The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Culture Conditions
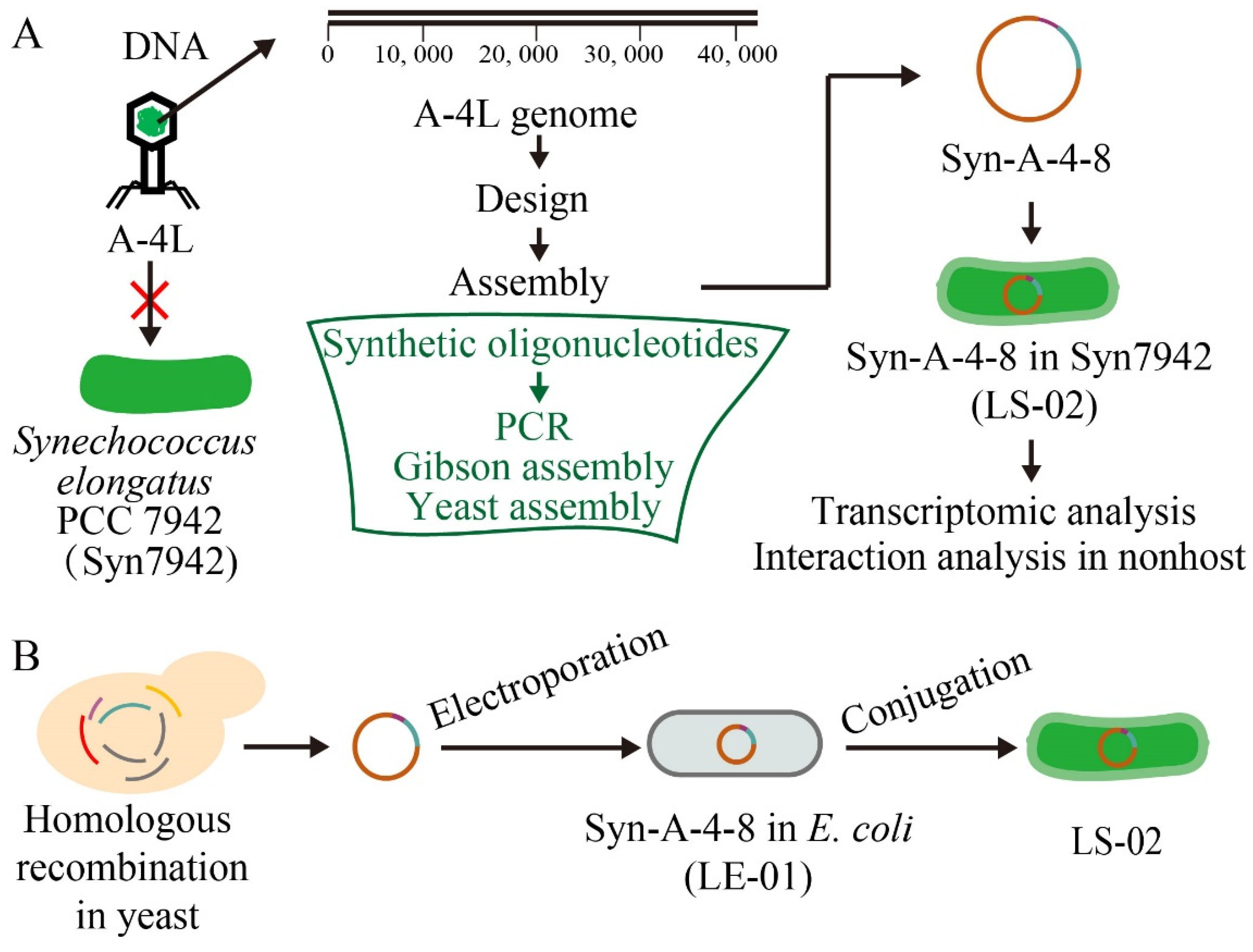
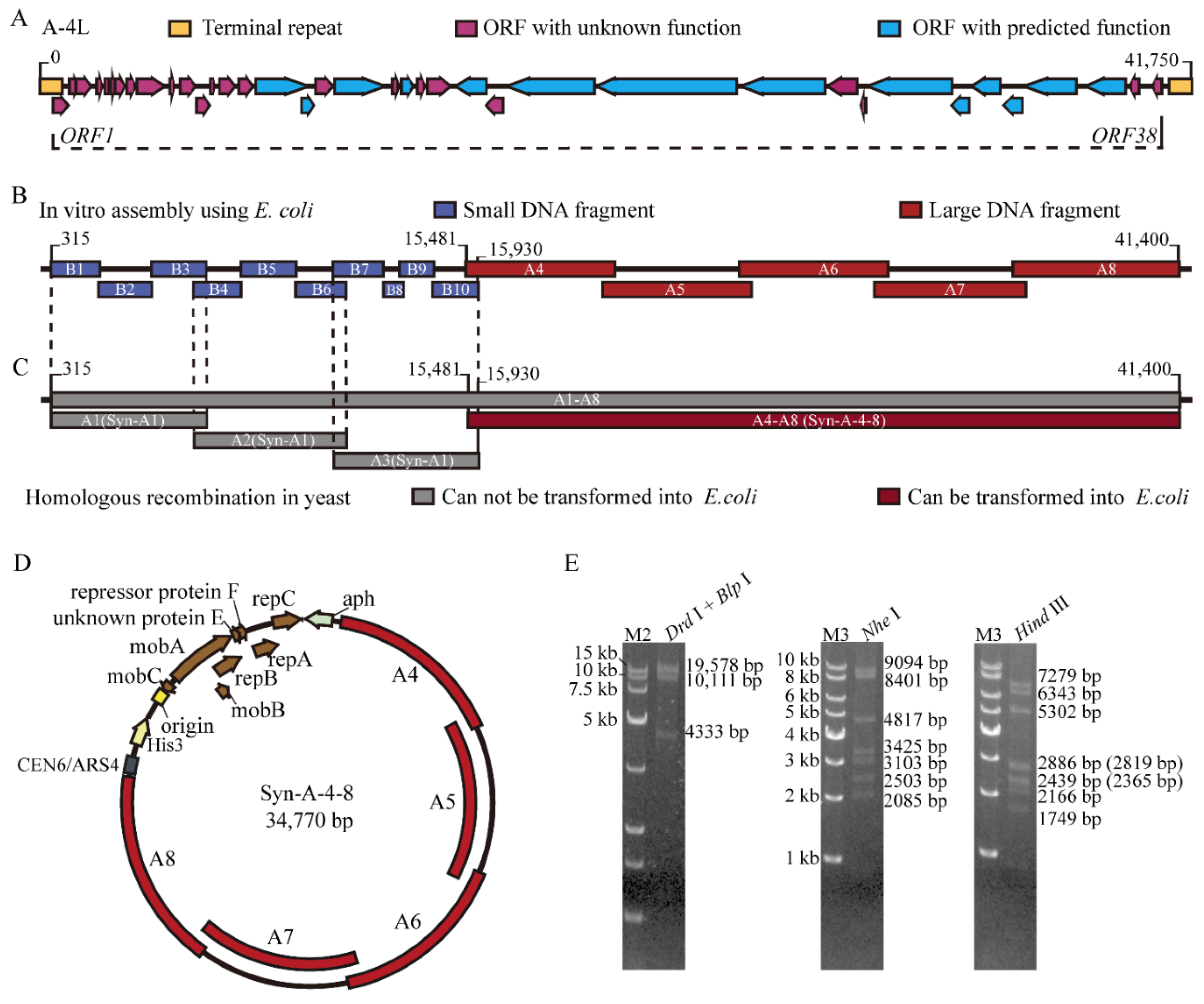
2.2. Construction of Truncated Cyanophage Genome and Plasmid
2.3. Transformation of Syn7942
2.4. Transcriptomic Analysis
2.5. Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Synthesis and Assembly of a Truncated Cyanophage Genome Syn-A-4-8
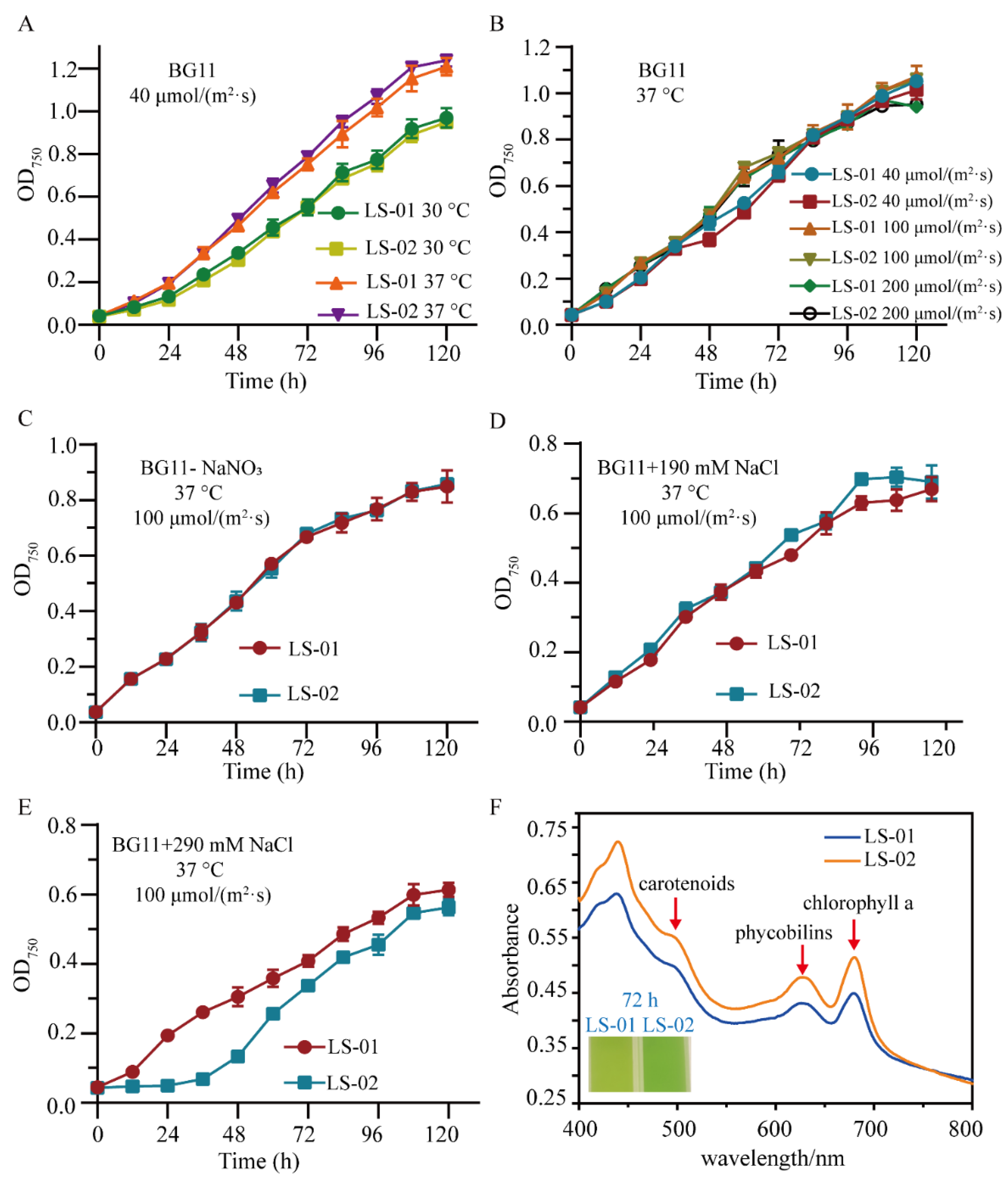
3.2. Heterogeneous Expression and Investigation of Syn-A-4-8 in Syn7942
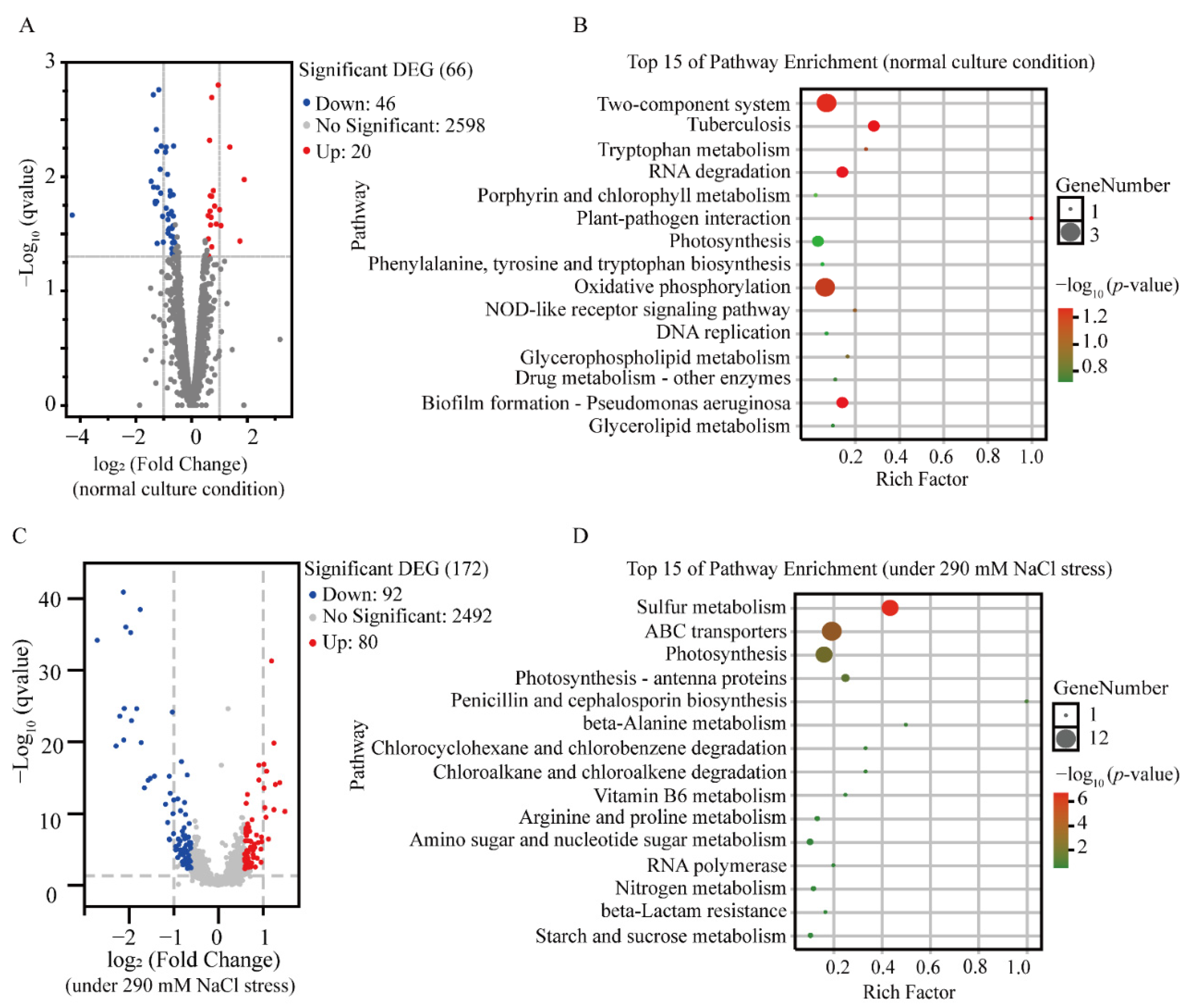
3.3. Transcriptional Analysis Revealing the Effect of Syn-A-4-8 on Syn7942
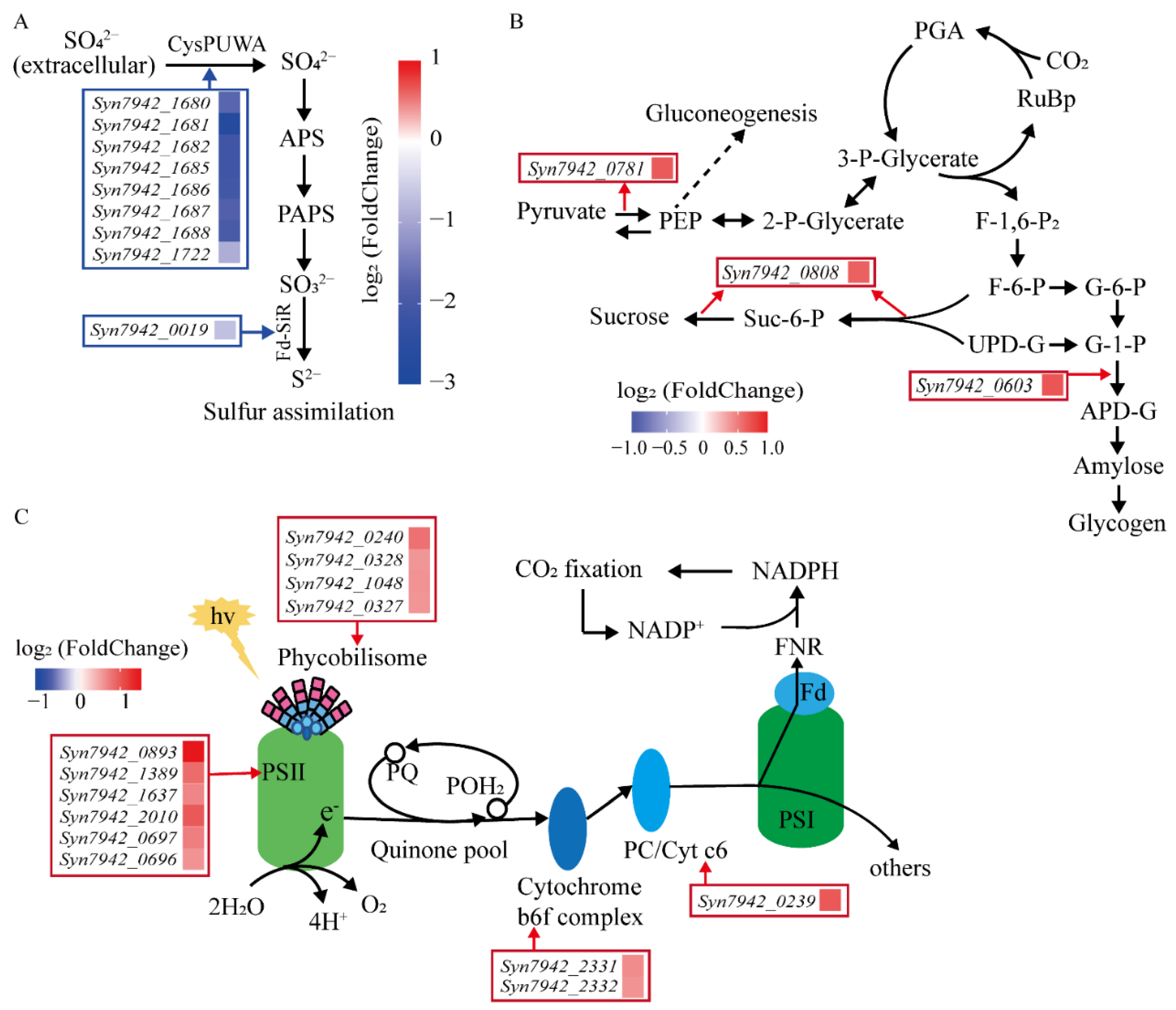
3.3.1. Transcriptomic Comparations of LS-02 and LS-01 under Normal Conditions
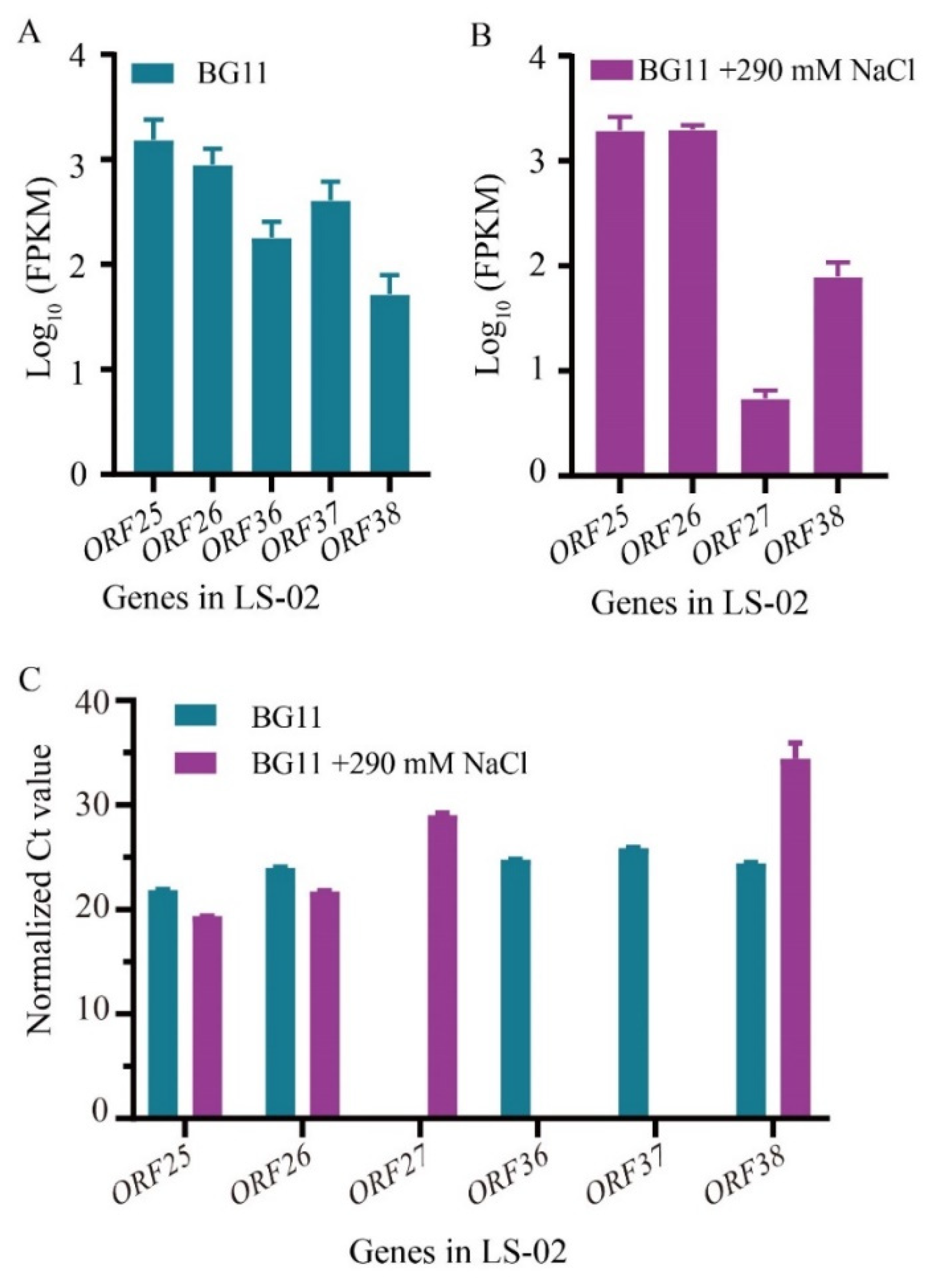
3.3.2. Transcriptomic Response of LS-02 to Salt Stress
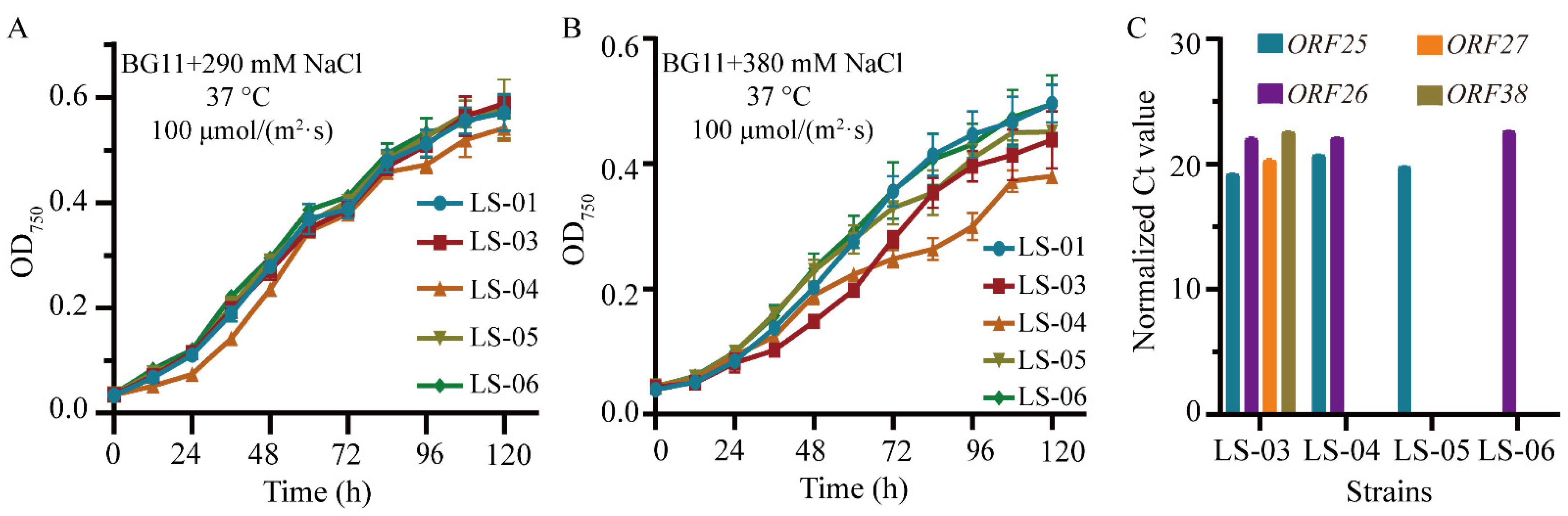
3.4. Identification of the Genes in Syn-A-4-8 Causing the Prolonged Lag Phase of Syn7942
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paerl, H.W.; Fulton, R.S.; Moisander, P.H.; Dyble, J. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. Sci. World J. 2001, 1, 139109. [Google Scholar] [CrossRef] [PubMed]
- Conley, D.J.; Paerl, H.W.; Howarth, R.W.; Boesch, D.F.; Seitzinger, S.P.; Havens, K.E.; Lancelot, C.; Likens, G.E. Controlling eutrophication: Nitrogen and phosphorus. Science 2009, 323, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.A. Is the future blue-green? A review of the current model predictions of how climate change could affect pelagic freshwater cyanobacteria. Water Res. 2012, 46, 1364–1371. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L.; Zheng, L.; Dai, G.; Ma, H.; Shan, K.; Wu, H.; Zhou, Q.; Song, L. Patterns of succession between bloom-forming cyanobacteria Aphanizomenon flos-aquae and Microcystis and related environmental factors in large, shallow Dianchi Lake, China. Hydrobiologia 2016, 765, 1–13. [Google Scholar] [CrossRef]
- Paerl, H.W.; Otten, T.G. Duelling ‘CyanoHABs’: Unravelling the environmental drivers controlling dominance and succession among diazotrophic and non-N2-fixing harmful cyanobacteria. Environ. Microbiol. 2016, 18, 316–324. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.B.; Waterbury, J.B.; Chisholm, S.W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 2003, 424, 1047–1051. [Google Scholar] [CrossRef]
- Dekel-Bird, N.P.; Sabehi, G.; Mosevitzky, B.; Lindell, D. Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ. Microbiol. 2015, 17, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Solonenko, S.A.; Ignacio-Espinoza, J.C.; LaButti, K.; Copeland, A.; Sudek, S.; Maitland, A.; Chittick, L.; dos Santos, F.; Weitz, J.S.; et al. Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genom. 2016, 17, 930. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C.A.; Marston, M.F.; Martiny, J.B.H. Biogeographic variation in host range phenotypes and taxonomic composition of marine cyanophage isolates. Front. Microbiol. 2016, 7, 983. [Google Scholar] [CrossRef]
- Nishimura, Y.; Watai, H.; Honda, T.; Mihara, T.; Omae, K.; Roux, S.; Blanc-Mathieu, R.; Yamamoto, K.; Hingamp, P.; Sako, Y.; et al. Environmental viral genomes shed new light on virus-host interactions in the ocean. mSphere 2017, 2, e00359-16. [Google Scholar] [CrossRef]
- Šulčius, S.; Šimoliūnas, E.; Alzbutas, G.; Gasiūnas, G.; Jauniškis, V.; Kuznecova, J.; Miettinen, S.; Nilsson, E.; Meškys, R.; Roine, E.; et al. Genomic characterization of cyanophage vB_AphaS-CL131 infecting filamentous diazotrophic cyanobacterium Aphanizomenon flos-aquae reveals novel insights into virus-bacterium interactions. Appl. Environ. Microbiol. 2019, 85, e01311-18. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jin, H.; Wang, X.-Q.; Li, Q.; Zhang, J.-T.; Cui, N.; Jiang, Y.-L.; Chen, Y.; Wu, Q.-F.; Zhou, C.-Z.; et al. Genomic analysis of Mic1 reveals a novel freshwater long-tailed cyanophage. Front. Microbiol. 2020, 11, 484. [Google Scholar] [CrossRef]
- Xia, H.; Li, T.; Deng, F.; Hu, Z. Freshwater cyanophages. Virol. Sin. 2013, 28, 253–259. [Google Scholar] [CrossRef]
- Le Moal, M.; Gascuel-Odoux, C.; Ménesguen, A.; Souchon, Y.; Étrillard, C.; Levain, A.; Moatar, F.; Pannard, A.; Souchu, P.; Lefebvre, A.; et al. Eutrophication: A new wine in an old bottle? Sci. Total Environ. 2019, 651, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Otten, T.G. Blooms bite the hand that feeds them. Science 2013, 342, 433–434. [Google Scholar] [CrossRef]
- Bartosiewicz, M.; Przytulska, A.; Deshpande, B.N.; Antoniades, D.; Cortes, A.; MacIntyre, S.; Lehmann, M.F.; Laurion, I. Effects of climate change and episodic heat events on cyanobacteria in a eutrophic polymictic lake. Sci. Total Environ. 2019, 693, 133414. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.P.; Madamwar, D.; Incharoensakdi, A. Bloom dynamics of cyanobacteria and their toxins: Environmental health impacts and mitigation strategies. Front. Microbiol. 2015, 6, 1254. [Google Scholar] [CrossRef]
- Glibert, P.M. Harmful algae at the complex nexus of eutrophication and climate change. Harmful Algae 2020, 91, 101583. [Google Scholar] [CrossRef] [PubMed]
- Puxty, R.J.; Millard, A.D.; Evans, D.J.; Scanlan, D.J. Shedding new light on viral photosynthesis. Photosynth. Res. 2015, 126, 71–97. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Gui, J.-F. Diversity, evolutionary contribution and ecological roles of aquatic viruses. Sci. China-Life Sci. 2018, 61, 1486–1502. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.W.; Carberry, M.J.; Eldridge, M.L.; Poorvin, L.; Saxton, M.A.; Doblin, M.A. Marine and freshwater cyanophages in a Laurentian Great Lake: Evidence from infectivity assays and molecular analyses of g20 genes. Appl. Environ. Microbiol. 2006, 72, 4957–4963. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, F. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ. Microbiol. 2008, 10, 300–312. [Google Scholar] [CrossRef]
- Zborowsky, S.; Lindell, D. Resistance in marine cyanobacteria differs against specialist and generalist cyanophages. Proc. Natl. Acad. Sci. USA 2019, 116, 16899–16908. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Hu, M.; Zhao, G.; Du, Y.; Xu, N.; Chen, X.; Jiao, X.a. The “fighting wisdom and bravery” of tailed phage and host in the process of adsorption. Microbiol. Res. 2020, 230, 126344. [Google Scholar] [CrossRef]
- Xiong, Z.; Wang, Y.; Dong, Y.; Zhang, Q.; Xu, X.; Mullineaux, C.W. Cyanophage A-1(L) adsorbs to lipopolysaccharides of Anabaena sp. Strain PCC 7120 via the tail protein lipopolysaccharide-interacting protein (ORF36). J. Bacteriol. 2019, 201, e00516-18. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.-Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef]
- Hutchison, C.A.; Chuang, R.-Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef]
- Ostrov, N.; Landon, M.; Guell, M.; Kuznetsov, G.; Teramoto, J.; Cervantes, N.; Zhou, M.; Singh, K.; Napolitano, M.G.; Moosburner, M.; et al. Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353, 819–822. [Google Scholar] [CrossRef]
- Annaluru, N.; Muller, H.; Mitchell, L.A.; Ramalingam, S.; Stracquadanio, G.; Richardson, S.M.; Dymond, J.S.; Kuang, Z.; Scheifele, L.Z.; Cooper, E.M.; et al. Total synthesis of a functional designer eukaryotic chromosome. Science 2014, 344, 55–58. [Google Scholar] [CrossRef]
- Wu, Y.; Li, B.-Z.; Zhao, M.; Mitchell, L.A.; Xie, Z.-X.; Lin, Q.-H.; Wang, X.; Xiao, W.-H.; Wang, Y.; Zhou, X.; et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science 2017, 355, eaaf4706. [Google Scholar] [CrossRef]
- Xie, Z.-X.; Li, B.-Z.; Mitchell, L.A.; Wu, Y.; Qi, X.; Jin, Z.; Jia, B.; Wang, X.; Zeng, B.-X.; Liu, H.-M.; et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science 2017, 355, eaaf4704. [Google Scholar] [CrossRef]
- Chen, W.; Han, M.; Zhou, J.; Ge, Q.; Wang, P.; Zhang, X.; Zhu, S.; Song, L.; Yuan, Y. An artificial chromosome for data storage. Natl. Sci. Rev. 2021, 8, nwab028. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, C.; Wei, R.; Han, M.; Wang, S.; Yang, K.; Zhang, L.; Chen, W.; Wen, M.; Li, C.; et al. Exogenous artificial DNA forms chromatin structure with active transcription in yeast. Sci. China-Life Sci. 2021, 65, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.O.; Hutchison, C.A.; Pfannkoch, C.; Venter, J.C. Generating a synthetic genome by whole genome assembly: φX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 15440–15445. [Google Scholar] [CrossRef]
- Jaschke, P.R.; Lieberman, E.K.; Rodriguez, J.; Sierra, A.; Endy, D. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 2012, 434, 278–284. [Google Scholar] [CrossRef]
- Chan, L.Y.; Kosuri, S.; Endy, D. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005, 1, 2005.0018. [Google Scholar] [CrossRef]
- Yeom, H.; Ryu, T.; Lee, A.C.; Noh, J.; Lee, H.; Choi, Y.; Kim, N.; Kwon, S. Cell-free bacteriophage genome synthesis using low-cost sequence-verified array-synthesized oligonucleotides. ACS Synth. Biol. 2020, 9, 1376–1384. [Google Scholar] [CrossRef]
- Hu, N.-T.; Thiel, T.; Giddings, T.H.; Wolk, C.P. New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology 1981, 114, 236–246. [Google Scholar] [CrossRef]
- Ou, T.; Liao, X.-Y.; Gao, X.-C.; Xu, X.-D.; Zhang, Q.-Y. Unraveling the genome structure of cyanobacterial podovirus A-4L with long direct terminal repeats. Virus Res. 2015, 203, 4–9. [Google Scholar] [CrossRef]
- Holtman, C.K.; Chen, Y.; Sandoval, P.; Gonzales, A.; Nalty, M.S.; Thomas, T.L.; Youderian, P.; Golden, S.S. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 Genome. DNA Res. 2005, 12, 103–115. [Google Scholar] [CrossRef]
- Shestakov, S.V.; Khyen, N.T. Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol. Gen. Genet. 1970, 107, 372–375. [Google Scholar] [CrossRef]
- Elhai, J.; Wolk, C.P. Conjugal Transfer of DNA to Cyanobacteria. In Methods Enzymol.; Academic Press: Cambridge, MA, USA, 1988; Volume 167, pp. 747–754. [Google Scholar]
- Li, S.; Sun, T.; Xu, C.; Chen, L.; Zhang, W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018, 48, 163–174. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, T.; Chen, Z.; Song, X.; Chen, L.; Zhang, W. Development of a new biocontainment strategy in model cyanobacterium Synechococcus strains. ACS Synth. Biol. 2019, 8, 2576–2584. [Google Scholar] [CrossRef] [PubMed]
- Anfelt, J.; Hallström, B.; Nielsen, J.; Uhlén, M.; Hudson, E.P. Using transcriptomics to improve butanol tolerance of Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2013, 79, 7419–7427. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Pei, G.; Wang, J.; Chen, L.; Zhang, W. A novel small RNA CoaR regulates coenzyme A biosynthesis and tolerance of Synechocystis sp. PCC6803 to 1-butanol possibly via promoter-directed transcriptional silencing. Biotechnol. Biofuels 2017, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Simons, R.W.; Houman, F.; Kleckner, N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 1987, 53, 85–96. [Google Scholar] [CrossRef]
- Wendt, K.E.; Pakrasi, H.B. Genomics approaches to deciphering natural transformation in cyanobacteria. Front. Microbiol. 2019, 10, 1259. [Google Scholar] [CrossRef]
- Taton, A.; Ecker, A.; Diaz, B.; Moss, N.A.; Anderson, B.; Reher, R.; Leão, T.F.; Simkovsky, R.; Dorrestein, P.C.; Gerwick, L.; et al. Heterologous expression of cryptomaldamide in a cyanobacterial host. ACS Synth. Biol. 2020, 9, 3364–3376. [Google Scholar] [CrossRef]
- Glazer, A.N. Light guides: Directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 1989, 264, 1–4. [Google Scholar] [CrossRef]
- Liu, L.-N. Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 256–265. [Google Scholar] [CrossRef]
- Mullineaux, C.W. Co-existence of photosynthetic and respiratory activities in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.E.; Schlarb-Ridley, B.G.; Bendall, D.S.; Howe, C.J. Terminal oxidases of cyanobacteria. Biochem. Soc. Trans. 2005, 33, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Schmetterer, G.; Valladares, A.; Pils, D.; Steinbach, S.; Pacher, M.; Muro-Pastor, A.M.; Flores, E.; Herrero, A. The coxBAC operon encodes a cytochrome c oxidase required for heterotrophic growth in the cyanobacterium Anabaena variabilis Strain ATCC 29413. J. Bacteriol. 2001, 183, 6429–6434. [Google Scholar] [CrossRef]
- Niwa, S.; Takeda, K.; Kosugi, M.; Tsutsumi, E.; Mogi, T.; Miki, K. Crystal structure of heme A synthase from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, 11953–11957. [Google Scholar] [CrossRef]
- Lewin, A.; Hederstedt, L. Heme A synthase in bacteria depends on one pair of cysteinyls for activity. Biochim. Biophys. Acta 2016, 1857, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Shimakawa, G.; Miyake, C. Respiratory terminal oxidases alleviate photo-oxidative damage in photosystem I during repetitive short-pulse illumination in Synechocystis sp. PCC 6803. Photosynth. Res. 2018, 137, 241–250. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.; Li, J.; Song, X.; Tong, Y.; An, W. Host cyanobacteria killing by novel lytic cyanophage YongM: A protein profiling analysis. Microorganisms 2022, 10, 257. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, N.; Xia, L.; Li, Q.; Shao, J.; Shen, Q.; Zhang, R.; Kivisaar, M. ResDE two-component regulatory system mediates oxygen limitation-induced biofilm formation by Bacillus amyloliquefaciens SQR9. Appl. Environ. Microbiol. 2018, 84, e02744-17. [Google Scholar] [CrossRef]
- Huang, Z.; Pan, X.; Xu, N.; Guo, M. Bacterial chemotaxis coupling protein: Structure, function and diversity. Microbiol. Res. 2019, 219, 40–48. [Google Scholar] [CrossRef]
- Luu, R.A.; Schomer, R.A.; Brunton, C.N.; Truong, R.; Ta, A.P.; Tan, W.A.; Parales, J.V.; Wang, Y.-J.; Huo, Y.-W.; Liu, S.-J.; et al. Hybrid two-component sensors for identification of bacterial chemoreceptor function. Appl. Environ. Microbiol. 2019, 85, e01626-19. [Google Scholar] [CrossRef]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50. [Google Scholar] [CrossRef]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef]
- Yoshihara, S.; Geng, X.; Ikeuchi, M. pilG gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2002, 43, 513–521. [Google Scholar] [CrossRef]
- Chen, Z.; Li, X.; Tan, X.; Zhang, Y.; Wang, B. Recent advances in biological functions of thick pili in the cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Baneyx, F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 2000, 36, 1360–1370. [Google Scholar] [CrossRef]
- Zhao, L.; Vecchi, G.; Vendruscolo, M.; Körner, R.; Hayer-Hartl, M.; Hartl, F.U. The Hsp70 chaperone system stabilizes a thermo-sensitive subproteome in E. coli. Cell Rep. 2019, 28, 1335–1345.e6. [Google Scholar] [CrossRef] [PubMed]
- Miwa, T.; Chadani, Y.; Taguchi, H. Escherichia coli small heat shock protein IbpA is an aggregation-sensor that self-regulates its own expression at posttranscriptional levels. Mol. Microbiol. 2021, 115, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Buchner, J. Bacterial Hsp90-desperately seeking clients. Mol. Microbiol. 2010, 76, 540–544. [Google Scholar] [CrossRef]
- Nakamoto, H.; Kojima, K. Non-housekeeping, non-essential GroEL (chaperonin) has acquired novel structure and function beneficial under stress in cyanobacteria. Physiol. Plant. 2017, 161, 296–310. [Google Scholar] [CrossRef]
- Naithani, S.; Hou, J.-M.; Chitnis, P.R. Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of Photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 2000, 63, 225–236. [Google Scholar] [CrossRef]
- Fridman, S.; Flores-Uribe, J.; Larom, S.; Alalouf, O.; Liran, O.; Yacoby, I.; Salama, F.; Bailleul, B.; Rappaport, F.; Ziv, T.; et al. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2017, 2, 1350–1357. [Google Scholar] [CrossRef]
- Karandashova, I.V.; Elanskaya, I.V. Genetic control and mechanisms of salt and hyperosmotic stress resistance in cyanobacteria. Russ. J. Genet. 2005, 41, 1311–1321. [Google Scholar] [CrossRef]
- Yang, W.; Wang, F.; Liu, L.-N.; Sui, N. Responses of membranes and the photosynthetic apparatus to salt stress in cyanobacteria. Front. Plant Sci. 2020, 11, 713. [Google Scholar] [CrossRef]
- Aguilar-Barajas, E.; Díaz-Pérez, C.; Ramírez-Díaz, M.I.; Riveros-Rosas, H.; Cervantes, C. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 2011, 24, 687–707. [Google Scholar] [CrossRef] [PubMed]
- Nazar, R.; Iqbal, N.; Masood, A.; Syeed, S.; Khan, N.A. Understanding the significance of sulfur in improving salinity tolerance in plants. Environ. Exp. Bot. 2011, 70, 80–87. [Google Scholar] [CrossRef]
- Gisselmann, G.; Klausmeier, P.; Schwenn, J.D. The ferredoxin:sulphite reductase gene from Synechococcus PCC7942. Biochim. Et Biophys. Acta BBA-Bioenerg. 1993, 1144, 102–106. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, M.; Wang, M.; Zhang, W.; Qiao, C.; Luo, Q.; Lu, X.; Kelly, R.M. Freshwater cyanobacterium Synechococcus elongatus PCC 7942 adapts to an environment with salt stress via ion-induced enzymatic balance of compatible solutes. Appl. Environ. Microbiol. 2020, 86, e02904-19. [Google Scholar] [CrossRef] [PubMed]
- Niersbach, M.; Kreuzaler, F.; Geerse, R.H.; Postma, P.W.; Hirsch, H.J. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol. Gen. Genet. 1992, 231, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.R.; Patel, M.V.; Lloyd, C.R.; Vanderpool, C.K. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J. Bacteriol. 2013, 195, 4816–4825. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Noël, G.M.A.; Cumino, A.C.; de los Angeles Kolman, M.; Salerno, G.L. First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett. 2013, 587, 1669–1674. [Google Scholar] [CrossRef]
- Ghosh, P.; Meyer, C.; Remy, E.; Peterson, D.; Preiss, J. Cloning, expression, and nucleotide sequence of glgC gene from an allosteric mutant of Escherichia coli B. Arch. Biochem. Biophys. 1992, 296, 122–128. [Google Scholar] [CrossRef]
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev. 2003, 67, 213–225. [Google Scholar] [CrossRef]
- Qiao, C.; Duan, Y.; Zhang, M.; Hagemann, M.; Luo, Q.; Lu, X.; Atomi, H. Effects of reduced and enhanced glycogen pools on salt-induced sucrose production in a sucrose-secreting strain of Synechococcus elongatus PCC 7942. Appl. Environ. Microbiol. 2018, 84, e02023-17. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.J.; Shan, J.; Bailey, S.; Jia, Y.; Krisch, H.M.; West, S.; Mann, N.H. Transcription of a ‘photosynthetic’ T4-type phage during infection of a marine cyanobacterium. Environ. Microbiol. 2006, 8, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Koksharova, O.; Wolk, C. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 2002, 58, 123–137. [Google Scholar] [CrossRef]
- Roucourt, B.; Lavigne, R. The role of interactions between phage and bacterial proteins within the infected cell: A diverse and puzzling interactome. Environ. Microbiol. 2009, 11, 2789–2805. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, F.; Klähn, S.; Hagemann, M. Salt-regulated accumulation of the compatible solutes sucrose and glucosylglycerol in cyanobacteria and its biotechnological potential. Front. Microbiol. 2019, 10, 2139. [Google Scholar] [CrossRef] [PubMed]
| Strains/Plasmids | Characteristics 1,2 |
|---|---|
| Plasmids (Size/bp) | |
| pJA2 (6537) | KmR, E. coli-cyanobacteria shuttle vector |
| Syn-A-4-8 (34,770) | pJA2 harboring the A4-A8 cassette |
| pSJ03 (20,889) | pJA2 harboring the ORF25, ORF26, ORF27 and ORF38 of the A-4L genome |
| pSJ04 (14,845) | pJA2 harboring the ORF25 and ORF26 of the A-4L genome |
| pSJ05 (12,253) | pJA2 harboring the ORF25 of the A-4L genome |
| pSJ06 (14,243) | pJA2 harboring the ORF26 of the A-4L genome |
| Strains | |
| LS-01 | Syn7942 harboring the plasmid pJA2, KmR |
| LS-02 | Syn7942 harboring genome Syn-A-4-8, KmR |
| LS-03 | Syn7942 harboring the plasmid pSJ03, KmR |
| LS-04 | Syn7942 harboring the plasmid pSJ04, KmR |
| LS-05 | Syn7942 harboring the plasmid pSJ05, KmR |
| LS-06 | Syn7942 harboring the plasmid pSJ06, KmR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Feng, J.; Sun, T.; Xu, B.; Zhang, J.; Li, G.; Zhou, J.; Jiang, J. The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host. Life 2022, 12, 1234. https://doi.org/10.3390/life12081234
Liu S, Feng J, Sun T, Xu B, Zhang J, Li G, Zhou J, Jiang J. The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host. Life. 2022; 12(8):1234. https://doi.org/10.3390/life12081234
Chicago/Turabian StyleLiu, Shujing, Jia Feng, Tao Sun, Bonan Xu, Jiabao Zhang, Guorui Li, Jianting Zhou, and Jianlan Jiang. 2022. "The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host" Life 12, no. 8: 1234. https://doi.org/10.3390/life12081234
APA StyleLiu, S., Feng, J., Sun, T., Xu, B., Zhang, J., Li, G., Zhou, J., & Jiang, J. (2022). The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host. Life, 12(8), 1234. https://doi.org/10.3390/life12081234







