First Molecular Evidence of Seewis Virus in Croatia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Shrew Specimen Sampling
2.2. RNA Extraction, cDNA Synthesis and Amplification of S, M, and L Segments
2.3. Sequence Data Analysis and Phylogenetic Inference
2.4. The Genetic Identification of Shrews
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yanagihara, R.; Gu, S.H.; Arai, S.; Kang, H.J.; Song, J.-W. Hantaviruses: Rediscovery and New Beginnings. Virus Res. 2014, 187, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Baek, L.J.; Schmaljohn, C.S.; Yanagihara, R. Thottapalayam Virus, a Prototype Shrewborne Hantavirus. Emerg. Infect. Dis. 2007, 13, 980–985. [Google Scholar] [CrossRef]
- Song, J.-W.; Gu, S.; Bennett, S.N.; Arai, S.; Puorger, M.; Hilbe, M.; Yanagihara, R. Seewis Virus, a Genetically Distinct Hantavirus in the Eurasian Common Shrew (Sorex araneus). Virol. J. 2007, 4, 114. [Google Scholar] [CrossRef] [PubMed]
- Gavrilovskaya, I.N.; Apekina, N.S.; Myasnikov, Y.A.; Bernshtein, A.D.; Ryltseva, E.V.; Gorbachkova, E.A.; Chumakov, M.P. Features of Circulation of Hemorrhagic Fever with Renal Syndrome (HFRS) Virus among Small Mammals in the European U.S.S.R. Arch. Virol. 1983, 75, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Gligic, A.; Stojanovic, R.; Obradovic, M.; Hlaca, D.; Dimkovic, N.; Diglisic, G.; Lukac, V.; Ler, Z.; Bogdanovic, R.; Antonijevic, B.; et al. Hemorrhagic Fever with Renal Syndrome in Yugoslavia: Epidemiologic and Epizootiologic Features of a Nationwide Outbreak in 1989. Eur. J. Epidemiol. 1992, 8, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Arai, S.; Hope, A.G.; Song, J.-W.; Cook, J.A.; Yanagihara, R. Genetic Diversity and Phylogeography of Seewis Virus in the Eurasian Common Shrew in Finland and Hungary. Virol. J. 2009, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Yashina, L.N.; Abramov, S.A.; Gutorov, V.V.; Dupal, T.A.; Krivopalov, A.V.; Panov, V.V.; Danchinova, G.A.; Vinogradov, V.V.; Luchnikova, E.M.; Hay, J.; et al. Seewis Virus: Phylogeography of a Shrew-Borne Hantavirus in Siberia, Russia. Vector-Borne Zoonotic Dis. 2010, 10, 585–591. [Google Scholar] [CrossRef]
- Schlegel, M.; Radosa, L.; Rosenfeld, U.M.; Schmidt, S.; Triebenbacher, C.; Löhr, P.-W.; Fuchs, D.; Heroldová, M.; Jánová, E.; Stanko, M.; et al. Broad Geographical Distribution and High Genetic Diversity of Shrew-Borne Seewis Hantavirus in Central Europe. Virus Genes 2012, 45, 48–55. [Google Scholar] [CrossRef]
- Ling, J.; Sironen, T.; Voutilainen, L.; Hepojoki, S.; Niemimaa, J.; Isoviita, V.-M.; Vaheri, A.; Henttonen, H.; Vapalahti, O. Hantaviruses in Finnish Soricomorphs: Evidence for Two Distinct Hantaviruses Carried by Sorex araneus Suggesting Ancient Host-Switch. Infect. Genet. Evol. 2014, 27, 51–61. [Google Scholar] [CrossRef]
- Zelena, H.; Strakova, P.; Heroldova, M.; Mrazek, J.; Kastl, T.; Zakovska, A.; Ruzek, D.; Smetana, J.; Rudolf, I. Molecular Epidemiology of Hantaviruses in the Czech Republic. Emerg. Infect. Dis. 2019, 25, 2133–2135. [Google Scholar] [CrossRef]
- Lwande, O.W.; Mohamed, N.; Bucht, G.; Ahlm, C.; Olsson, G.; Evander, M. Seewis Hantavirus in Common Shrew (Sorex Araneus) in Sweden. Virol. J. 2020, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, F.; Arai, S.; Hejduk, J.; Hayashi, A.; Markowski, J.; Markowski, M.; Rychlik, L.; Khodzinskyi, V.; Kamiya, H.; Mizutani, T.; et al. Phylogeny of Shrew- and Mole-Borne Hantaviruses in Poland and Ukraine. Viruses 2023, 15, 881. [Google Scholar] [CrossRef] [PubMed]
- Resman, K.; Korva, M.; Fajs, L.; Zidarič, T.; Trilar, T.; Županc, T.A. Molecular Evidence and High Genetic Diversity of Shrew-Borne Seewis Virus in Slovenia. Virus Res. 2013, 177, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.H.; Hejduk, J.; Markowski, J.; Kang, H.J.; Markowski, M.; Połatyńska, M.; Sikorska, B.; Liberski, P.P.; Yanagihara, R. Co-Circulation of Soricid- and Talpid-Borne Hantaviruses in Poland. Infect. Genet. Evol. 2014, 28, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Markotić, A.; Krajinović, L.C.; Margaletić, J.; Turk, N.; Miletić-Medved, M.; Zmak, L.; Janković, M.; Kurolt, I.C.; Soprek, S.; Daković Rode, O.; et al. Zoonoses and vector-borne diseases in Croatia—A multidisciplinary approach. Vet. Ital. 2009, 45, 55–66. [Google Scholar] [PubMed]
- Markotić, A.; Nichol, S.T.; Kuzman, I.; Sanchez, A.J.; Ksiazek, T.G.; Gagro, A.; Rabatić, S.; Zgorelec, R.; Avšič-Županc, T.; Beus, I.; et al. Characteristics of Puumala and Dobrava Infections in Croatia. J. Med. Virol. 2002, 66, 542–551. [Google Scholar] [CrossRef]
- Cvetko, L.; Markotić, A.; Plyusnina, A.; Margaletić, J.; Miletić-Medved, M.; Turk, N.; Milas, Z.; Avšič-Županc, T.; Plyusnin, A. Puumala Virus in Croatia in the 2002 HFRS Outbreak. J. Med. Virol. 2005, 77, 290–294. [Google Scholar] [CrossRef]
- Scharninghausen, J.J.; Pfeffer, M.; Meyer, H.; Davis, D.S.; Honeycutt, R.L.; Faulde, M. Genetic Evidence for Tula Virus in Microtus arvalis and Microtus agrestis Populations in Croatia. Vector-Borne Zoonotic Dis. 2002, 2, 19–27. [Google Scholar] [CrossRef]
- Gannon, W.L.; Sikes, R.S.; Banack, S.; Burt, M.S. Guidelines of the American Society of Mammalogists for the use of wildmammals in research. J. Mammal. 2007, 88, 809–823. [Google Scholar] [CrossRef]
- Klempa, B.; Fichet-Calvet, E.; Lecompte, E.; Auste, B.; Aniskin, V.; Meisel, H.; Denys, C.; Koivogui, L.; Ter Meulen, J.; Krüger, D.H. Hantavirus in African Wood Mouse, Guinea. Emerg. Infect. Dis. 2006, 12, 838–840. [Google Scholar] [CrossRef]
- Radosa, L.; Schlegel, M.; Gebauer, P.; Ansorge, H.; Heroldová, M.; Jánová, E.; Stanko, M.; Mošanský, L.; Fričová, J.; Pejčoch, M.; et al. Detection of Shrew-Borne Hantavirus in Eurasian Pygmy Shrew (Sorex minutus) in Central Europe. Infect. Genet. Evol. 2013, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.50. 2019. Available online: http://www.mesquiteproject.org (accessed on 12 October 2023).
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinform 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinform 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the Cytochrome b Gene of Mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree, Version 1.4.4. (Computer Program). 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 September 2023).
- Hugot, J.-P.; Gu, S.H.; Feliu, C.; Ventur, J.; Ribas, A.; Dormion, J.; Yanagihara, R.; Nicolas, V. Genetic Diversity of Talpa europaea and Nova Hanta Virus (NVAV) in France. Bull. Acad. Vet. Fr. 2014, 167, 277–284. [Google Scholar] [CrossRef]
- Igea, J.; Aymerich, P.; Bannikova, A.A.; Gosálbez, J.; Castresana, J. Multilocus species trees and species delimitation in a temporal context: Application to the water shrews of the genus Neomys. BMC Evol. Biol. 2015, 15, 209. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P.; Dobler, G.; Markotić, A.; Kurolt, I.-C.; Speck, S.; Habuš, J.; Vucelja, M.; Krajinović, L.C.; Tadin, A.; Margaletić, J.; et al. Survey for Hantaviruses, Tick-Borne Encephalitis Virus, and Rickettsia spp. In Small Rodents in Croatia. Vector Borne Zoonotic Dis. 2014, 14, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Tadin, A.; Bjedov, L.; Margaletic, J.; Zibrat, B.; Krajinovic, L.C.; Svoboda, P.; Kurolt, I.C.; Majetic, Z.S.; Turk, N.; Rode, O.D.; et al. High infection rate of bank voles (Myodes glareolus) with Puumala virus is associated with a winter outbreak of haemorrhagic fever with renal syndrome in Croatia. Epidemiol. Infect. 2014, 142, 1945–1951. [Google Scholar] [CrossRef] [PubMed]
- Shchipanov, N.A.; Zima, J.; Churchfield, S. Introducing the Common Shrew. In Shrews, Chromosomes and Speciation; Zima, J., Searle, J.B., Polly, P.D., Eds.; Cambridge University Press: Cambridge, UK, 2019; pp. 19–67. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Bradfute, S.B.; Calisher, C.H.; Klempa, B.; Klingström, J.; Laenen, L.; Palacios, G.; Schmaljohn, C.S.; Tischler, N.D.; Maes, P. Pending Reorganization of Hantaviridae to Include Only Completely Sequenced Viruses: A Call to Action. Viruses 2023, 15, 660. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Bennett, S.N.; Sumibcay, L.; Arai, S.; Hope, A.G.; Mocz, G.; Song, J.-W.; Cook, J.A.; Yanagihara, R. Evolutionary Insights from a Genetically Divergent Hantavirus Harbored by the European Common Mole (Talpa europaea). PLoS ONE 2009, 4, e6149. [Google Scholar] [CrossRef]
- Kang, H.J.; Bennett, S.N.; Hope, A.G.; Cook, J.A.; Yanagihara, R. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 2011, 85, 7496–7503. [Google Scholar] [CrossRef]
- Guo, W.P.; Lin, X.D.; Wang, W.; Tian, J.H.; Cong, M.L.; Zhang, H.L.; Wang, M.R.; Zhou, R.H.; Wang, J.B.; Li, M.H.; et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013, 9, e1003159. [Google Scholar] [CrossRef]
- Bennett, S.N.; Gu, S.H.; Kang, H.J.; Arai, S.; Yanagihara, R. Reconstructing the evolutionary origins and phylogeography of hantaviruses. Trends Microbiol. 2014, 22, 473–482. [Google Scholar] [CrossRef]
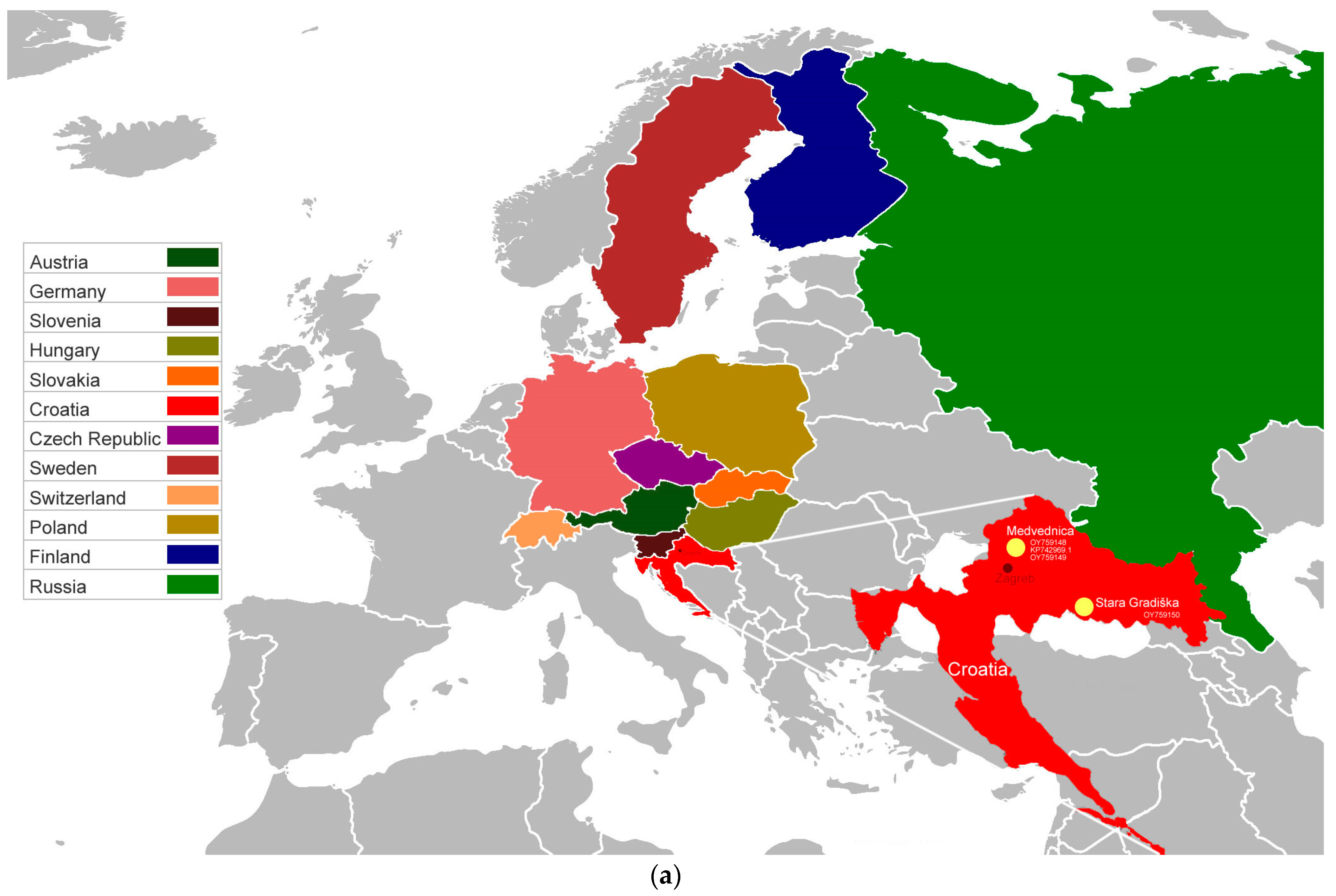
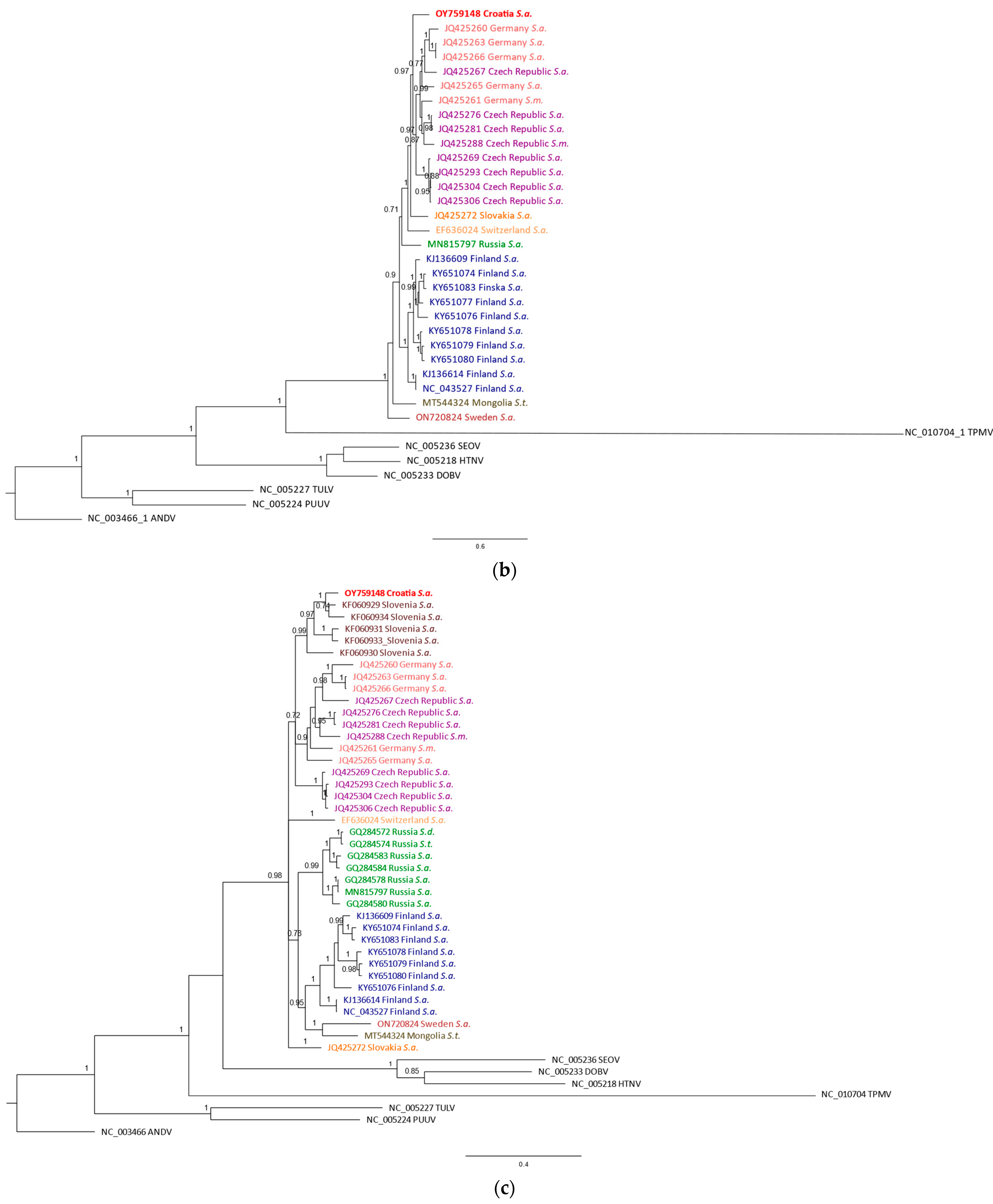
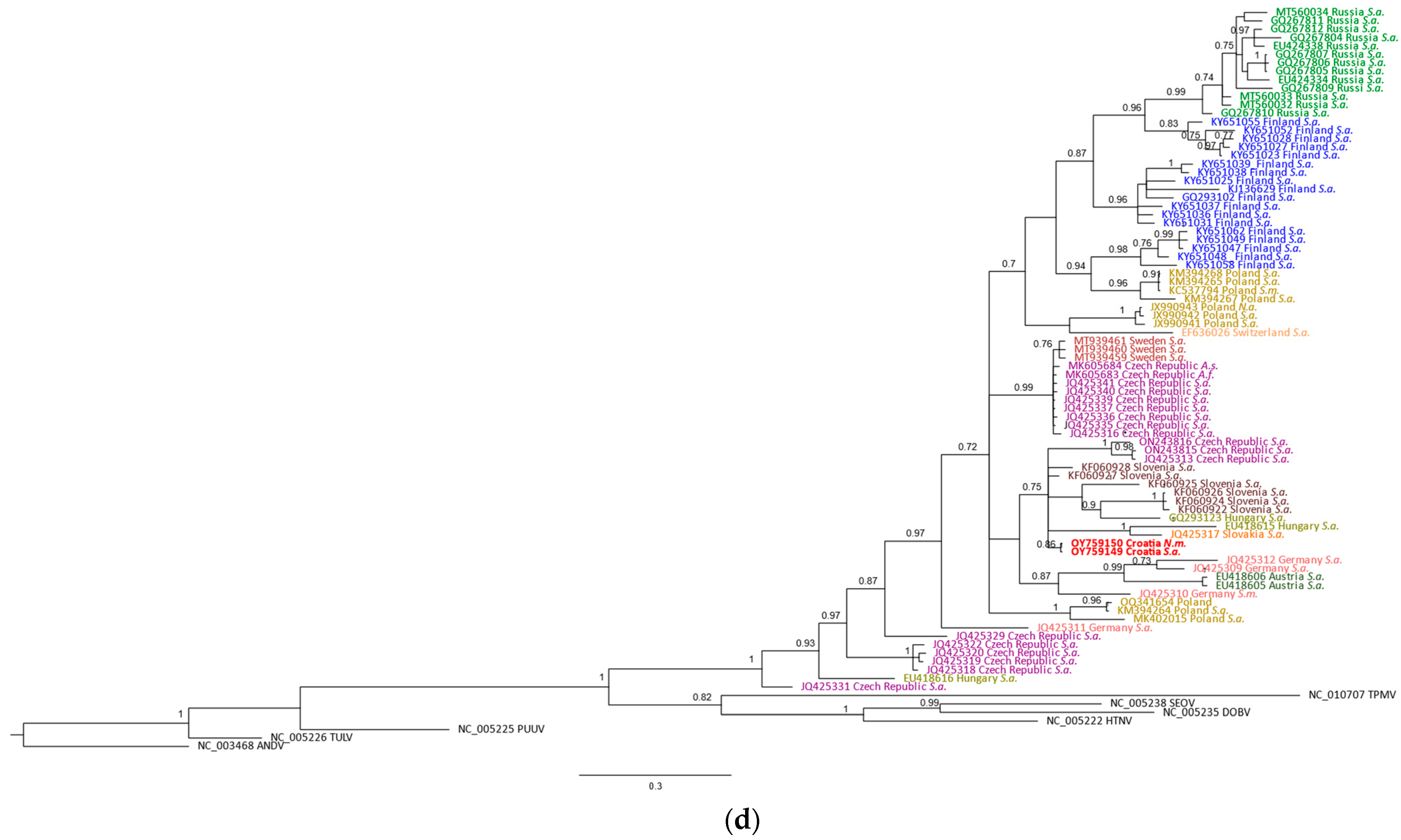
| Sample | Species 1 | Sampling Site | Coordinates | Trapping Year | SWSV RNA 2 | Organs Examined 3 |
|---|---|---|---|---|---|---|
| M-1390 | Sorex araneus | Medvednica | N45 54.453 E15 58.058 | 2013 | KP742969.1, OY759148, OY759149 | lungs (L), kidney, liver (S, L), intestines |
| M-2115 | Koprivnica | N46 11.405 E16 50.826 | 2017 | kidney, liver, spleen | ||
| M-528 | Neomys milleri 4 | Ivanić Grad | N45 38.134 E16 26.011 | 2003 | lungs, kidney | |
| M-1824 | Stara Gradiška | N45 11.866 E17 09.268 | 2016 | kidney, liver, spleen | ||
| M-1825 | Stara Gradiška | N45 11.866 E17 09.354 | 2016 | OY759150 | kidney (L), liver, spleen intestines | |
| M-2150 | Lipovljani | N45 22.044 E16 52.631 | 2017 | lungs, kidney, liver, spleen | ||
| M-1971 | Neomys fodiens | Sunja | N45 24.363 E16 43.719 | 2016 | lungs, kidney, liver | |
| M-1993 | Crocidura suaveolens | Čakovec | N46 21.087 E16 23.277 | 2016 | lungs, kidney, liver, spleen | |
| M-1650 | Crocidura leucodon | Plitvice | N44 52.186 E15 36.017 | 2014 | lungs, kidney, spleen |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svoboda Karić, P.; Anđelić Dmitrović, B.; Mrmić, S.; Paić, A.; Bjedov, L.; Štritof, Z.; Margaletić, J.; Kurolt, I.-C. First Molecular Evidence of Seewis Virus in Croatia. Life 2023, 13, 2359. https://doi.org/10.3390/life13122359
Svoboda Karić P, Anđelić Dmitrović B, Mrmić S, Paić A, Bjedov L, Štritof Z, Margaletić J, Kurolt I-C. First Molecular Evidence of Seewis Virus in Croatia. Life. 2023; 13(12):2359. https://doi.org/10.3390/life13122359
Chicago/Turabian StyleSvoboda Karić, Petra, Barbara Anđelić Dmitrović, Stella Mrmić, Antonia Paić, Linda Bjedov, Zrinka Štritof, Josip Margaletić, and Ivan-Christian Kurolt. 2023. "First Molecular Evidence of Seewis Virus in Croatia" Life 13, no. 12: 2359. https://doi.org/10.3390/life13122359
APA StyleSvoboda Karić, P., Anđelić Dmitrović, B., Mrmić, S., Paić, A., Bjedov, L., Štritof, Z., Margaletić, J., & Kurolt, I.-C. (2023). First Molecular Evidence of Seewis Virus in Croatia. Life, 13(12), 2359. https://doi.org/10.3390/life13122359








