Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Embryos
2.2. Isolation of Ovarian Follicles
2.3. RNA Extraction
2.4. Cloning and Sequence Analysis of Medaka Igf1r
2.5. RT-PCR and Real-Time PCR Analysis
2.6. RNA In Situ Hybridization
2.7. Microscopic Observation
3. Results
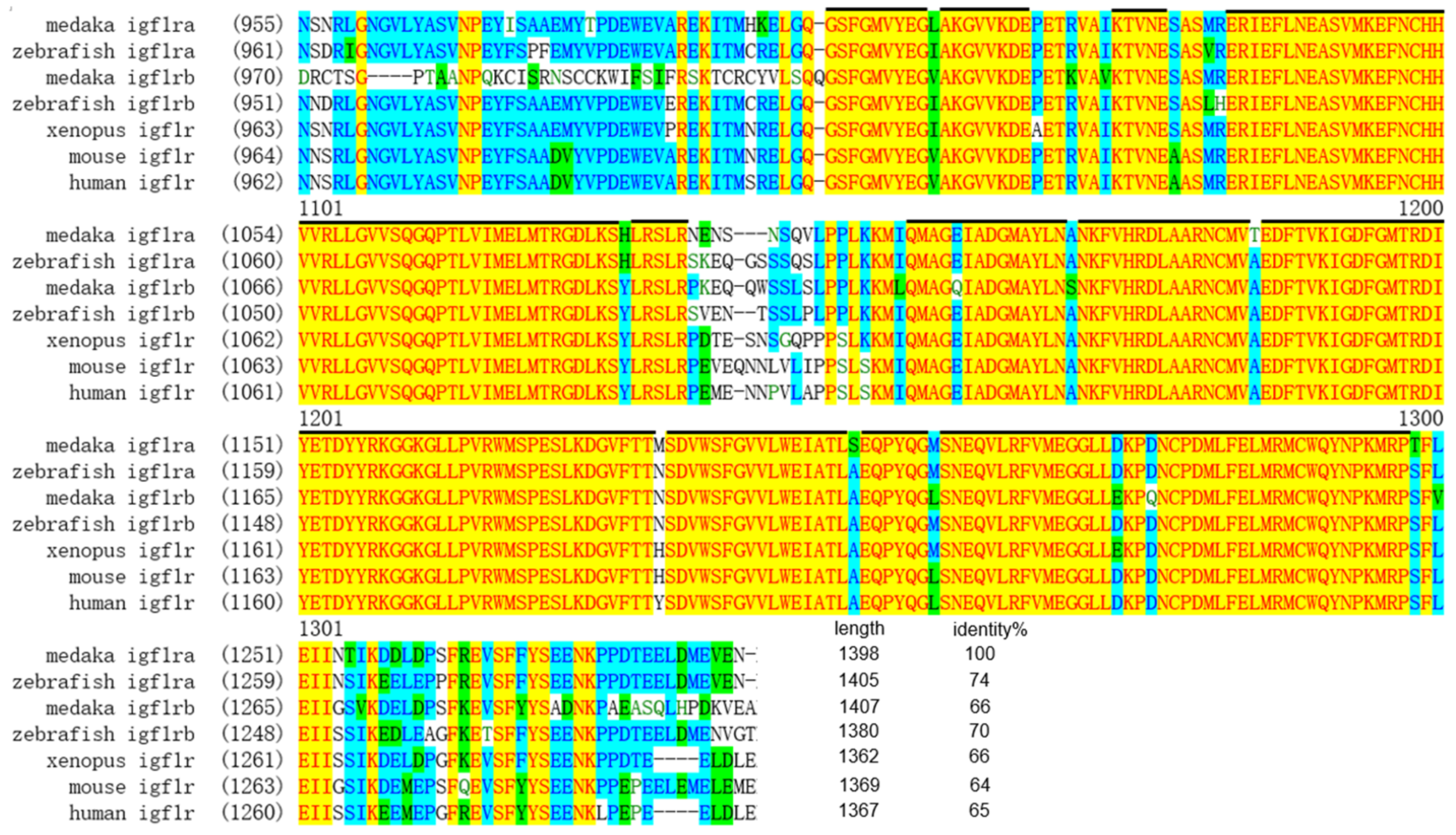
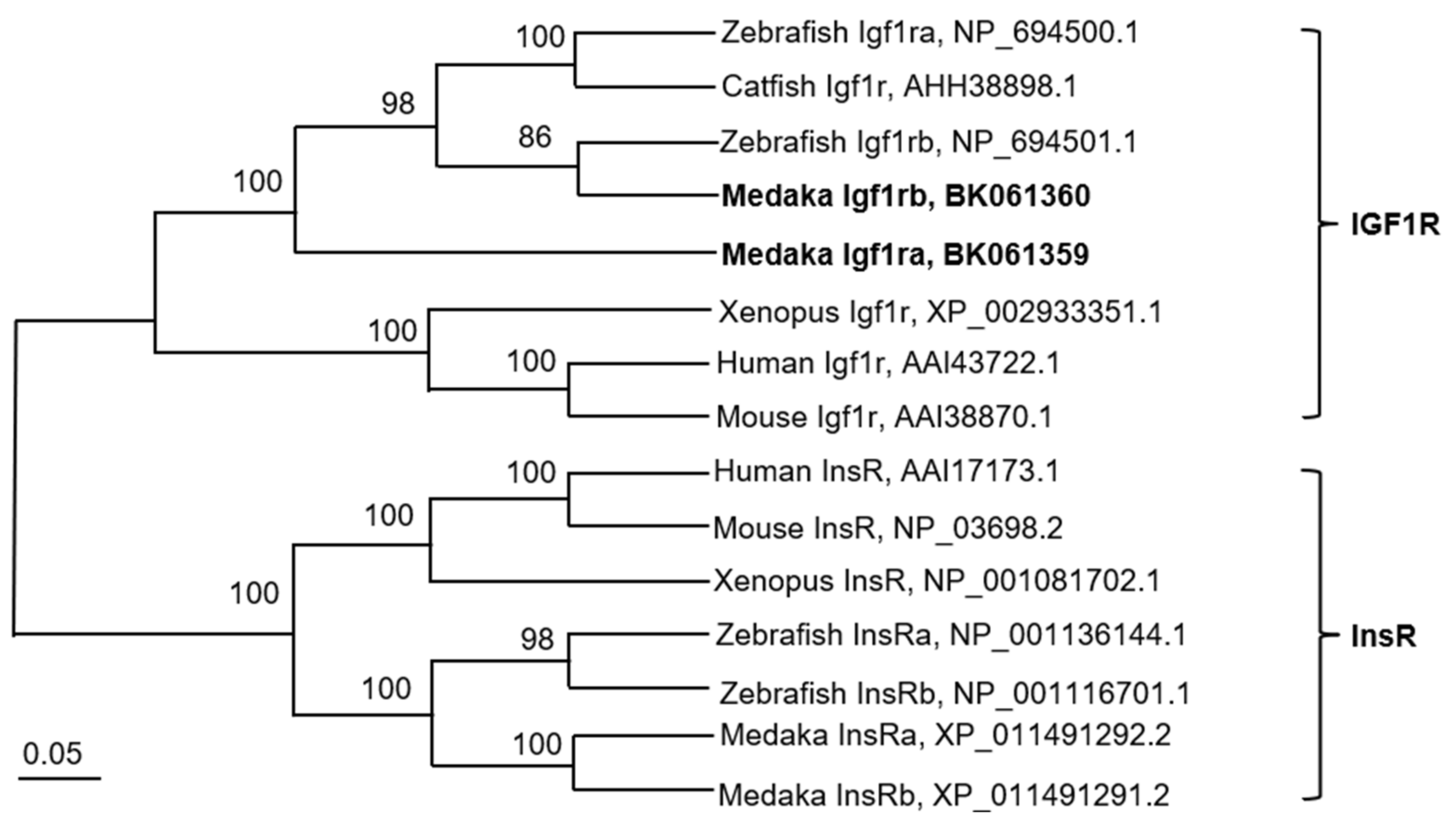
3.1. Cloning and Sequence Analysis of Medaka Igf1r
3.2. RT-PCR Analysis of Igf1r RNA Expression
3.3. Gonadal Expression of Igf1ra RNA and Igf1rb RNA by ISH
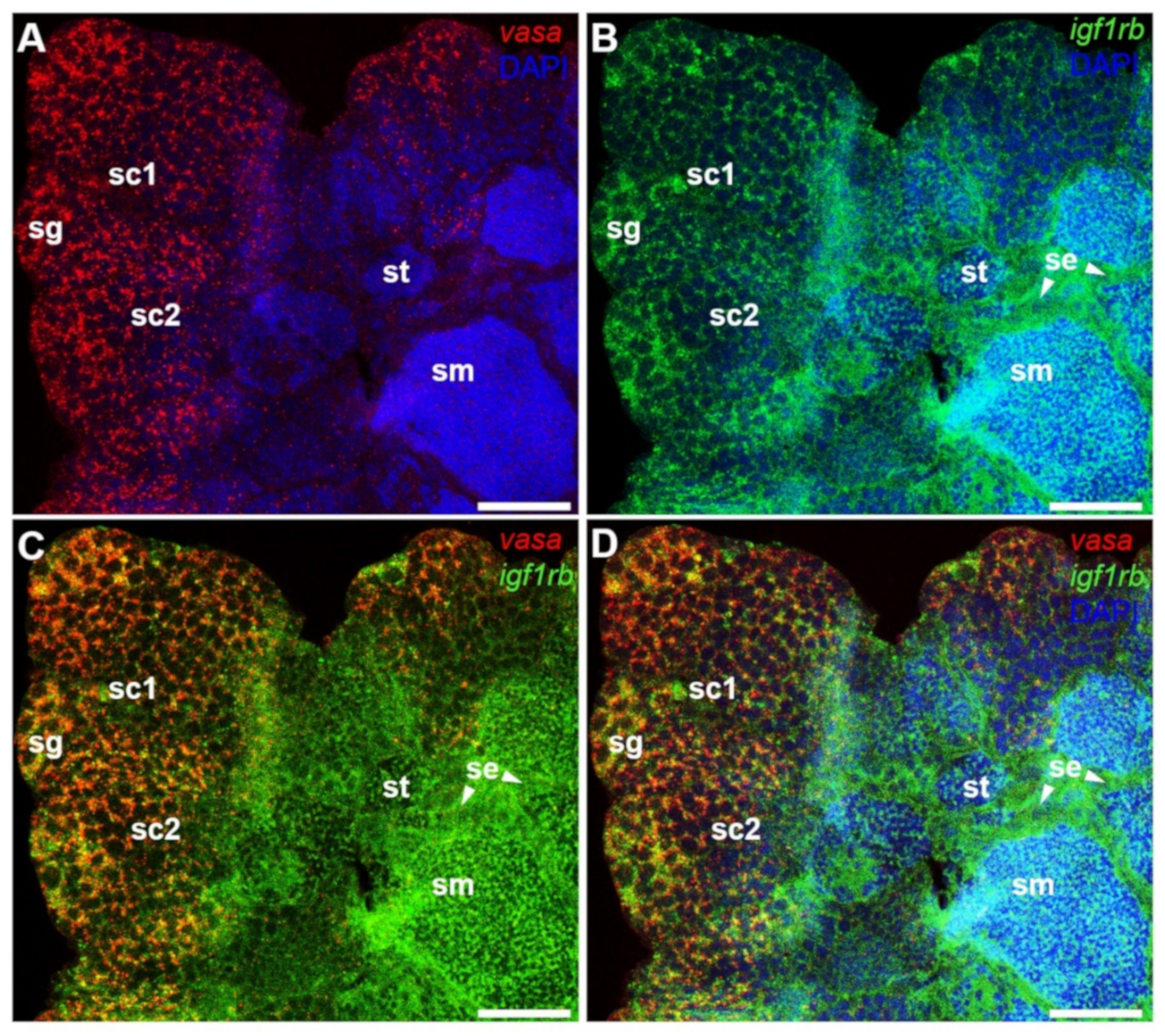
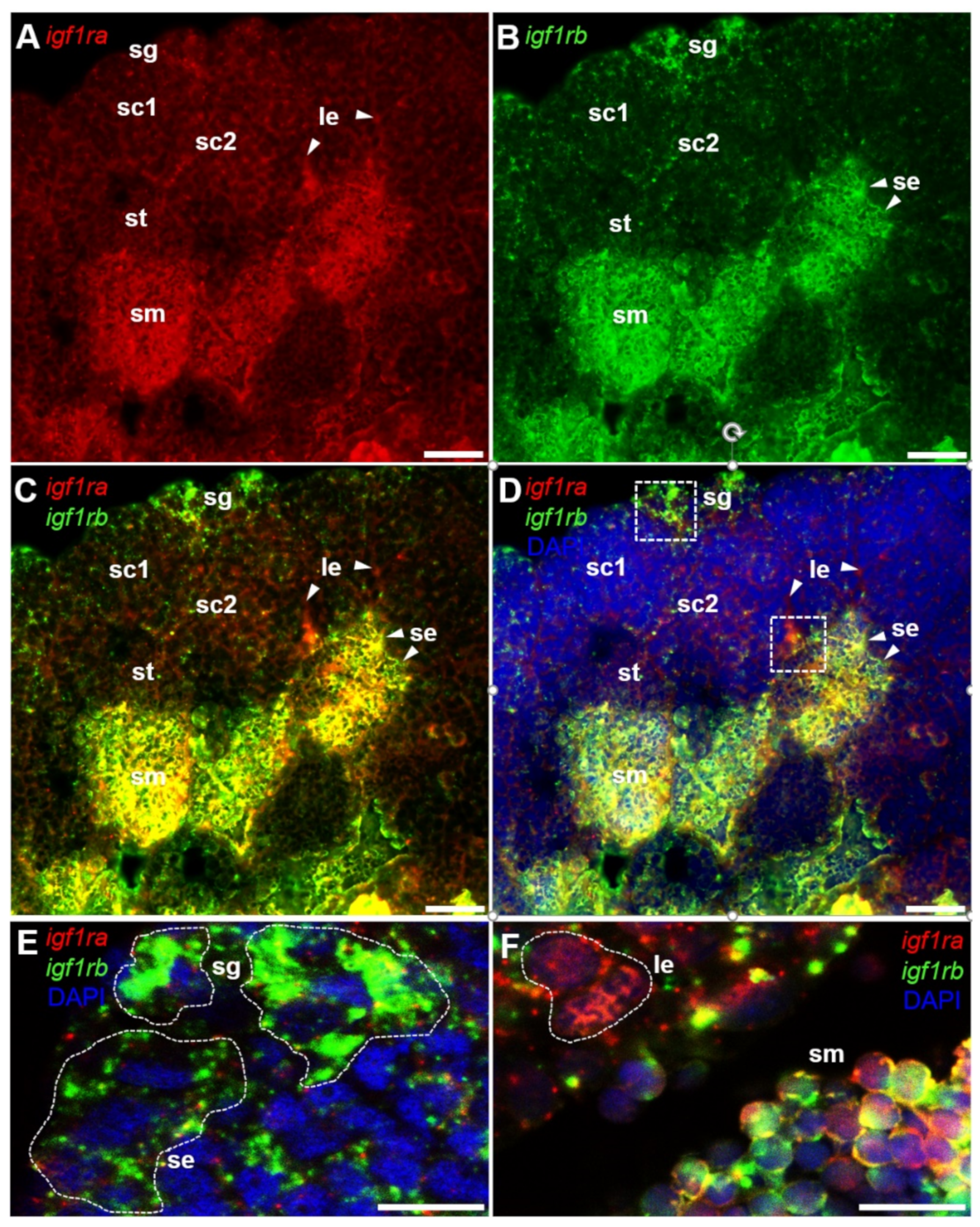
3.4. Ovarian Differential Expression of Igf1ra and Igf1rb RNAs by Fluorescence ISH
3.5. Testicular Differential Expression of Igf1ra and Igf1rb RNAs by Fluorescence ISH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wood, A.W.; Duan, C.; Bern, H.A. Insulin-like growth factor signaling in fish. Int. Rev. Cytol. 2005, 243, 215–285. [Google Scholar] [PubMed]
- Liu, J.P.; Baker, J.; Perkins, A.S.; Robertson, E.J.; Efstratiadis, A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 1993, 75, 59–72. [Google Scholar] [CrossRef]
- Berishvili, G.; D’Cotta, H.; Baroiller, J.F.; Segner, H.; Reinecke, M. Differential expression of IGF-I mRNA and peptide in the male and female gonad during early development of a bony fish, the tilapia Oreochromis niloticus. Gen. Comp. Endocrinol. 2006, 146, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.P.; Melo, R.M.C.; Weber, A.A.; Rizzo, E. Insulin-like growth factors 1 and 2 are associated with testicular germ cell proliferation and apoptosis during fish reproduction. Reprod. Fertil. Dev. 2020, 32, 988–998. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; Kang, T.; Li, M.; Wang, D.; Cheng, C.H.K. Igf3: A novel player in fish reproductiondagger. Biol. Reprod. 2021, 104, 1194–1204. [Google Scholar] [CrossRef]
- Li, M.; Liu, X.; Dai, S.; Xiao, H.; Qi, S.; Li, Y.; Zheng, Q.; Jie, M.; Cheng, C.H.K.; Wang, D. Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia. Cell. Mol. Life Sci. 2020, 77, 4921–4938. [Google Scholar] [CrossRef]
- Fan, R.; Ran, M.; Rui, X.; Jie, M. m6A reader Igf2bp3 enables germ plasm assembly by m6A-dependent regulation of gene expression in zebrafish. Sci. Bull. 2021, 66, 1119–1128. [Google Scholar]
- Mei, J.; Yan, W.; Fang, J.; Yuan, G.; Chen, N.; He, Y. Identification of a gonad-expression differential gene insulin-like growth factor-1 receptor (Igf1r) in the swamp eel (Monopterus albus). Fish Physiol. Biochem. 2014, 40, 1181–1190. [Google Scholar] [CrossRef]
- Perrot, V.; Moiseeva, E.B.; Gozes, Y.; Chan, S.J.; Funkenstein, B. Insulin-like growth factor receptors and their ligands in gonads of a hermaphroditic species, the gilthead seabream (Sparus aurata): Expression and cellular localization. Biol. Reprod. 2000, 63, 229–241. [Google Scholar] [CrossRef][Green Version]
- Kineman, R.D.; Del Rio-Moreno, M.; Sarmento-Cabral, A. 40 YEARS of IGF1: Understanding the tissue-specific roles of IGF1/IGF1R in regulating metabolism using the Cre/loxP system. J. Mol. Endocrinol. 2018, 61, T187–T198. [Google Scholar] [CrossRef]
- Cannarella, R.; Condorelli, R.A.; La Vignera, S.; Bellucci, C.; Luca, G.; Calafiore, R.; Calogero, A.E. IGF2 and IGF1R mRNAs are Detectable in Human Spermatozoa. World J. Mens Health 2020, 38, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Gallelli, M.F.; Bianchi, C.; Lombardo, D.; Rey, F.; Rodriguez, F.M.; Castillo, V.A.; Miragaya, M. Leptin and IGF1 receptors in alpaca (Vicugna pacos) ovaries. Anim. Reprod. Sci. 2019, 200, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Le Gac, F.; Loir, M.; le Bail, P.Y.; Ollitrault, M. Insulin-like growth factor (IGF-I) mRNA and IGF-I receptor in trout testis and in isolated spermatogenic and Sertoli cells. Mol. Reprod. Dev. 1996, 44, 23–35. [Google Scholar] [CrossRef]
- Schlueter, P.J.; Royer, T.; Farah, M.H.; Laser, B.; Chan, S.J.; Steiner, D.F.; Duan, C. Gene duplication and functional divergence of the zebrafish insulin-like growth factor 1 receptors. FASEB J. 2006, 20, 1230–1232. [Google Scholar] [CrossRef]
- Gu, W.; Yang, Y.; Ning, C.; Wang, Y.; Hu, J.; Zhang, M.; Kuang, S.; Sun, Y.; Li, Y.; Zhang, Y.; et al. Identification and characteristics of insulin-like growth factor system in the brain, liver, and gonad during development of a seasonal breeding teleost, Pampus argenteus. Gen. Comp. Endocrinol. 2021, 300, 113645. [Google Scholar] [CrossRef]
- Maures, T.; Chan, S.J.; Xu, B.; Sun, H.; Ding, J.; Duan, C. Structural, biochemical, and expression analysis of two distinct insulin-like growth factor I receptors and their ligands in zebrafish. Endocrinology 2002, 143, 1858–1871. [Google Scholar] [CrossRef]
- Schlueter, P.J.; Sang, X.; Duan, C.; Wood, A.W. Insulin-like growth factor receptor 1b is required for zebrafish primordial germ cell migration and survival. Dev. Biol. 2007, 305, 377–387. [Google Scholar] [CrossRef]
- Wittbrodt, J.; Shima, A.; Schartl, M. Medaka—A model organism from the far East. Nat. Rev. Genet. 2002, 3, 53–64. [Google Scholar] [CrossRef]
- Yi, M.; Hong, N.; Hong, Y. Generation of medaka fish haploid embryonic stem cells. Science 2009, 326, 430–433. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, T.; Zhao, H.; Xu, H.; Wang, W.; Liu, R.; Chen, T.; Deng, J.; Gui, J. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 8011–8016. [Google Scholar] [CrossRef]
- Hong, N.; Li, M.; Yuan, Y.; Wang, T.; Yi, M.; Xu, H.; Zeng, H.; Song, J.; Hong, Y. Dnd is a critical specifier of primordial germ cells in the medaka fish. Stem Cell Rep. 2016, 6, 411–421. [Google Scholar] [CrossRef]
- Li, M.; Hong, N.; Gui, J.; Hong, Y. Medaka piwi is essential for primordial germ cell migration. Curr. Mol. Med. 2012, 12, 1040–1049. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Nishimura, T.; Sato, T.; Yamamoto, Y.; Watakabe, I.; Ohkawa, Y.; Suyama, M.; Kobayashi, S.; Tanaka, M. Sex determination. foxl3 is a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka. Science 2015, 349, 328–331. [Google Scholar]
- Yuan, C.; Chen, K.; Zhu, Y.; Yuan, Y.; Li, M. Medaka igf1 identifies somatic cells and meiotic germ cells of both sexes. Gene 2018, 642, 423–429. [Google Scholar] [CrossRef]
- Yuan, Y.; Hong, Y. Medaka insulin-like growth factor-2 supports self-renewal of the embryonic stem cell line and blastomeres in vitro. Sci. Rep. 2017, 7, 78. [Google Scholar] [CrossRef]
- Xie, J.; Zhong, Y.; Zhao, Y.; Xie, W.; Guo, J.; Gui, L.; Li, M. Characterization and expression analysis of gonad specific igf3 in the medaka ovary. Aquac. Fish. 2020, 7, 259–268. [Google Scholar] [CrossRef]
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004, 121, 605–618. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Shved, N.; Berishvili, G.; D’Cotta, H.; Baroiller, J.F.; Segner, H.; Eppler, E.; Reinecke, M. Ethinylestradiol differentially interferes with IGF-I in liver and extrahepatic sites during development of male and female bony fish. J. Endocrinol. 2007, 195, 513–523. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, Y.; Zhu, T.; Song, P.; Guo, J.; Zhong, Y.; Gui, L.; Li, M. Vasa identifies germ cells in embryos and gonads of Oryzias celebensis. Gene 2022, 823, 146369. [Google Scholar] [CrossRef]
- Song, P.; Sun, B.; Zhu, Y.; Zhong, Y.; Guo, J.; Gui, L.; Li, M. Bucky ball induces primordial germ cell increase in medaka. Gene 2021, 768, 145317. [Google Scholar] [CrossRef]
- Li, M.; Zhao, H.; Wei, J.; Zhang, J.; Hong, Y. Medaka vasa gene has an exonic enhancer for germline expression. Gene 2015, 555, 403–408. [Google Scholar] [CrossRef]
- Schulz, R.W.; de Franca, L.R.; Lareyre, J.J.; Le Gac, F.; Chiarini-Garcia, H.; Nobrega, R.H.; Miura, T. Spermatogenesis in fish. Gen. Comp. Endocrinol. 2010, 165, 390–411. [Google Scholar] [CrossRef]
- Guo, L.; Yang, S.; Li, M.M.; Meng, Z.N.; Lin, H.R. Divergence and polymorphism analysis of IGF1Ra and IGF1Rb from orange-spotted grouper, Epinephelus coioides (Hamilton). Genet. Mol. Res. 2016, 15, gmr15048768. [Google Scholar] [CrossRef]
- Wu, J.; Li, W.; Craddock, B.P.; Foreman, K.W.; Mulvihill, M.J.; Ji, Q.S.; Miller, W.T.; Hubbard, S.R. Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J. 2008, 27, 1985–1994. [Google Scholar] [CrossRef]
- Hakuno, F.; Takahashi, S.I. IGF1 receptor signaling pathways. J. Mol. Endocrinol. 2018, 61, T69–T86. [Google Scholar] [CrossRef]
- Kuang, Y.M.; Li, W.S.; Lin, H.R. Molecular cloning and mRNA profile of insulin-like growth factor type 1 receptor in orange-spotted grouper, Epinephelus coioides. Acta Biochim. Biophys. Sin. 2005, 37, 327–334. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Z.; Cheng, Q.; Chen, X. Expression of insulin-like growth factor I receptors at mRNA and protein levels during metamorphosis of Japanese flounder (Paralichthys olivaceus). Gen. Comp. Endocrinol. 2011, 173, 78–85. [Google Scholar] [CrossRef]
- Weber, G.M.; Sullivan, C.V. Effects of insulin-like growth factor-I on in vitro final oocyte maturation and ovarian steroidogenesis in striped bass, Morone saxatilis. Biol. Reprod. 2000, 63, 1049–1057. [Google Scholar] [CrossRef]
- Neirijnck, Y.; Calvel, P.; Kilcoyne, K.R.; Kuhne, F.; Stevant, I.; Griffeth, R.J.; Pitetti, J.L.; Andric, S.A.; Hu, M.C.; Pralong, F.; et al. Insulin and IGF1 receptors are essential for the development and steroidogenic function of adult Leydig cells. FASEB J. 2018, 32, 3321–3335. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.X.; Huang, D.; Chu, L.H.; Liu, Y.; Sun, X.; Li, J.Z.; Cheng, C.H.K. Igf3 is essential for ovary differentiation in zebrafish. Biol. Reprod. 2021, 104, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hong, N.; Xu, H.; Yi, M.; Li, C.; Gui, J.; Hong, Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009, 126, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Maestro, M.A.; Planas, J.V.; Moriyama, S.; Gutierrez, J.; Planas, J.; Swanson, P. Ovarian receptors for insulin and insulin-like growth factor I (IGF-I) and effects of IGF-I on steroid production by isolated follicular layers of the preovulatory coho salmon ovarian follicle. Gen. Comp. Endocrinol. 1997, 106, 189–201. [Google Scholar] [CrossRef]
- Baumgarten, S.C.; Armouti, M.; Ko, C.; Stocco, C. IGF1R expression in ovarian granulosa cells is essential for steroidogenesis, follicle survival, and fertility in female mice. Endocrinology 2017, 158, 2309–2318. [Google Scholar] [CrossRef]
- Safian, D.; Bogerd, J.; Schulz, R.W. Igf3 activates beta-catenin signaling to stimulate spermatogonial differentiation in zebrafish. J. Endocrinol. 2018, 238, 245–257. [Google Scholar] [CrossRef]



| Primer | Sequence (5′ to 3′ Direction) | Purpose |
|---|---|---|
| igf1ra-F | ATGGACATCCGAAACGAC | RT-PCR |
| igf1ra-R | TCGTCTGTACAGGCCAGCTTG | |
| igf1rb-F | TGCTTCGGGAATGAGGCCTCC | RT-PCR |
| igf1rb-R | TGATGTGCAGGTTTCCTTTGAT | |
| igf1ra-F1 | ATGGGCAGGCTGACCTTGTTTTGG | CDS cloning |
| igf1ra-R1 | TTCGTCTGGAGTGTACATCTC | |
| igf1ra-F2 | ATGTACACTCCAGACGAATGG | CDS cloning |
| igf1ra-R2 | TCAGCAGGCCGACGACTGGGGCAG | |
| igf1rb-F1 | ATGAGGCCTCCAGCGGAAACGAGG | CDS cloning |
| igf1rb-R1 | GGCCACGCCCTCGTACACCAT | |
| igf1rb-F2 | ATGGTGTACGAGGGCGTGGCC | CDS cloning |
| igf1rb-R2 | CCCTTCAGCAGGCTGAG | |
| igf1ra-QF | CGCCTGCTTGGTGTAGTCT | Real-time PCR |
| igf1ra-QR | GGACCTGGCTGTTGGAGTT | |
| igf1rb-QF | GGTCTGATGCTGGCTCTGT | Real-time PCR |
| igf1rb-QR | ACTTCCTGGTTGGCGTTGT | |
| Actin-F | TTCAACAGCCCTGCCATGTA | Internal control |
| Actin-R | CCTCCAATCCAGACAGTAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Zhu, Y.; Yuan, C.; Zhao, Y.; Zhou, W.; Li, M. Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads. Life 2022, 12, 859. https://doi.org/10.3390/life12060859
Wei W, Zhu Y, Yuan C, Zhao Y, Zhou W, Li M. Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads. Life. 2022; 12(6):859. https://doi.org/10.3390/life12060859
Chicago/Turabian StyleWei, Wenbo, Yefei Zhu, Cancan Yuan, Yuli Zhao, Wenzong Zhou, and Mingyou Li. 2022. "Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads" Life 12, no. 6: 859. https://doi.org/10.3390/life12060859
APA StyleWei, W., Zhu, Y., Yuan, C., Zhao, Y., Zhou, W., & Li, M. (2022). Differential Expression of Duplicate Insulin-like Growth Factor-1 Receptors (igf1rs) in Medaka Gonads. Life, 12(6), 859. https://doi.org/10.3390/life12060859







