Abstract
Listerias of the phylogenetic lineage II (PLII) are common in the European environment and are hypovirulent. Despite this, they caused more than a third of the sporadic cases of listeriosis and multi-country foodborne outbreaks. L. monocytogenes ST37 is one of them. During the COVID-19 pandemic, ST37 appeared in clinical cases and ranked second in occurrence among food isolates in the Moscow region. The aim of this study was to describe the genomic features of ST37 isolates from different sources. All clinical cases of ST37 were in the cohort of male patients (age, 48–81 years) with meningitis–septicemia manifestation and COVID-19 or Influenza in the anamnesis. The core genomes of the fish isolates were closely related. The clinical and meat isolates revealed a large diversity. Prophages (2–4/genome) were the source of the unique genes. Two clinical isolates displayed pseudolysogeny, and excided prophages were A006-like. In the absence of plasmids, the assortment of virulence factors and resistance determinants in the chromosome corresponded to the hypovirulent characteristics. However, all clinical isolates caused severe disease, with deaths in four cases. Thus, these studies allow us to speculate that a previous viral infection increases human susceptibility to listeriosis.
1. Introduction
Listeria monocytogenes is ubiquitously distributed in the environment as a saprophyte. The population of L. monocytogenes consists of four divergent lineages (I–IV) [1]. L. monocytogenes of the phylogenetic lineages (PL) I and II are the actual foodborne pathogens and cause the majority of human illness. L. monocytogenes of the PLI, which was introduced into Europe (for example, clonal complex 1 (CC1) around 1868) [2], has hypervirulent clones CC1, CC2, CC4, and CC6 with high clinical frequency [3]. At the same time, the clones of L. monocytogenes of the PLII are common for the European environment, are hypovirulent, and infect mostly highly immunocompromised individuals [3]. In the largest European collection of L. monocytogenes collected in France between January 2005 and October 2013, isolates of the PLI accounted for 66%, and isolates of the PLII accounted for 34% of the total clinical isolates [4]. The isolates of ST37 accounted for 5% of the clinical isolates in the PLII [4]. It was in seventh place after CC8-16, CC9, CC121, CC7, CC155, and CC101-90 [4]. The first registered clinical isolates of ST37 were mentioned by Ragon et al. in two cases of the maternal–neonatal (MN) infection in 1991 [5]. In the LiSEQ (Listeria SEQuencing) study of the 1143 L. monocytogenes isolates collected during the EU-wide baseline survey in 2010 and 2011, the isolates of ST37 accounted for 2% of the clinical, sporadic isolates [6].
There are some sources of L. monocytogenes ST37. First, it is the environment. So Linke et al. demonstrated that L. monocytogenes ST37, along with ST517, ST91, and ST101, could be repeatedly isolated from soil samples in two regions near the Schwarza and Danube rivers over several months [7]. L. monocytogenes fecal carriage occurs in clinically healthy ruminants (domestic and wild), swine, ground game, and birds [8,9,10]. These animals are potentially important reservoirs for L. monocytogenes and shed bacteria with feces in the environment. ST37 was one of the most frequent STs found in different sample types of hunted game and game meat in Finland between 2012 and 2020 [10]. L. monocytogenes ST37 was the third most common on Finnish dairy farms from 2013 to 2016 [11]. On 27 meat and dairy cattle farms in Latvia, CC37 was one of the predominant L. monocytogenes clones from March 2019 to August 2020 [12].
In Slovakia, L. monocytogenes ST37 was isolated from animals with clinical manifestations of listeriosis from FPE (food processing environment), meat, and dairy products, with CC37 being one of the most prevalent in the milk sector [13]. On the food production plants, L. monocytogenes ST37 was isolated in Germany (2008–2016) [14], in Austria (the meat processing facility, 2013–2018) [15], and in Spain (the new meat processing facility, 2017) [16]. In Russia, L. monocytogenes ST37 was earlier isolated from meat and dairy products [17], but clinical isolates of this genotype have never been reported in invasive listeriosis.
The crucial change in the spectrum of the L. monocytogenes ST causing invasive listeriosis was indicated during the COVID-19 pandemic [18]. Now patients who have had COVID-19 have become sensitive to L. monocytogenes ST8, ST21, ST37, ST391, and ST425. The range of products from which L. monocytogenes ST37 was isolated has been expanded [18].
Thus, L. monocytogenes ST37 required special attention. The present study is devoted to the analysis of isolates of this genotype from different sources.
2. Materials and Methods
2.1. Materials
Isolates of L. monocytogenes ST37 were collected during the multicenter monitoring of L. monocytogenes in the Moscow region between November 2018 and February 2023 and were submitted to the Bacterial Isolate Genome Sequence Database for L. monocytogenes (BIGSdb-Lm) (https://bigsdb.pasteur.fr/listeria/ (accessed on 5 September 2023)), ID: 76308; 78656; 82484; 98284; 98285; 98295; 102091 (clinical isolates); 42997; 42998; 49371; 49372; 76388; 82493; 98277; 98278; and 98280 (foodborne isolates).
2.2. Methods
2.2.1. DNA Isolation
For genotyping, listeria isolates were grown on BHI agar overnight at 37 °C. One loopful of cells was suspended in 20 µL of the lysis buffer (0.25% SDS and 0.05 M NaOH), warmed up for 15 min at 95 °C, and 180 µL of the bidistilled water was added. The resulting lysate was stored at a temperature of 4 °C. For the whole genome sequencing, listeria isolates grown on BHI agar overnight at 37 °C were suspended in the BHI broth and grown overnight at the same temperature. The cells were centrifuged, and the washed pellet was used for DNA isolation according to the Monarch Kit instructions (New England Biolabs, Ipswich, MA, USA).
2.2.2. MultiLocus Sequence Typing
MultiLocus Sequence Typing (MLST) and Multi-Virulent-Locus Sequence Typing (MvLST) were performed as described earlier [18]. The MvLST scheme included four internalin genes (inlABCE). The combination of four detected alleles (in order, inlABCE) received an internalin profile (IP) number.
2.2.3. Whole Genome Sequencing (WGS)
The genomes of the strains with ST37 were sequenced on the Illumina platform. The Nextera DNA Flex Library Prep (Illumina, San Diego, CA, USA) protocol and the NadPrep EZ DNA Library Preparation protocol (Nanodigmbio (Nanjing) Biotechnology Co. Ltd., Nanjing, China) were used for the library’s preparation. Sequencing was performed on MiSeq and NextSeq 500/550 (Illumina, San Diego, CA, USA).
2.2.4. Data Analysis
CLC Genomic Workbench v.21.0.1 (QIAGEN, Germantown, MD, USA) and SPAdes v.3.13.0 (St. Petersburg genome assembler, Russia, URL: http://cab.spbu.ru/software/spades/ (accessed on 5 September 2023)) were used for genome assembling. CGView Server (http://stothard.afns.ualberta.ca/cgview_server/ (accessed on 5 September 2023)) was applied for the visualization of assembling results and for the genome comparison [19]. The software Rapid Annotations Subsystems Technology (RAST) and SEED [20] and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [21] were used for genome annotation. Prophage sequences were revealed with the help of PHASTER (PHAge Search Tool Enhanced Release, https://phaster.ca/ (accessed on 5 September 2023)) [22].
WGS data are available in GenBank: Bio Project PRJNA605697, accession numbers: CP127186–CP127191, CP127193, CP127240, CP127241, CP127377, and CP129425–CP129430.
The core genome MLST scheme of 1748 loci (cgMLST) was used for L. monocytogenes genome characterization [23] on the basis of an open bioinformatics platform (https://bigsdb.pasteur.fr/listeria/ (accessed on 5 September 2023)). Alleles that were not characterized with the computational tool due to their differences from the registered sequences were searched by comparing genomic data with alleles of the reference EGD-e strain using BLAST (2.10.0+) [24]. The identified sequences of the corresponding alleles of the isolates of the same ST were compared by the number of differences with the closest match. New alleles were considered identical if there were the same substitutions.
The phylogenetic tree on the base of the concatenated core genes was constructed using the neighbor-joining method with the help of the Whole Genome Alignment Plugin in CLC Genomic Workbench v.21.0.1 (QIAGEN, Germantown, MD, USA) and was represented in the MEGA7 program [25].
Pan genomic studies of the ST37 isolates were made with BPGA: Bacterial Pan Genome Analysis Pipeline (BPGA-Version-1.3) and included determining core (conserved), accessory (dispensable), and unique (strain-specific) genes with the KEGG (Kyoto Encyclopedia of Genes and Genomes) and COG (Clusters of Orthologous Genes) mapping [26]. The analysis was performed with the following parameters: clustering was made with USEARCH, amino acid identity cutoff = 50%, and the type of phylogeny tree was neighbor joining.
Prophages regions were identified using PHASTER (an extended version of the PHAge search tool, https://phaster.ca/ (accessed on 5 September 2023)) [27].
CRISPR (clustered regularly interspaced short palindromic repeats) arrays and their associated (Cas) proteins were identified with the help of the CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/ (accessed on 5 September 2023)) [28].
The virulence factor database (VFDB) (http://www.mgc.ac.cn/VFs/ (accessed on 5 September 2023)), an extensive collection of VFs from the important bacterial pathogens [29], as well as the BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/ (accessed on 5 September 2023)) were used for the VFs analysis of L. monocytogenes ST37.
The resistome of the L. monocytogenes ST37 isolates was analyzed using the Antibiotic Resistance BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/ (accessed on 5 September 2023) and the data from the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/ (accessed on 5 September 2023)) [30].
We used all bioinformation resources with default parameters.
2.2.5. Ethical Approval
The study was carried out with the informed consent of the patients. The research protocol was approved by the Biomedical Ethics Committee of the N.F. Gamaleya National Research Center for Epidemiology and Microbiology (protocol No. 14, 4 July 2018).
3. Results
The COVID-19 pandemic revealed a large diversity of L. monocytogenes of the PLII in clinical cases (Figure 1) [18]. L. monocytogenes ST37 that appeared during the pandemic took first place and shared 13% among all clinical isolates (Figure 1) and 21% among isolates of the PLII.
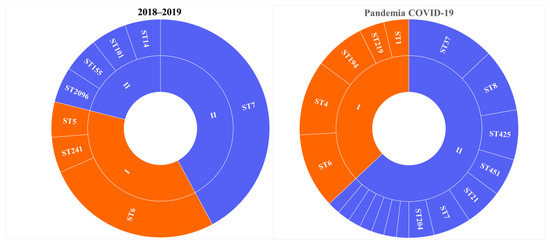
Figure 1.
Genetic diversity of L. monocytogenes isolated from patients with invasive listeriosis (A) before and (B) during the COVID-19 pandemic. I - phylogenetic lineage I, II - phylogenetic lineage II.
3.1. Prevalence of L. monocytogenes ST37
The greatest contribution to the diversity of the PLII genotypes was made by patients with meningitis and septicemia. A total of 72% of patients in this cohort were male. All cases of L. monocytogenes ST37 were noted in this group. The male patients were mostly infected with L. monocytogenes of the PLII (Figure 2), and ST37 shared 20% in this group.
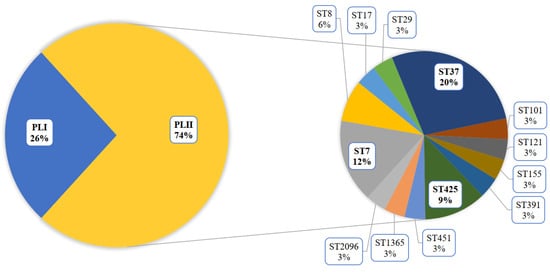
Figure 2.
Genotype diversity of L. monocytogenes isolated from male patients with invasive listeriosis.
The male patients infected by L. monocytogenes ST37 were aged 48–81 years (mean age—66 years) and had COVID-19 (the most cases) or Influenza A (one case) in their anamnesis. Only two patients survived.
During the monitoring period, 2018–2023, in addition to clinical specimens, the sources of L. monocytogenes ST37 were fish and meat products and restaurant cooking environments (Figure 3). Overall, among food isolates, ST37 ranked second in occurrence after ST121.
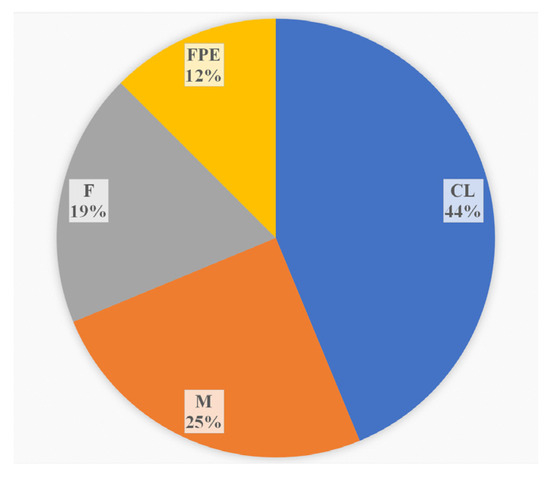
Figure 3.
The sources of L. monocytogenes ST37 in the study. CL—clinical specimens, M—meat products, F—fish products, and FPE—restaurant cooking environments.
3.2. Genomic Analysis of the L. monocytogenes ST37 Isolates
All isolates of L. monocytogenes ST37 were analyzed together via whole genome sequencing (WGS).
3.2.1. Core Genome including 1748 Genes
The cgMLST was performed on 16 genomes of L. monocytogenes ST37. The results of the comparison are shown in Figure 4 (Table S1). Only tree pairs of the genomes had null or one locus of difference. The pair of clinical isolates came from two samples of one patient: blood and brain membranes (postmortem). These isolates had the same core genomes.

Figure 4.
Phylogenetic tree at the base of the concatenated 1748 core genes. Colored circles for the source of isolates are indicated as follows: red—clinical, blue—fish, cream—meat, and green—the restaurant cooking environments. The number next to the bracket shows the loci of difference between a pair of core genomes. The numbers at the nodes indicate bootstrap values.
The pair of FPE isolates had one locus of difference. The samples for their isolation were collected in one place and over one day. At last, the pair of fish isolates were from salmon fillets from one manufacturer, delivered two weeks apart. The third fish isolate differed from the isolates of the pair by 11–12 loci. Clinical isolates, all but two mentioned above, had 16–37 loci of differences. They differed from the pair of one patient’s isolates by 74–84 loci. The same differences were observed from this pair of clinical isolates from food and FPE. So, the pair of clinical isolates had an unknown source that was significantly different from the sources of other isolates.
3.2.2. Pan Genomic Studies of the ST37 Isolates
The pan genome and core genome sizes determined with the BPGA were 3189 and 2611, respectively. An increase in the core to 2611 genes changed the topology of some branches on the tree (Figure 5). In the phylogenetic group of the fish isolates, the branch of the clinical isolates appeared. Meat isolates, all but one, and two clinical isolates formed a separate phylogenetic group.
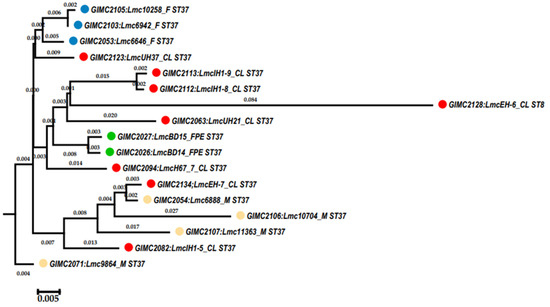
Figure 5.
Phylogenetic tree generated with BPGA using 16 isolates of ST37 and 1 isolate of ST8 based on concatenated core genes. Colored circles for the source of isolates are indicated as follows: red—clinical, blue—fish, cream—meat, and green—the restaurant cooking environments. The numbers at the nodes indicate the length of the branches.
The central phylogenetic group included the most different clinical isolates, the pair of the FPE isolates, and two other clinical isolates—one of which is LmcUH21 and has the smallest number of ORFs (open reading frames, 2840) and homologous genes (72) but also the largest number of exclusively absent genes (38) (Figure 6).
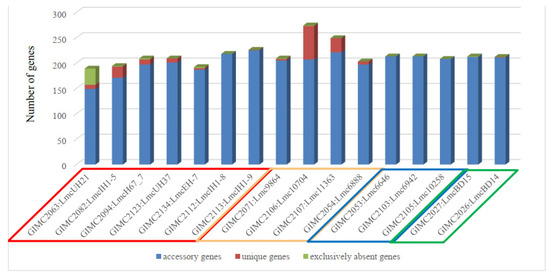
Figure 6.
The genes outside the core genome. Colored frames for the source of isolates are indicated as follows: red—clinical, cream—meat, blue—fish, and green—the restaurant cooking environments.
Meat isolates were the leaders in the number of unique genes, followed by some clinical isolates. Unique genes were absent in a group of fish and FPE isolates as well as in a pair of very different clinical isolates.
The distribution of the COG groups was different for core, accessory, and unique genes (Table 1). Since the core genes are responsible for the basic aspects of the biology of the species [26], it is not surprising that 40% of the core genes of L. monocytogenes ST37 belong to the “Metabolism” group. The genes responsible for “Cellular processes” and “Information storage” were the most abundant in the unique and accessory groups. Analysis of the location of unique genes in the genomes of L. monocytogenes ST37 showed that they are localized in the regions of prophages. Isolates Lmc10704 and Lmc11363 with the most prophages, — 4 (Table 2), had the highest number of unique genes (Figure 6), which confirms that prophages as mobile genetic elements are the sources of bacterial diversity [31].

Table 1.
COG distribution in Pan-genome L. monocytogenes ST37.

Table 2.
Prophages and phages in L. monocytogenes ST37 genomes.
3.2.3. Prophages and Phages in the ST37 Isolates
There are some “hot spots” of genome evolution for Listeria spp. that are affected by the transduction processes of bacteriophages [32]. We found seven such regions in the genomes of L. monocytogenes ST37 (Table 2). Two of them were downstream loci of the regulators of transcription (lmo0112 and comK), and five regions were located adjacent to the tRNA genes. The lmo0112 questionable prophage, which is conserved across all lineages of L. monocytogenes [33], has been revealed in all genomes of L. monocytogenes ST37 (14.4 Kb) and harbors a complete lma operon (lmaDCBA) known as the monocin locus. The tRNAArg(tct) intact prophage was in all but one isolate (42.6–46.1 Kb). The next intact prophage, tRNASer(cga), was 34.2Kb and has been found in all meats and one clinical isolate. The tRNALeu(gag) prophage inserted only in the meat isolate had the highest number of unique genes (questionable, 51.1 Kb). The tRNAArg(ccg) prophage was revealed in four isolates from different sources (intact 41.7 Kb or questionable 41.3 Kb). At last, the tRNAThr(ggt) prophage was found in all fish (intact 49.3 Kb) and in three clinical isolates (questionable, 53.1–57.9 Kb). The comK locus was disrupted via phage integration in two genomes (clinical and meat). Inserted prophages were intact (40.3 Kb) or questionable (49.9 Kb).
The tRNAThr(ggt) prophages of the one patient’s isolates (GIMC2112:LmcIH1-8 and GIMC2113:LmcIH1-9) had 59 ORF and were A006-like. WGS of these isolates demonstrated that genomes consisted of two replicons: chromosomes and bacteriophages of the class Caudoviricetes. The bacteriophages LP-LmcIH1-8 (OR234009) and LP-LmcIH1-9 (OR234010) had 66 and 65 ORF and were A006-like too. The tRNAThr(ggt) prophages and bacteriophages were organized from common functional modules [34]: (1) “late genes” coding for structural proteins, DNA packaging, and the lysis systems (holin and endolysin); (2) genes for lysogeny functions (integrase, repressor, and similar) and the prophage attachment and integration locus attP; and (3) “early genes” for the early state of phage reproduction encoding products for replication, recombination, and modification of the viral DNA. The tRNAThr(ggt) prophage and bacteriophage of one isolate had 19 identical ORF: 8 in the left arm of the bacteriophage genome, 6 in the right arm, and 5 ORF for phage proteins in the third module. The amino acid sequences of integrase, recombinase, holin, transcriptional regulators, and some phage proteins had the least similarity between prophage and phage.
The WGS data indicated non-synchronous replication of the phage and the bacterial chromosome since the coverage of the phage replicon was 4–5 times greater than the coverage of the chromosome. Thus, we can assume there is pseudolysogeny, i.e., a type of phage–bacteria interaction in which the phage nucleic acid neither integrates into the host chromosome as a prophage (lysogeny) nor elicits a lytic response but resides within the cell in a non-active form [35,36]. Pseudolysogeny has been described for several tailed dsDNA phages [37], which include phages of the class Caudoviricetes.
Note that the phage LP-LmcIH1-9 of L. monocytogenes isolated from the brain membranes was somewhat different from the phage LP-LmcIH1-8 of L. monocytogenes isolated from the blood. The deletion was revealed in the gene for the major capsid protein and in the third module, which led to the loss of the ORF of one of the phage proteins.
3.2.4. The CRISPR System
All L. monocytogenes ST37 isolates had one CRISPR locus with level of the evidence 4. The locus contained repeating sequences of length of 29 bp with nine spacers. No cas genes were found to be associated with this locus, which was located downstream of the ORF of ribose-phosphate pyrophosphokinase in the intergenic region of 1244 bp.
3.2.5. Virulence Factors
A total of 54 virulence factor genes were found in the genomes of L. monocytogenes ST37; most of them had the same alleles. In 10 genes, individual isolates had SNV (single nucleotide variant) of the common allele sequences (Table S2). Two isolates (clinical and meat) had no differences in these loci. Two isolates had disrupted the gene comK mentioned above. The remaining meat and fish isolates and one clinical isolate had one different locus. Three clinical samples and two isolates from the restaurant cooking environment had two loci with SNV. An increase in the sample size of the ST37 isolates confirmed the previously identified absence of the vip gene in Listeria of this genotype [18].
3.2.6. Antibiotic Resistance
All ST37 isolates had antibiotic resistance genes only on chromosomes. These genes, fosX (lmo1702), lmo0919 (lin), lmo1695 (mprF), and norB (lmo2818), encode resistance to fosfomycin, lincosamides, against cationic antimicrobial peptides, and quinolones. All but one isolate had the same allele of fosX (4); the fish isolate GIMC2053:Lmc6646 had an allele with an SNV of allele 4. The meat isolate GIMC2054:Lmc6888 had another allele of the norB (31), which was SNV of the common allele 8. All ST37 isolates acquired additional resistance to quinolones via mutations Ser83 and Glu87 in ParC (the subunit of topoisomerase IV).
Antibiograms provided by clinical laboratories showed resistance of clinical isolates of ST37 to ampicillin and meropenem. So, we analyzed the genes of the penicillin-binding protein (PBP) and beta-lactamases in the genomes of the isolates. All of them had two genes for PBP (1A and 3, COG0768 and COG0744), two genes for beta-lactamase (class C, COG1680), the gene for metallo-beta-lactamase (COG2333), and the gene for metal-dependent hydrolase with the domain of the beta-lactamase (COG1234). On the one hand, PBP3 is the primary target for beta-lactams, and the blocking of these enzymes is lethal for the bacterial cell [38]. However, the affinity of beta-lactams to the other PBPs is variable and does not immediately have a deleterious effect [38]. On the other hand, beta-lactamases could hydrolase beta-lactams, which explains the phenotypic resistance.
4. Discussion
In a COVID-19 pandemic, from March 2020 to February 2023, the number of food isolates of L. monocytogenes ST37 markedly increased, and cases of invasive listeriosis with manifestations of meningitis and septicemia caused by L. monocytogenes ST37 have appeared in the Moscow region. Only male patients 48–81 years old were infected with L. monocytogenes ST37. Four of them died, and two survived. In Europe, apart from the sporadic clinical cases mentioned above [4,5,6], one of three large outbreaks in Denmark in 2022 was caused by ST37 (FUD2080) [39]. An additional outbreak of L. monocytogenes ST37 was reported, with 8–14 cases accumulating over the last 3–5 years [39]. The sources of these outbreaks remain unknown. Note that other outbreaks in Denmark in 2022 were also caused by L. monocytogenes of the PLII, and the number of listeriosis cases per year noticeably increased [39]. The European Centre for Disease Prevention and Control (ECDC) also reported that the number of foodborne outbreaks in 2021 was the highest since the European Food Safety Authority (EFSA) first started collecting data [40]. Two of the four multi-country listeriosis outbreak clusters were caused by L. monocytogenes of the PLII (ST7 and ST451) [40].
Analyzing the distribution of invasive listeriosis with manifestations of meningitis and septicemia by gender, we saw that before the COVID-19 pandemic, the male-to-female ratio was 1.2:1 in the Moscow region, but during pandemic, this ratio reached 4:1. In the ECDC 2021 report, male-to-female ratio for confirmed listeriosis cases with known sex in all age categories was 1.2:1. However, in the categories 45–64 years and 65+, this ratio was 1.6:1 and 1.8:1, respectively [39]. Thus, men over the age of 45 are more susceptible to listeriosis than women.
In the Moscow region, L. monocytogenes ST37 was isolated from meat and fish products. In European countries, L. monocytogenes ST37 was reported in meat and dairy products [11,12,13,14,15,16]. In Austria in 2017, ST37 was the fifth of the ten most frequent STs among the category of food, which included meat, dairy products, and vegetables [41]. The fish products were mentioned as the source of L. monocytogenes ST37 in Poland (RTE smoked mackerel paste, 2017). Interesting that core genome of this isolate was identical to the genome of the clinical isolate of 2013, so a possible source of infection was proposed four years later [42].
Pan-genomic studies of the ST37 isolates showed that a different repertoire of accessory, unique, and exclusively absent genes did not affect Listeria’s ability to cause disease. The number of unique genes correlated with the number of prophages. Dorscht et al. have suggested that prophages can improve the competitive fitness of the lysogenized host strains [34], but experimentally, the role of prophages was shown only for those that are integrated into the regions of transcription regulators: lmo0112 and comK. Hain et al. demonstrated an upregulation of the expression of phage-derived genes when Listeria proliferates intracellularly [43], and deletions in the monocin locus of prophages led to severe virulence attenuation [43]. Rabinovich et al. studied the role of the comK-integrated prophage in the regulation of phagolysosome escape by Listeria during infection. Prophage excision during intracellular growth within phagosomes led to the formation of a functional comK gene, and the production of progeny virions was prevented [44].
We described the pseudolysogeny for the strains GIMC2112:LmcIH1-8 and GIMC2113:LmcIH1-9. The locus of integration of the excised prophage is unknown; however, the data of Rabinovich et al. give reason to believe that the excision of the prophage occurred in the stationary phase of growth of Listeria cells [44].
The role of the prophages in bacterial adaptation is important when Listeria proliferates intracellularly, but this stage is preceded by invasion into enterocytes. The interaction between Listeria and enterocytes is prevented by the mucus if it renews and is enriched with health-related bacteria. When the mucus as the first line of defense is damaged, for example, after a viral infection, invasion of enterocytes becomes possible. Therefore, even hypovirulent Listeria of the PLII becomes dangerous for people who have had COVID-19 or Influenza.
5. Conclusions
The increase in the population’s susceptibility to hypovirulent L. monocytogenes raises the importance of molecular-epidemiological studies of Listeria of the PLII for the timely identification of the acquired virulence and resistance factors in actively evolving bacteria.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/life13112167/s1, Table S1: The differences in the core genomes of ST37 isolates; Table S2: Virulence factors that differ among ST37 isolates; and Table S3: Alleles of resistance genes.
Author Contributions
Conceptualization, O.L.V., E.A.K., O.A.G., A.R.M. and I.S.T.; methodology, O.L.V., N.N.R., M.S.K. and E.I.A.; software, N.N.R., M.S.K. and E.I.A.; validation, O.L.V., N.N.R., M.S.K. and E.I.A.; formal analysis, N.N.R., E.I.A. and M.A.K.; investigation, O.L.V., N.N.R., M.S.K., E.I.A., M.A.K. and T.I.K.; resources, O.L.V., E.A.K., O.A.G., A.R.M. and I.S.T.; data curation, M.S.K., N.N.R. and O.L.V.; writing—original draft preparation, O.L.V., N.N.R., M.S.K. and E.I.A.; writing—review and editing, O.L.V., N.N.R., M.S.K. and E.I.A.; visualization, M.S.K., N.N.R. and O.L.V.; supervision, O.L.V.; project administration, I.S.T.; funding acquisition, O.L.V., E.A.K., O.A.G. and A.R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The research protocol was approved by the Biomedical Ethics Committee of the N.F. Gamaleya Nation-al Research Center for Epidemiology and Microbiology (protocol No. 14, 4 July 2018).
Informed Consent Statement
The study was carried out with the informed consent of the patients.
Data Availability Statement
The data supporting the reported results can be found in BIGSdb-Lm (https://bigsdb.pasteur.fr/listeria/), ID: 76308; 78656; 82484; 98284; 98285; 98295; 102091 (clinical isolates); 42997; 42998; 49371; 49372; 76388; 82493; 98277; 98278; and 98280 (foodborne isolates), and in GenBank NCBI (https://www.ncbi.nlm.nih.gov/genome/, accessed on 14–19 July 2023), Bio Project PRJNA605697, accession numbers: CP127186–CP127191, CP127193, CP127240, CP127241, CP127377, and CP129425–CP129430.
Acknowledgments
The authors are grateful to the laboratory assistants of the microbiological laboratories of the Moscow laboratory diagnostic centers for providing L. monocytogenes isolates for the study: Burmistrova E.N., Dudnikova S.V., Nechaeva S.A., Pokidysheva A.Y. and Pronina T.V.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Moura, A.; Lefrancq, N.; Wirth, T.; Leclercq, A.; Borges, V.; Gilpin, B.; Dallman, T.J.; Frey, J.; Franz, E.; Nielsen, E.M.; et al. Emergence and global spread of Listeria monocytogenes main clinical clonal complex. Sci. Adv. 2021, 7, eabj9805. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Painset, A.; Björkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Félix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. LiSEQ-whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019, 5, e000257. [Google Scholar] [CrossRef]
- Linke, K.; Rückerl, I.; Brugger, K.; Karpiskova, R.; Walland, J.; Muri-Klinger, S.; Tichy, A.; Wagner, M.; Stessl, B. Reservoirs of listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 2014, 80, 5583–5592. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.J.; Ivanek, R.; Gröhn, Y.T.; Nightingale, K.K.; Wiedmann, M. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L Monocytogenes Subtypes. Prev. Vet. Med. 2007, 80, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.I.; Oporto, B.; Aduriz, G.; Juste, R.A.; Hurtado, A. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet. Res. 2009, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Sauvala, M.; Kurittu, P.; Heljanko, V.; Heikinheimo, A.; Paulsen, P. Characterisation of Listeria monocytogenes Isolates from Hunted Game and Game Meat from Finland. Foods 2022, 11, 3679. [Google Scholar] [CrossRef]
- Castro, H.; Douillard, F.P.; Korkeala, H.; Lindström, M. Mobile Elements Harboring Heavy Metal and Bacitracin Resistance Genes Are Common among Listeria monocytogenes Strains Persisting on Dairy Farms. mSphere 2021, 6, e0038321. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Šteingolde, Ž.; Meistere, I.; Elferts, D.; Avsejenko, J.; Streikiša, M.; Gradovska, S.; Alksne, L.; Ķibilds, J.; Bērziņš, A. Prevalence, Genetic Diversity and Factors Associated with Distribution of Listeria monocytogenes and Other Listeria spp. in Cattle Farms in Latvia. Pathogens 2021, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Kubicová, Z.; Roussel, S.; Félix, B.; Cabanová, L. Genomic diversity of Listeria monocytogenes isolates from Slovakia (2010 to 2020). Front. Microbiol. 2021, 12, 729050. [Google Scholar] [CrossRef]
- Roedel, A.; Dieckmann, R.; Brendebach, H.; Hammerl, J.A.; Kleta, S.; Noll, M.; Al Dahouk, S.; Vincze, S. Biocide-Tolerant Listeria monocytogenes Isolates from German Food Production Plants Do Not Show Cross-Resistance to Clinically Relevant Antibiotics. Appl. Environ. Microbiol. 2019, 85, e01253-19. [Google Scholar] [CrossRef] [PubMed]
- Stessl, B.; Szakmary-Brändle, K.; Vorberg, U.; Schoder, D.; Wagner, M. Temporal analysis of the Listeria monocytogenes population structure in floor drains during reconstruction and expansion of a meat processing plant. Int. J. Food Microbiol. 2020, 314, 108360. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the emergence and population diversity of Listeria monocytogenes in a newly built meat facility through whole genome sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef]
- Psareva, E.K.; Liskova, E.A.; Razheva, I.V.; Yushina, Y.K.; Grudistova, M.A.; Gladkova, N.A.; Potemkin, E.A.; Zhurilov, P.A.; Sokolova, E.V.; Andriyanov, P.A.; et al. Diversity of Listeria monocytogenes strains isolated from food products in the Central European part of Russia in 2000–2005 and 2019–2020. Foods 2021, 10, 2790. [Google Scholar] [CrossRef]
- Voronina, O.L.; Kunda, M.S.; Ryzhova, N.N.; Aksenova, E.I.; Kutuzova, A.V.; Tikulmina, A.N.; Karpova, T.I.; Melkumyan, A.R.; Klimova, E.A.; Gruzdeva, O.A.; et al. Genomic characteristics of listeria that caused invasive listeriosis during the COVID-19 pandemic. Lond. J. Res. Sci. Nat. Form. (LJRS) 2023, 23, 33–61. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33 (Suppl. S1), D325–D328. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Hain, T.; Steinweg, C.; Kuenne, C.T.; Billion, A.; Ghai, R.; Chatterjee, S.S.; Domann, E.; Kärst, U.; Goesmann, A.; Bekel, T.; et al. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 2006, 188, 7405–7415. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Allnutt, T.; Bradbury, M.I.; Fanning, S.; Chandry, P.S. Comparative Genomics of the Listeria monocytogenes ST204 Subgroup. Front. Microbiol. 2016, 7, 2057. [Google Scholar] [CrossRef] [PubMed]
- Dorscht, J.; Klumpp, J.; Bielmann, R.; Schmelcher, M.; Born, Y.; Zimmer, M.; Calendar, R.; Loessner, M.J. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 2009, 191, 7206–7215. [Google Scholar] [CrossRef]
- Ripp, S.; Miller, R.V. The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology 1997, 143, 2065–2070. [Google Scholar] [CrossRef]
- Ripp, S.; Miller, R.V. Dynamics of the pseudolysogenic response in slowly growing cells of Pseudomonas aeruginosa. Microbiology (Reading) 1998, 144 Pt 8, 2225–2232. [Google Scholar] [CrossRef]
- Mäntynen, S.; Laanto, E.; Oksanen, H.M.; Poranen, M.M.; Díaz-Muñoz, S.L. Black box of phage-bacterium interactions: Exploring alternative phage infection strategies. Open Biol. 2021, 11, 210188. [Google Scholar] [CrossRef]
- Hof, H.; Nichterlein, T.; Kretschmar, M. Management of listeriosis. Clin. Microbiol. Rev. 1997, 10, 345–357. [Google Scholar] [CrossRef]
- Espenhain, L.; Takeuchi-Storm, N.; Munch, P.K.; Hansen, L.T.; Nielsen, N.L.; Andersen, J.K.; Schjørring, S. Listeria monocytogenes. In Annual Report on Zoonoses in Denmark 2022; Lassen, B., Olsen, A., Sandberg, M., Müller, L., Torpdahl, M., Petersen, C.K., Eds.; Annual Report on Zoonoses in Denmark; National Food Institute, Technical University of Denmark: Kongens Lyngby, Denmark, 2023; pp. 10–14. [Google Scholar]
- European Centre for Disease Prevention and Control. Listeriosis. In ECDC. Annual Epidemiological Report for 2021; ECDC: Stockholm, Sweden, 2022. [Google Scholar]
- Cabal, A.; Pietzka, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; Schmid, D. Isolate-Based Surveillance of Listeria monocytogenes by Whole Genome Sequencing in Austria. Front. Microbiol. 2019, 10, 2282. [Google Scholar] [CrossRef]
- Maćkiw, E.; Korsak, D.; Kowalska, J.; Felix, B.; Stasiak, M.; Kucharek, K.; Antoszewska, A.; Postupolski, J. Genetic diversity of Listeria monocytogenes isolated from ready-to-eat food products in retail in Poland. Int. J. Food Microbiol. 2021, 358, 109397. [Google Scholar] [CrossRef] [PubMed]
- Hain, T.; Ghai, R.; Billion, A.; Kuenne, C.T.; Steinweg, C.; Izar, B.; Mohamed, W.; Mraheil, M.A.; Domann, E.; Schaffrath, S.; et al. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genom. 2012, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).