The Joint Link of the rs1051730 and rs1902341 Polymorphisms and Cigarette Smoking to Peripheral Artery Disease and Atherosclerotic Lesions of Different Arterial Beds
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Patients
2.2. Genetic Analysis
2.3. Statistical and Bioinformatics Analysis
3. Results
3.1. Associations of Polymorphisms rs1051730 and rs1902341 with Peripheral Artery Disease
3.2. Join Effects of the Studied Polymorphisms on the Risk of Peripheral Artery Disease
3.3. Relationship between the Polymorphisms and Plasma Lipids
3.4. The Impact of the Studied Polymorphisms on Other Atherosclerotic Lesions in PAD Patients
3.5. Functional Annotation of the Studied Polymorphisms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aday, A.W.; Matsushita, K. Epidemiology of Peripheral Artery Disease and Polyvascular Disease. Circ. Res. 2021, 128, 1818–1832. [Google Scholar] [CrossRef]
- Gerhard-Herman, M.D.; Gornik, H.L.; Barrett, C.; Barshes, N.R.; Corriere, M.A.; Drachman, D.E.; Fleisher, L.A.; Flowkes, F.G.R.; Hamburg, N.M.; Kinlay, S.; et al. 2016 AHA/ACC Guideline on the Management of Patients with Lower Extremity Peripheral Artery Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2017, 135, e726–e779. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Santulli, G. Update on peripheral artery disease: Epidemiology and evidence-based facts. Atherosclerosis 2018, 275, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.W.; Assimes, T.L.; Li, J.; Quertermous, T.; Cooke, J.P. Genetic susceptibility to peripheral arterial disease: A dark corner in vascular biology. Arter. Thromb. Vasc. Biol. 2007, 27, 2068–2078. [Google Scholar] [CrossRef] [PubMed]
- Leeper, N.J.; Kullo, I.J.; Cooke, J.P. Genetics of peripheral artery disease. Circulation 2012, 125, 3220–3228. [Google Scholar] [CrossRef]
- Zhestovskaja, A.S.; Kukes, V.G.; Sychev, D.A. Personalized medicine: Myth or reality? The position of Russian clinical pharmacologists. EPMA J. 2013, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Kontorovich, A.R.; Hao, K.; Ma, L.; Iyegbe, C.; Björkegren, J.L.; Kovacic, J.C. Precision Medicine Approaches to Vascular Disease: JACC Focus Seminar 2/5. J. Am. Coll. Cardiol. 2021, 77, 2531–2550. [Google Scholar] [CrossRef]
- Klarin, D.; Program, V.M.V.; Lynch, J.; Aragam, K.; Chaffin, M.; Assimes, T.L.; Huang, J.; Lee, K.M.; Shao, Q.; Huffman, J.E.; et al. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat. Med. 2019, 25, 1274–1279. [Google Scholar] [CrossRef]
- Van Zuydam, N.R.; Stiby, A.; Abdalla, M.; Austin, E.; Dahlström, E.H.; McLachlan, S.; Vlachopoulou, E.; Ahlqvist, E.; Di Liao, C.; Sandholm, N.; et al. Genome-Wide Association Study of Peripheral Artery Disease. Circ. Genom. Precis. Med. 2021, 14, e002862. [Google Scholar] [CrossRef]
- Ioannidis, J.P. Non-replication and inconsistency in the genome-wide association setting. Hum. Hered. 2007, 64, 203–213. [Google Scholar] [CrossRef]
- Lazarenko, V.; Churilin, M.; Azarova, I.; Klyosova, E.; Bykanova, M.; Ob'Edkova, N.; Churnosov, M.; Bushueva, O.; Mal, G.; Povetkin, S.; et al. Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines 2022, 10, 259. [Google Scholar] [CrossRef]
- Cooley, P.; Clark, R.; Folsom, R. Assessing Gene-Environment Interactions in Genome Wide Association Studies: Statistical Approaches; RTI Press: Triangle Park, NC, USA, 2014. [Google Scholar] [CrossRef]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers 2018, 2018, 5812802. [Google Scholar] [CrossRef]
- Azarova, I.E.; Klyosova, E.Y.; Sakali, S.Y.; Kovalev, A.P. Contribution of rs11927381 polymorphism of the IGF2BP2 gene to the pathogenesis of type 2 diabetes. Res. Results Biomed. 2020, 6, 9–19. [Google Scholar] [CrossRef]
- Azarova, I.; Klyosova, E.; Polonikov, A. The Link between Type 2 Diabetes Mellitus and the Polymorphisms of Glutathione-Metabolizing Genes Suggests a New Hypothesis Explaining Disease Initiation and Progression. Life 2021, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Klyosova, E.Y.; Azarova, I.E.; Sunyaykina, O.A.; Polonikov, A.V. Validity of a brief screener for environmental risk factors of age-related diseases using type 2 diabetes and coronary artery disease as examples. Res. Results Biomed. 2022, 8, 130–137. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef]
- Veiga-da-Cunha, M.; Hadi, F.; Balligand, T.; Stroobant, V.; Van Schaftingen, E. Molecular identification of hydroxylysine kinase and of ammoniophospholyases acting on 5-phosphohydroxy-L-lysine and phosphoethanolamine. J. Biol. Chem. 2012, 287, 7246–7255. [Google Scholar] [CrossRef]
- Rut, W.; Drag, M. Human 20S proteasome activity towards fluorogenic peptides of various chain lengths. Bioll Chem. 2016, 397, 921–926. [Google Scholar] [CrossRef]
- Zhou, A.X.; Tabas, I. The UPR in atherosclerosis. Seminl Immunopathol. 2013, 35, 321–332. [Google Scholar] [CrossRef]
- Ivanova, E.A.; Orekhov, A.N. The Role of Endoplasmic Reticulum Stress and Unfolded Protein Response in Atherosclerosis. Int J. Mol. Sci. 2016, 17, 193. [Google Scholar] [CrossRef]
- Mann, N.; Kause, F.; Henze, E.K.; Gharpure, A.; Shril, S.; Connaughton, D.M.; Nakayama, M.; Klämbt, V.; Majmundar, A.J.; Wu, C.-H.W.; et al. CAKUT and Autonomic Dysfunction Caused by Acetylcholine Receptor Mutations. Am. J. Hum. Genet. 2019, 105, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandarajah, A.; Chan, Y.L.; Chen, H.; Machaalani, R. Prenatal cigarette smoke exposure effects on apoptotic and nicotinic acetylcholine receptor expression in the infant mouse brainstem. Neurotoxicology 2016, 53, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Wu, Y.; Wu, Q.; Muskhelishvili, L.; Davis, K.; Tripathi, P.; Chen, Y.; Chen, T.; Bryant, M.; Rosenfeldt, H.; et al. Integration of transcriptome analysis with pathophysiological endpoints to evaluate cigarette smoke toxicity in an in vitro human airway tissue model. Arch. Toxicol. 2021, 95, 1739–1761. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Mummalaneni, S.; Grider, J.R.; Damaj, M.I.; Lyall, V. Nicotinic acetylcholine receptors (nAChRs) are expressed in Trpm5 positive taste receptor cells (TRCs). PLoS ONE 2018, 13, e0190465. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.; Böhmert, L.; Alhalabi, D.; Buhrke, T.; Lampen, A.; Braeuning, A. Identification of a transcriptomic signature of food-relevant genotoxins in human HepaRG hepatocarcinoma cells. Food Chem. Toxicol. 2020, 140, 111297. [Google Scholar] [CrossRef]
- Lutz, S.M.; Cho, M.; Kinney, G.; Young, K.; Pratte, K.; Duca, L.; Foreman, M.; Beaty, T.; Silverman, E.; Budoff, M.; et al. The Effect of Nicotinic Acetylcholine Receptor Genes CHRNA3/5 on Thoracic Aortic Calcium is Mediated by Smoking. Circulation 2016, 133 (Suppl. 1), AMP82. [Google Scholar] [CrossRef]
- Saccone, N.L.; Emery, L.S.; Sofer, T.; Gogarten, S.M.; Becker, D.M.; Bottinger, E.P.; Chen, L.-S.; Culverhouse, R.C.; Duan, W.; Hancock, D.B.; et al. Genome-Wide Association Study of Heavy Smoking and Daily/Nondaily Smoking in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Nicotine Tob. Res. 2018, 20, 448–457. [Google Scholar] [CrossRef]
- Erzurumluoglu, A.M.; Liu, M.; Jackson, V.E.; Barnes, D.R.; Datta, G.; Melbourne, C.A.; Young, R.; Batini, C.; Surendran, P.; Jiang, T.; et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol. Psychiatry 2020, 25, 2392–2409. [Google Scholar] [CrossRef]
- Busse, R.; Trogisch, G.; Bassenge, E. The role of endothelium in the control of vascular tone. Basic Res. Cardiol. 1985, 80, 475–490. [Google Scholar] [CrossRef]
- Pavlov, V.A.; Tracey, K.J. The cholinergic anti-inflammatory pathway. Brain Behav. Immun. 2005, 19, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Alves, I.; Coimbra-Campos, L.M.C.; Sancho, M.; da Silva, R.F.; Cortes, S.F.; Lemos, V.S. Role of the α7 Nicotinic Acetylcholine Receptor in the Pathophysiology of Atherosclerosis. Front. Physiol. 2020, 11, 621769. [Google Scholar] [CrossRef] [PubMed]
- Bauer, R.C.; Tohyama, J.; Cui, J.; Cheng, L.; Yang, J.; Zhang, X.; Ou, K.; Paschos, G.K.; Zheng, X.L.; Parmacek, M.S.; et al. Knockout of Adamts7, a Novel Coronary Artery Disease Locus in Humans, Reduces Atherosclerosis in Mice. Circulation 2015, 131, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Koriyama, H.; Nakagami, H.; Katsuya, T.; Sugimoto, K.; Yamashita, H.; Takami, Y.; Maeda, S.; Kubo, M.; Takahashi, A.; Nakamura, Y.; et al. Identification of Evidence Suggestive of an Association with Peripheral Arterial Disease at the OSBPL10 Locus by Genome-Wide Investigation in the Japanese Population. J. Atheroscler. Thromb. 2010, 17, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Anand, K.; Chiapparino, A.; Kumar, A.; Poletto, M.; Kaksonen, M.; Gavin, A.-C. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 2013, 501, 257–261. [Google Scholar] [CrossRef]
- Perttilä, J.; Merikanto, K.; Naukkarinen, J.; Surakka, I.; Martin, N.W.; Tanhuanpää, K.; Grimard, V.; Taskinen, M.-R.; Thiele, C.; Salomaa, V.; et al. OSBPL10, a novel candidate gene for high triglyceride trait in dyslipidemic Finnish subjects, regulates cellular lipid metabolism. J. Mol. Med. 2009, 87, 825–835. [Google Scholar] [CrossRef]
- Nissilä, E.; Ohsaki, Y.; Weber-Boyvat, M.; Perttilä, J.; Ikonen, E.; Olkkonen, V.M. ORP10, a cholesterol binding protein associated with microtubules, regulates apolipoprotein B-100 secretion. Biochim. Biophys. Acta. 2012, 1821, 1472–1484. [Google Scholar] [CrossRef]
- Olofsson, S.O.; Borén, J. Apolipoprotein B secretory regulation by degradation. Arter. Thromb Vasc Biol. 2012, 32, 1334–1338. [Google Scholar] [CrossRef]
- Tabas, I.; Williams, K.J.; Borén, J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation 2007, 116, 1832–1844. [Google Scholar] [CrossRef]
- Rada-Iglesias, A. Is H3K4me1 at enhancers correlative or causative? Nat. Genet. 2018, 50, 4–5. [Google Scholar] [CrossRef]
- Russell, D.W. Oxysterol biosynthetic enzymes. Biochim. Biophys. Acta 2000, 1529, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Zmysłowski, A.; Szterk, A. Oxysterols as a biomarker in diseases. Clin. Chim. Acta 2019, 491, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zmysłowski, A.; Szterk, A. Current knowledge on the mechanism of atherosclerosis and pro-atherosclerotic properties of oxysterols. Lipids Health Dis. 2017, 16, 188. [Google Scholar] [CrossRef] [PubMed]
- Umetani, M.; Ghosh, P.; Ishikawa, T.; Umetani, J.; Ahmed, M.; Mineo, C.; Shaul, P.W. The Cholesterol Metabolite 27-Hydroxycholesterol Promotes Atherosclerosis via Proinflammatory Processes Mediated by Estrogen Receptor Alpha. Cell Metab. 2014, 20, 172–182. [Google Scholar] [CrossRef]
- Brown, R.B. Phospholipid packing defects and oxysterols in atherosclerosis: Dietary prevention and the French paradox. Biochimie 2019, 167, 145–151. [Google Scholar] [CrossRef]
- Sato, T.; Sako, Y.; Sho, M.; Momohara, M.; Suico, M.A.; Shuto, T.; Nishitoh, H.; Okiyoneda, T.; Kokame, K.; Kaneko, M.; et al. STT3B-Dependent Posttranslational N-Glycosylation as a Surveillance System for Secretory Protein. Mol. Cell 2012, 47, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, X.; Yu, W.; Chen, J.; Li, Q.; Jiao, Y.; He, P.; Shen, C. Effects of chemokine-like factor 1 on vascular smooth muscle cell migration and proliferation in vascular inflammation. Atherosclerosis 2013, 226, 49–57. [Google Scholar] [CrossRef]
- Thorgeirsson, T.E.; Geller, F.; Sulem, P.; Rafnar, T.; Wiste, A.; Magnusson, K.P.; Manolescu, A.; Thorleifsson, G.; Stefansson, H.; Ingason, A.; et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008, 452, 638–642. [Google Scholar] [CrossRef]
| Characteristics | Healthy Controls (N = 643) | PAD Patients (N = 620) | p-Value | |
|---|---|---|---|---|
| Age, mean ± standard deviation | 60.66 ± 7.93 | 62.18 ± 9.13 | 0.001 | |
| Sex | Males, N (%) | 391 (60.81) | 532 (85.81) | <0.0001 |
| Females, N (%) | 252 (39.19) | 88 (14.19) | ||
| Body mass index (kg/m2). mean ± standard deviation | 27.07 ± 4.54 | 26.59 ± 6.36 | 0.12 | |
| Smokers a, N (%) | 295 (47.2) | 324 (52.3) | 0.07 | |
| Coronary artery disease, N (%) | - | 193 (30.63) | - | |
| Hypertension, N (%) | - | 369 (58.57) | - | |
| TC (mmol/L), Me (Q1; Q3) | NA | 5.07 (4.40; 5.60) | - | |
| LDL-C (mmol/L), Me (Q1; Q3) | NA | 3.20 (2.10; 4.50) | - | |
| HDL-C (mmol/L), Me (Q1; Q3) | NA | 1.18 (1.00; 1.57) | - | |
| TG (mmol/L), Me (Q1; Q3) | NA | 1.50 (1.11; 2.20) | - | |
| SNP ID(Gene) | Geno-Type, Allele | Genotype/Allele Frequencies. N (%) | p(q) a | OR (95% CI) b | |
|---|---|---|---|---|---|
| Healthy Controls (N = 643) | PAD Patients (N = 620) | ||||
| Entire groups | |||||
| rs1902341 (OSBPL10) | C/C | 196 (30.5) | 204 (32.9) | 0.70 (0.70) | 1.00 |
| C/T | 342 (53.2) | 302 (48.7) | 0.91 (0.63–1.32) | ||
| T/T | 105 (16.3) | 114 (18.4) | 1.10 (0.68–1.77) | ||
| T c | 0.429 | 0.427 | 0.93 (0.93) | 0.99 (0.85–1.16) | |
| rs1051730 (CHRNA3) | G/G | 309 (48.1) | 228 (36.8) | 5.1 × 10−6 (3.1 × 10−5) | 1.00 |
| G/A | 285 (44.3) | 302 (48.7) | 1.94 (1.36–2.75) | ||
| A/A | 49 (7.6) | 90 (14.5) | 2.75 (1.59–4.77) | ||
| A c | 0.325 | 0.389 | 0.0005 (0.0015) | 1.32 (1.13–1.55) | |
| Smokers | |||||
| rs1902341 (OSBPL10) | C/C | 96 (32.5) | 89 (27.5) | 0.05 (0.06) | 1.00 |
| C/T | 157 (53.2) | 167 (51.5) | 1.38 (0.83–2.28) | ||
| T/T | 42 (14.2) | 68 (21.0) | 2.28 (1.16–4.48) | ||
| T c | 0.408 | 0.468 | 0.03 (0.043) | 1.27 (1.02–1.59) | |
| rs1051730 (CHRNA3) | G/G | 141 (47.8) | 114 (35.2) | 0.008 (0.024) | 1.00 |
| G/A | 134 (45.4) | 165 (50.9) | 1.67 (1.04–2.66) | ||
| A/A | 20 (6.8) | 45 (13.9) | 2.97 (1.38–6.40) | ||
| A c | 0.295 | 0.394 | 0.0003 (0.0015) | 1.55 (1.22–1.97) | |
| Non-smokers | |||||
| rs1902341 (OSBPL10) | C/C | 96 (29.1) | 115 (38.9) | 0.05 (0.06) | 1.00 |
| C/T | 175 (53.0) | 135 (45.6) | 0.52 (0.28–0.95) | ||
| T/T | 59 (17.9) | 46 (15.5) | 0.45 (0.18–1.10) | ||
| T c | 0.444 | 0.383 | 0.03 (0.043) | 0.78 (0.62–0.98) | |
| rs1051730 (CHRNA3) | G/G | 155 (47.0) | 114 (38.5) | 0.013 (0.026) | 1.00 |
| G/A | 149 (45.1) | 137 (46.3) | 2.48 (1.30–4.72) | ||
| A/A | 26 (7.9) | 45 (15.2) | 2.49 (0.87–7.07) | ||
| A c | 0.305 | 0.383 | 0.003 (0.006) | 1.42 (1.12–1.80) | |
| № | Genotype Combination (Diplotype) | PAD Patients | Healthy Controls | OR (95% CI) a | p b | q c | ||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||
| G1 | rs1051730G/G × rs1902341C/C | 74 | 11.9 | 104 | 16.2 | 0.70 (0.51–0.97) | 0.03 | 0.07 |
| G2 | rs1051730G/G × rs1902341C/T | 112 | 18.1 | 158 | 24.6 | 0.68 (0.52–0.89) | 0.005 | 0.02 |
| G3 | rs1051730G/G × rs1902341T/T | 42 | 6.8 | 47 | 7.3 | 0.92 (0.60–1.42) | 0.71 | 0.77 |
| G4 | rs1051730G/A × rs1902341C/C | 98 | 15.8 | 78 | 12.1 | 1.36 (0.99–1.87) | 0.06 | 0.11 |
| G5 | rs1051730G/A × rs1902341C/T | 149 | 24.0 | 159 | 24.7 | 0.96 (0.74–1.25) | 0.77 | 0.77 |
| G6 | rs1051730G/A × rs1902341T/T | 55 | 8.9 | 48 | 7.5 | 1.21 (0.81–1.81) | 0.36 | 0.46 |
| G7 | rs1051730A/A × rs1902341C/C | 32 | 5.2 | 14 | 2.2 | 2.45 (1.29–4.63) | 0.005 | 0.02 |
| G8 | rs1051730A/A × rs1902341C/T | 41 | 6.6 | 25 | 3.9 | 1.75 (1.05–2.92) | 0.03 | 0.07 |
| G9 | rs1051730A/A × rs1902341T/T | 17 | 2.7 | 10 | 1.6 | 1.78 (0.81–3.93) | 0.14 | 0.21 |
| № | Genotype Combination (Diplotypes) | PAD Patients | Healthy Controls | OR (95% CI) a | P b | Q c | ||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||
| Smokers | ||||||||
| G1 | rs1051730GG × rs1902341CC | 28 | 8.6 | 47 | 15.9 | 0.50 (0.30–0.82) | 0.006 | 0.05 |
| G2 | rs1051730GG × rs1902341CT | 60 | 18.5 | 73 | 24.7 | 0.69 (0.47–1.02) | 0.06 | 0.14 |
| G3 | rs1051730GG × rs1902341TT | 26 | 8.0 | 21 | 7.1 | 1.14 (0.63–2.07) | 0.67 | 0.86 |
| G4 | rs1051730GA × rs1902341CC | 47 | 14.5 | 42 | 14.2 | 1.02 (0.65–1.60) | 0.92 | 0.92 |
| G5 | rs1051730GA × rs1902341CT | 82 | 25.3 | 73 | 24.7 | 1.03 (0.72–1.48) | 0.87 | 0.92 |
| G6 | rs1051730GA × rs1902341TT | 36 | 11.1 | 19 | 6.4 | 1.82 (1.02–3.24) | 0.04 | 0.12 |
| G7 | rs1051730AA × rs1902341CC | 14 | 4.3 | 7 | 2.4 | 1.80 (0.73–4.40) | 0.27 | 0.49 |
| G8 | rs1051730AA × rs1902341CT | 25 | 7.7 | 11 | 3.7 | 2.16 (1.04–4.47) | 0.03 | 0.12 |
| G9 | rs1051730AA × rs1902341TT | 6 | 1.9 | 2 | 0.7 | 2.40 (0.55–10.39) | 0.35 | 0.53 |
| Non-smokers | ||||||||
| G1 | rs1051730GG × rs1902341CC | 46 | 15.5 | 54 | 16.4 | 0.94 (0.61–1.44) | 0.78 | 0.78 |
| G2 | rs1051730GG × rs1902341CT | 52 | 17.6 | 78 | 23.6 | 0.69 (0.46–1.02) | 0.06 | 0.18 |
| G3 | rs1051730GG × rs1902341TT | 16 | 5.4 | 23 | 7.0 | 0.76 (0.39–1.47) | 0.42 | 0.54 |
| G4 | rs1051730GA × rs1902341CC | 51 | 17.2 | 36 | 10.9 | 1.70 (1.07–2.69) | 0.02 | 0.09 |
| G5 | rs1051730GA × rs1902341CT | 67 | 22.6 | 85 | 25.8 | 0.84 (0.58–1.22) | 0.36 | 0.54 |
| G6 | rs1051730GA × rs1902341TT | 19 | 6.4 | 28 | 8.5 | 0.74 (0.40–1.35) | 0.33 | 0.54 |
| G7 | rs1051730AA × rs1902341CC | 18 | 6.1 | 6 | 1.8 | 3.32 (1.34–8.93) | 0.01 | 0.09 |
| G8 | rs1051730AA × rs1902341CT | 16 | 5.4 | 12 | 3.6 | 1.51 (0.70–3.26) | 0.29 | 0.54 |
| G9 | rs1051730AA × rs1902341TT | 11 | 3.7 | 8 | 2.4 | 1.53 (0.62–3.76) | 0.48 | 0.54 |
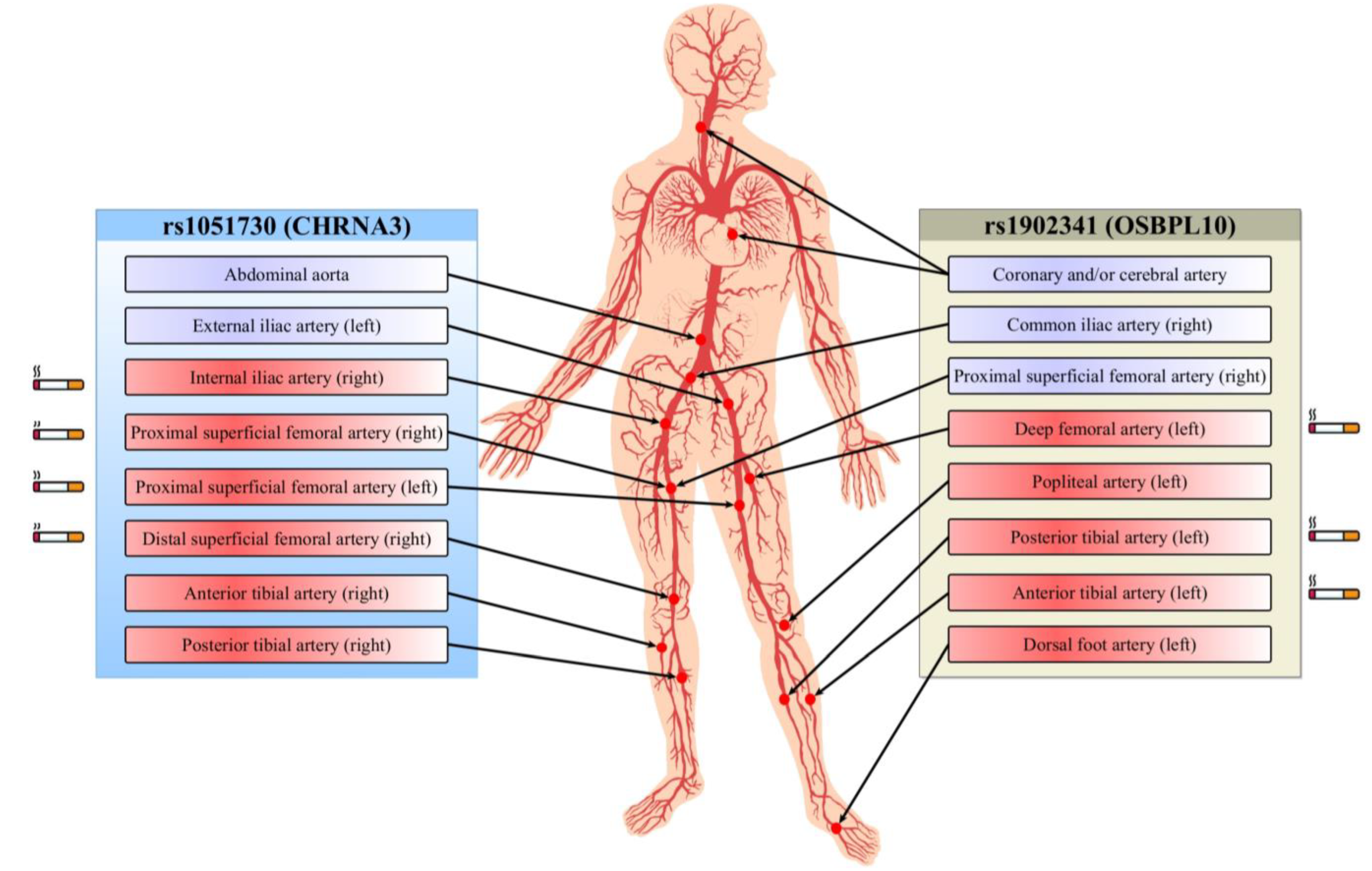
| Artery with Stenosis | rs1051730 | rs1902341 | ||
|---|---|---|---|---|
| Ultrasound | Angiography | Ultrasound | Angiography | |
| Multifocal atherosclerosis (coronary and/or cerebral artery atherosclerosis) | 1.89 (1.10–3.25) A p = 0.019 | - | - | - |
| Aorta (abdominal part) | 5.30 (1.85–15.19) A p = 7.0×10−4 | - | - | - |
| Common iliac artery (right) | - | - | 3.14 (1.05–9.35) D p = 0.02 | - |
| External iliac artery (left) | 7.19 (2.22–23.24) R p = 0.002 | - | - | - |
| Proximal superficial femoral artery (right) | - | 8.52 (1.25–57.83) R p = 0.03 | 5.41 (1.60–18.31) D p = 0.001 | - |
| Internal iliac artery (right) | - | 10.37 (1.39–77.61) D p = 0.02 | - | - |
| Deep femoral artery (left) | - | - | - | 4.73 (1.66–13.52) R p = 0.04 |
| Proximal superficial femoral artery (left) | - | 8.25 (2.16–31.46) Rp = 0.003 | - | - |
| Distal superficial femoral artery (right) | - | 9.82 (1.42–67.81) R p = 0.01 | - | - |
| Popliteal artery (left) | - | - | - | 2.07 (1.09–3.95) A p = 0.02 |
| Posterior tibial artery (right) | - | 4.02 (1.06–15.28) OD p = 0.024 | - | - |
| Posterior tibial artery (left) | - | - | - | 5.95 (1.21–29.37) R p = 0.03 |
| Anterior tibial artery (right) | - | 5.81 (1.09–30.97) OD p = 0.018 | - | - |
| Anterior tibial artery (left) | - | - | - | 7.90 (1.84–33.98) R p = 0.02 |
| Dorsal foot artery (left) | - | - | 8.67 (1.03–72.86) A p = 0.01 | |
| SNP ID | Allele Assessed | Changes in Gene Expression | Whole Blood a | Whole Blood b | Tibial Artery b | |||
|---|---|---|---|---|---|---|---|---|
| q | Z | p | NES | p | NES | |||
| rs1902341 | C | STT3B | 0.0015 * | −0.0183 * | - | - | - | - |
| T | OSBPL10-AS1 | 0.0027 * | −3.0015 * | - | - | - | - | |
| T | CMTM8 | 0.0089 * | −0.1641 * | - | - | - | - | |
| - | SUGT1P2 | 0.0289 * | 0.2276 * | - | - | - | - | |
| rs1051730 | A | CHRNA3 | - | - | - | - | 3.1 × 10−11 | −0.35 |
| A | ADAMTS7 | - | - | - | - | 1.7 × 10−7 | 0.17 | |
| A | PSMA4 | 1.2 × 10−90 | 20.2 | 1.5 × 10−9 | 0.09 | 0.0002 | 0.08 | |
| A | IREB2 | 3.3 × 10−310 | −41.4 | 0.0001 | −0.07 | - | - | |
| A | CTSH | 1.4 × 10−26 | 10.7 | - | - | - | - | |
| A | AGPHD1 | 4.6 × 10−7 | −5.0 | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhabin, S.; Lazarenko, V.; Azarova, I.; Klyosova, E.; Bykanova, M.; Chernousova, S.; Bashkatov, D.; Gneeva, E.; Polonikova, A.; Churnosov, M.; et al. The Joint Link of the rs1051730 and rs1902341 Polymorphisms and Cigarette Smoking to Peripheral Artery Disease and Atherosclerotic Lesions of Different Arterial Beds. Life 2023, 13, 496. https://doi.org/10.3390/life13020496
Zhabin S, Lazarenko V, Azarova I, Klyosova E, Bykanova M, Chernousova S, Bashkatov D, Gneeva E, Polonikova A, Churnosov M, et al. The Joint Link of the rs1051730 and rs1902341 Polymorphisms and Cigarette Smoking to Peripheral Artery Disease and Atherosclerotic Lesions of Different Arterial Beds. Life. 2023; 13(2):496. https://doi.org/10.3390/life13020496
Chicago/Turabian StyleZhabin, Sergey, Victor Lazarenko, Iuliia Azarova, Elena Klyosova, Marina Bykanova, Svetlana Chernousova, Daniil Bashkatov, Ekaterina Gneeva, Anna Polonikova, Mikhail Churnosov, and et al. 2023. "The Joint Link of the rs1051730 and rs1902341 Polymorphisms and Cigarette Smoking to Peripheral Artery Disease and Atherosclerotic Lesions of Different Arterial Beds" Life 13, no. 2: 496. https://doi.org/10.3390/life13020496
APA StyleZhabin, S., Lazarenko, V., Azarova, I., Klyosova, E., Bykanova, M., Chernousova, S., Bashkatov, D., Gneeva, E., Polonikova, A., Churnosov, M., Solodilova, M., & Polonikov, A. (2023). The Joint Link of the rs1051730 and rs1902341 Polymorphisms and Cigarette Smoking to Peripheral Artery Disease and Atherosclerotic Lesions of Different Arterial Beds. Life, 13(2), 496. https://doi.org/10.3390/life13020496








