; Wani, T.A. Protective Role of Quercetin in Carbon Tetrachloride Induced Toxicity in Rat Brain: Biochemical, Spectrophotometric Assays and Computational Approach. Molecules 2021, 26, 7526. [Google Scholar] [CrossRef] [PubMed]
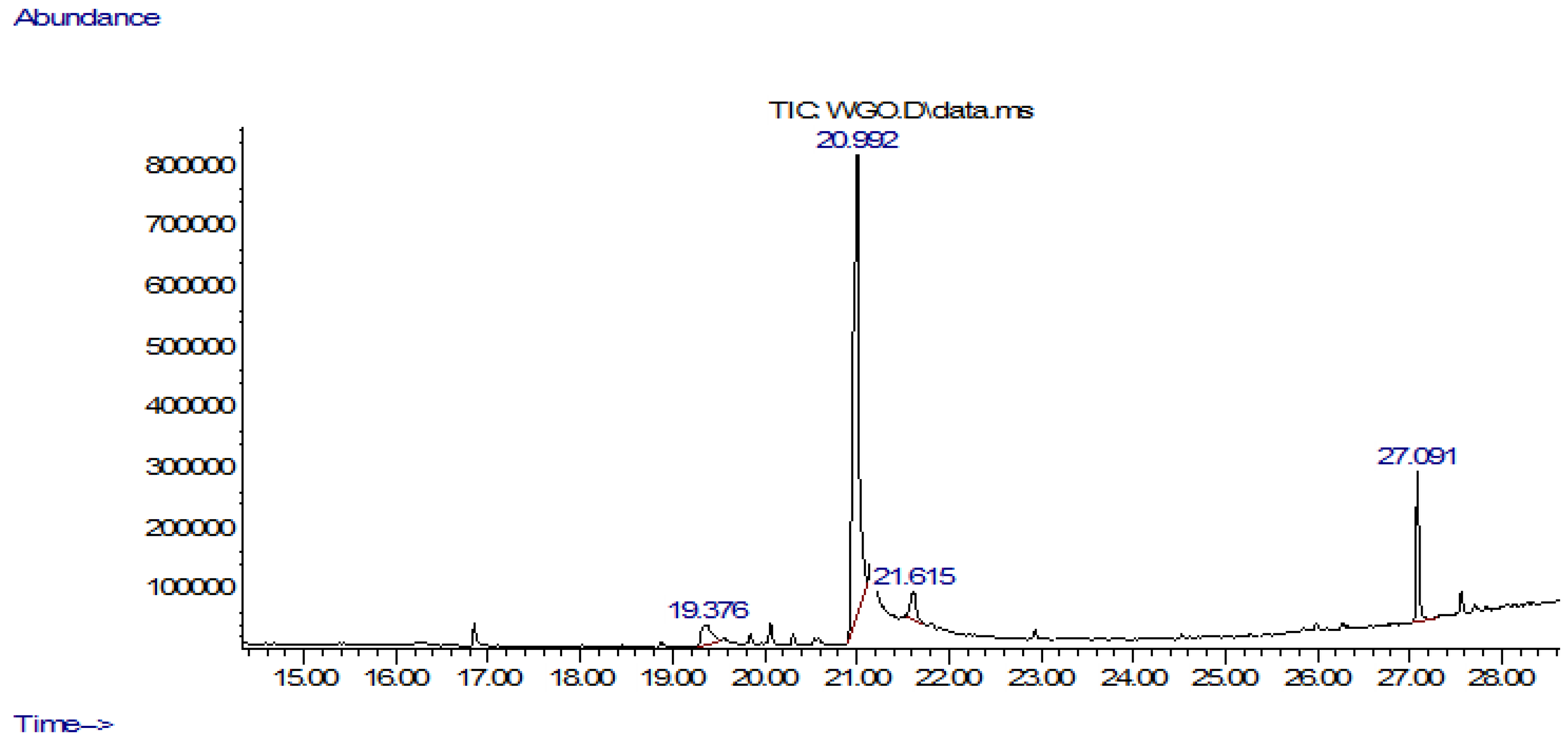




| S.No | RT (min) | Area (Ab*s) | Peak Width 50% (min) | Compound | Quality | Mol Weight (amu) |
|---|---|---|---|---|---|---|
| 1 | 12.136 | 765,017 | 0.463 | 2,4-Decadienal, (E,E)- | 80 | 152.12 |
| 2 | 16.863 | 95,170 | 0.189 | Cyclopentane, 1,1,3-trimethyl- | 53 | 112.125 |
| 3 | 18.906 | 43,909 | 0.189 | 1,4-Heptadiene, 3,3,6-trimethyl- | 12 | 138.141 |
| 4 | 19.397 | 337,266 | 0.265 | Pentadecanoic acid | 45 | 242.225 |
| 5 | 19.586 | 48,474 | 0.217 | n-Hexadecanoic acid | 60 | 256.24 |
| 6 | 19.842 | 43,328 | 0.123 | 2-Butyn-1-ol, 4-methoxy- | 10 | 100.052 |
| 7 | 20.314 | 42,111 | 0.132 | Cyclohexanol, 5-methyl-2-(1-methylethenyl)-[1R-(1.alpha.,2.beta.,5.alpha.)]-Cyclohexanol | 47 | 154.136 |
| 8 | 21.005 | 4,075,785 | 0.255 | trans-13-Octadecenoic acid | 99 | 282.256 |
| 9 | 21.288 | 724,675 | 0.369 | 9,12-Octadecadienoic acid (Z,Z)- | 99 | 280.24 |
| 10 | 22.933 | 42,472 | 0.113 | Piperazine, 1-[2-(2,5-dimethyl-1H-pyrrol-1-yl)ethyl]- | 30 | 207.174 |
| 11 | 27.103 | 593,470 | 0.265 | Squalene | 93 | 410.391 |
| 12 | 27.567 | 84,611 | 0.123 | Octadecane | 94 | 254.297 |
| NAME | C10H16O | C8H16 | C8H14 | C15H30O2 | C16H32O2 | C5H8O2 | C10H18O | C18H34O2 | C18H32O2 | C12H21N3 | C18H38 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight (100 g/mol) | 1.5223 | 1.1221 | 1.102 | 2.424 | 2.5742 | 1.0012 | 1.5425 | 2.8246 | 2.8045 | 2.0732 | 2.5449 |
| Num. heavy atoms | 11 | 8 | 8 | 17 | 18 | 7 | 11 | 20 | 20 | 15 | 18 |
| Num. arom. heavy atoms | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| Num. rotatable bonds | 6 | 0 | 3 | 13 | 14 | 1 | 1 | 15 | 14 | 3 | 15 |
| Num. H-bond acceptors | 1 | 0 | 0 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 0 |
| Num. H-bond donors | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Consensus Log Po/w | 2.85 | 3.07 | 2.87 | 4.84 | 5.2 | 0.18 | 2.42 | 5.64 | 5.45 | 1.28 | 7.18 |
| Log S (ESOL) | −2.44 | −2.81 | −2.49 | 0 | −5.03 | −0.05 | −2.59 | −5.41 | −5.05 | −1.67 | −6.33 |
| Solubility | 3.67 × 10−3 | 1.55 × 10−3 | 3.26 × 10−3 | 0 | 9.35 × 10−6 | 8.95 × 10−1 | 2.58 × 10−3 | 3.85 × 10−6 | 8.87 × 10−6 | 2.16 × 10−2 | 4.67 × 10−7 |
| Class | 1 | 1 | 1 | 0 | 0.5 | 2 | 1 | 0.5 | 0.5 | 2 | 0 |
| GI absorption | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| BBB permeant | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| P-gp substrate | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| CYP1A2 inhibitor | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 |
| CYP2C19 inhibitor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CYP2C9 inhibitor | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| CYP2D6 inhibitor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CYP3A4 inhibitor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Log Kp (skin permeation) | −4.92 | −4.42 | −4.54 | −3.07 | −2.78 | −7.3 | −5.15 | −2.6 | −3.05 | −7.01 | −1.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zargar, S.; Wani, T.A.; Rizwan Ahamad, S. An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections. Life 2023, 13, 526. https://doi.org/10.3390/life13020526
Zargar S, Wani TA, Rizwan Ahamad S. An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections. Life. 2023; 13(2):526. https://doi.org/10.3390/life13020526
Chicago/Turabian StyleZargar, Seema, Tanveer A. Wani, and Syed Rizwan Ahamad. 2023. "An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections" Life 13, no. 2: 526. https://doi.org/10.3390/life13020526
APA StyleZargar, S., Wani, T. A., & Rizwan Ahamad, S. (2023). An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections. Life, 13(2), 526. https://doi.org/10.3390/life13020526










